Abstract
Off-pump coronary artery bypass surgery reduces the myocardial injury associated with on pump surgery with cardiopulmonary bypass (CPB) and ischemic-cardioplegic arrest (CA). We sought to find a mechanistic explanation for this by comparing the transcriptomic changes in the myocardium of patients undergoing on- and off-pump surgery. Transcriptomic analyses were performed on left ventricular biopsies obtained from patients prior to (pre-op) and after completion of all coronary anastomoses (post-op). Microarray results were validated with real-time polymerase chain reaction. In on-pump group, 68 genes were upregulated in post-op vs. pre-op biopsies (P < 0.01, ≥2-fold). They included inflammatory genes CCL3 and CCL4, apoptotic gene GADD45B and prostaglandin synthesis gene PTGS2 (COX-2). In the off-pump group, 17 genes were upregulated in post-op vs. pre-op biopsies (P < 0.01, ≥2-fold), all shared with on-pump patients. To uncover the genes implicated in CPB and ischemic-CA response, we compared the postoperative gene profiles of the two groups. Thirty-eight genes were upregulated in the on-pump vs. off-pump patients (P < 0.01, ≥2-fold). On-pump surgery induces injury-related response, as demonstrated by the upregulation of apoptosis and remodeling markers, whereas off-pump surgery ameliorates that by mainly upregulating a cytoprotective genetic program. Blood levels of the identified cytokines and chemokines followed the same pattern obtained by transcriptomics, suggesting that the myocardium is a likely source for these proteomic changes. In conclusion, off-pump surgery is associated with fewer alterations in gene expression connected with inflammation, apoptosis, and remodeling seen after on-pump surgery with CPB and ischemic-CA.
Keywords: microarray, heart, cardiopulmonary bypass, inflammation
despite advances in perfusion, anesthesia, and surgical techniques, coronary artery bypass grafting (CABG) with cardiopulmonary bypass (CPB) and ischemic-cardioplegic arrest (CA) is still associated with a systemic inflammatory response, and reperfusion injury to the myocardium, which may lead to postoperative morbidity and transient organ dysfunction (4). The introduction of off-pump coronary artery bypass surgery in the 1990s was seen as a potential solution to reduce the complications associated with CPB and ischemic-CA. In previous work, we have shown that plasma biochemical markers of inflammatory response and myocardial reperfusion injury are significantly elevated at the end of on-pump CABG compared with off-pump surgery (5). Other groups have, however, showed increased serum levels of inflammatory mediators during coronary surgery with or without CPB (21).
Transcriptional profiling has emerged as a powerful tool for delineating complex patterns of gene expression in response to severe stimuli (18, 22). Recent studies have used microarray technology to examine the human myocardial stress response during cardiac surgery (3, 10, 11, 14, 16, 23). Ruel et al. (16) used a 12,625 gene array to examine the cardiac and peripheral gene expression responses to CPB and CA in human atrial myocardium and skeletal muscle. They showed an upregulation of inflammatory and transcription activators and apoptotic and stress genes (16). The same group also investigated the differences in gene expression profiles in atrial myocardium of diabetic and nondiabetic patients undergoing CPB and CA (23). However, to our knowledge, only one study investigated gene expression profiles in off-pump surgery. Tomic et al. (22) examined gene expression patterns in leukocytes and plasma proteomic response to on-pump and off-pump surgery. They showed that circulating leukocytes overexpress adhesion and signaling factors after contact with CPB, which potentially facilitates their trapping, e.g., in the lungs, and may promote a subsequent tissue-associated inflammatory response. Additionally, the blood leukocyte transcriptomics suggested that circulating leukocytes are not primarily responsible for the increase in blood cytokines and chemokines observed following surgery. The myocardium could, therefore, be the source for these circulating cytokines and chemokines.
The aim of our study was to investigate and compare the global transcriptomic ventricular myocardial alterations in patients undergoing off-pump and on-pump coronary artery bypass surgery.
MATERIALS AND METHODS
Patients.
This study analyses data from 10 patients with a diagnosis of three vessel ischemic heart disease undergoing elective primary myocardial revascularization with or without the use of CPB and ischemic-CA (both n = 5). The study was approved by the Local Research Ethics Committee, and all patients gave informed consent.
Anesthetic, cardiopulmonary bypass, and surgical technique for on-pump and off-pump surgery have been previously described (7, 24). In brief, for on-pump patients, intravenous heparin (300 IU/kg) was administered immediately before cannulation for CPB, and additional doses were given to maintain an activated clotting time of 480 s or greater. Cardiopulmonary bypass was instituted by cannulation of the distal ascending aorta and insertion of a single two-stage cannula into the right atrium. Nonpulsatile flow rates of 2.4 l·min−1·m−2 and normothermic temperatures (35–37°C) were used. The cardioplegic solution was prepared by the perfusionist by mixing whole blood with potassium chloride and magnesium sulphate using the methods described by Calafiore et al. (6).
The method of exposure and stabilization for performing off-pump surgery has been described previously (24). The left anterior descending coronary artery was grafted first in all patients, followed by the right coronary and circumflex coronary artery distal anastomoses. The proximal anastomoses onto the ascending aorta were constructed at the end. Postoperative management (fluid balance, transfusion requirements, and inotropic support, etc) were in accordance with unit protocols as previously described (7, 24).
ELISA protein analysis.
Whole blood was collected into heparinized tubes from five on-pump and five off-pump patients immediately after induction of anesthesia (preoperative) and 4, 24, and 48 h postoperatively. Each sample was immediately centrifuged at 4°C and 1,000 g for 15 min. The plasma samples were separated and multiple aliquots stored at −80°C until assayed. Human IL-6, IL-8, monocyte chemoattractant protein (MCP)1 (CCL2), MCP2 (CCL8), macrophage inflammatory protein (MIP)1α (CCL3), and MIP1β (CCL4) were measured in plasma samples using the e-Bioscience and R&D systems ELISA kits according to the manufacturer's instructions. Each sample was performed in duplicate. The plate was read using a microplate reader (μQuant, Bio-Tek) and the results were interpolated from the standard reference curve provided with each kit. Statistical analysis (t-test) was carried out using Instat 3 software. A P value of <0.05 was considered statistically significant.
Cardiac muscle biopsies.
Two biopsy specimens (10 mg net weight) were collected from the apex of the left ventricle using a “Trucut” needle as previously described (7). In the on-pump group, the first biopsy (pre-op) was taken immediately before institution of CPB and the second (post-op) 20 min after completion of all coronary anastomoses. For off-pump patients, the first biopsy was collected before starting the first anastomosis prior to any displacement of the heart (pre-op) and the second (post-op) 20 min after completion of all anastomoses. Each specimen was immediately snap-frozen in liquid nitrogen and stored at −80°C.
RNA extraction.
Tissue was mechanically homogenized in lysis reagent (Qiagen, Crawley, UK) and Total RNA was purified with RNeasy Micro Kit (Qiagen) and eluted into 12 μl of RNase-free water. The concentration and purity of the total RNA samples were assessed by spectrophotometry (Nanodrop, Wilmington, DE) and further analyzed for integrity with a Bioanalyzer 2100 with RNA 6000 Nano Assay (Agilent Technologies, Stockport, UK).
Gene microarrays.
Ventricular total RNAs (1 μg) from individual patients were used to generate biotinylated cRNAs. The quantity and size distribution of purified cRNA was assessed on a Bioanalyzer 2100 using RNA 6000 Nano Assay (Agilent Technologies) to ensure that the cRNA amplification was successful. Target fragmentation was achieved by incubation at 94°C for 35 min in fragmentation buffer (40 mM Tris-acetate, pH 8.1/100 mM KOAc/30 mM MgOAc). The size distribution of the fragmented labeled transcripts was assessed on the Agilent Technologies Bioanalyzer 2100 using the RNA 6000 Nano Assay. These cRNAs samples were used for hybridization to separate Affymetrix GeneChip arrays. For each experimental group, five samples from individual patients were processed. Hybridization of the labeled cRNA to the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array was carried out for 16 h in the Affymetrix GeneChip Hybridization Oven 640. Then GeneChip arrays were stained and washed on the GeneChip Fluidics Station 450 (Affymetrix). The fluorescent signals were detected with an Affymetrix GeneChip Scanner 3000 and stored as high-resolution fluorescence intensity data file. These data were initially analyzed with Affymetrix GeneChip operating software GCOS 1.2, which generates an expression report file that lists the quality control parameters. All of these parameters were scrutinized to ensure that array data had reached the necessary quality standards (scaling factor of <3-fold; average background values at 20–100 and the ratio of 3′:5′ signal no more than 3 for housekeeping genes GAPDH and β-actin). The complete MIAME-compliant datasets have been submitted and accepted by Gene Expression Omnibus at the National Center for Biotechnology Information (accession number GSE12504).
Microarray data analysis.
Separate microarrays were probed with independently generated target from each tissue. Raw data (CEL files) were uploaded into ArrayStar software version 2.1 (DNASTAR) for normalization and statistical analysis. The robust multichip analysis algorithm was used for background correction, quantile normalization and median polish summarization. The statistical analysis (ANOVA) was carried out using ArraStar software. P value was set to <0.01 and transcripts were filtered on the basis of ≥2-fold difference.
Gene annotations.
All gene annotations were checked by using online tools and databases: Entrez Gene (http://www.ncbi.nlm.nih.gov/sites/entrez). Note that the databases are in a constant state of flux and that annotations are subject to updates, redefinition, and correction. The DAVID (Database for Annotation, Visualization, and Integrated Discovery) resource (9) was used for functional annotation clustering. The software PathwayStudio and the ResNet database were used to explore the networks of interactions where the physiological regulated genes are potentially involved.
cDNA synthesis and real-time PCR.
Complementary DNA was reverse transcribed from 1 μg of total RNA using Superscript III cDNA first-strand synthesis kit (Invitrogen, UK), diluted twofold and 1 μl used in real-time PCR reactions. Optimized primers of human CCL2 (QT00212730), CCL3 (QT01008063), CCL4 (QT01008070), IL-8 (QT00000322), prostaglandin-endoperoxide synthase gene (PTGS2) (QT00040586), and SNF1-like kinase (SNF1LK) (QT00032270) were purchased from Qiagen (UK). Amplification and detection of specific products were carried out with Roche Lightcycler 1.5 detection System. Each sample was performed in duplicate. 18S mRNA was used as endogenous control transcript in each sample. Relative expression ratios were calculated by the relative quantification real-time-PCR method (12). Statistical analysis (one-way ANOVA followed by Bonferroni multiple comparison test) was carried out using Instat 3 software. A P value of <0.05 was considered statistically significant.
RESULTS
Patients characteristics and clinical outcomes are presented in Table 1. The two groups were comparable in terms of age and preoperative risk factors. There were no deaths and no major morbidity in either group. Patients in the off-pump group tended to have a shorter intensive care unit and hospital stay compared with patients in the on pump group.
Table 1.
Baseline characteristics and clinical outcomes
| On Pump (n = 5) | OPCAB (n = 5) | |
|---|---|---|
| Age | 68.0 ± 10.3 | 71.4 ± 10.2 |
| Male | 5 | 5 |
| Previous MI | 2 | 3 |
| Insulin-dependent DM | 0 | 0 |
| Hypertension | 4 | 5 |
| Body mass index | 31.1 ± 5.1 | 30.3 ± 2.6 |
| Euroscore | 2.9 ± 1.7 | 3.4 ± 2.7 |
| CPB time, min | 102.4 ± 8.4 | |
| Cross-clamp time, min | 51.4 ± 8.7 | |
| Number of grafts | 3.8 ± 0.4 | 2.8 ± 0.4 |
| In-hospital mortality | 0 | 0 |
| Postoperative ventilation time, hours | 11.6 ± 9.1 | 7.8 ± 4.9 |
| Postoperative ICU stay, hours | 60.4 ± 38.7 | 36.2 ± 15.7 |
| Postoperative hospital stay, days | 12.0 ± 10.7 | 5.0 ± 2.2 |
Data are means ± SD or numbers. OPCAB, off-pump coronary artery bypass; MI, myocardial infarction; DM, diabetes mellitus; CPB, cardiopulmonary bypass; ICU, intensive care unit.
Gene expression.
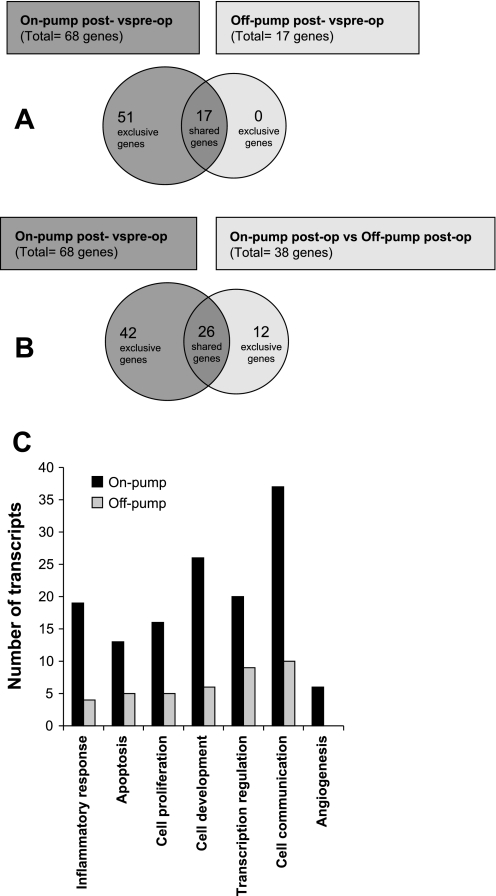
In the on-pump patients, 68 genes were identified as differentially upregulated in post-op vs. pre-op biopsies (P < 0.01; ≥2-fold; Table 2). In the off-pump patients, 17 genes were identified as differentially upregulated in post-op vs. pre-op biopsies (P < 0.01; ≥2-fold; Table 3). Interestingly all the 17 upregulated genes were shared with on-pump patients (Fig. 1A).
Table 2.
Genes differentially expressed in postversus preoperative left ventricular samples of on-pump patients
| Gene Title | Symbol | Fold | P Value | Gene ID |
|---|---|---|---|---|
| v-Fos FBJ murine osteosarcoma viral oncogene homolog | FOS | 14.875 up | 0.000117 | 2353 |
| Interleukin 8 | IL8 | 14.247 up | 0.000186 | 3576 |
| Suppressor of cytokine signaling 3 | SOCS3 | 12.611 up | 0.000169 | 9021 |
| v-Maf musculoaponeurotic fibrosarcoma oncogene homolog F | MAFF | 10.985 up | 0.00145 | 23764 |
| SNF1-like kinase | SNF1LK | 10.947 up | 0.0000204 | 150094 |
| Chemokine (C-C motif) ligand 3 | CCL3 | 10.728 up | 9.4E-10 | 414062 |
| Chemokine (C-X-C motif) ligand 2 | CXCL2 | 10.445 up | 0.00689 | 2920 |
| Activating transcription factor 3 | ATF3 | 10.229 up | 0.000107 | 467 |
| Early growth response 3 | EGR3 | 9.904 up | 0.000123 | 1960 |
| Early growth response 2 (Krox-20 homolog, Drosophila) | EGR2 | 9.170 up | 0.0000196 | 1959 |
| Nuclear receptor subfamily 4, group A, member 2 | NR4A2 | 8.912 up | 0.0000158 | 4929 |
| Solute carrier family 2 (facilitated glucose transporter), member 3 | SLC2A3 | 8.701 up | 0.0000242 | 6515 |
| Zinc finger protein 36, C3H type, homolog (mouse) | ZFP36 | 7.936 up | 0.0000034 | 7538 |
| FBJ murine osteosarcoma viral oncogene homolog B | FOSB | 7.881 up | 0.000772 | 2354 |
| Early growth response 1 | EGR1 | 7.438 up | 0.0000341 | 1958 |
| Cholesterol 25-hydroxylase | CH25H | 6.586 up | 0.000106 | 9023 |
| Chemokine (C-C motif) ligand 8 | CCL8 | 6.038 up | 0.000174 | 6355 |
| S100 calcium binding protein A12 | S100A12 | 5.443 up | 0.00008 | 6283 |
| Apolipoprotein L domain containing 1 | APOLD1 | 5.223 up | 0.00228 | 81575 |
| S100 calcium binding protein A8 | S100A8 | 5.072 up | 0.000505 | 6279 |
| AXIN1 upregulated 1 | AXUD1 | 5.044 up | 0.0000355 | 64651 |
| Kruppel-like factor 4 (gut) | KLF4 | 5.017 up | 0.000137 | 9314 |
| BTG family, member 2 | BTG2 | 4.956 up | 0.0003 | 7832 |
| Selectin E (endothelial adhesion molecule 1) | SELE | 4.895 up | 0.00115 | 6401 |
| Chemokine (C-C motif) ligand 4 | CCL4 | 4.707 up | 0.000000159 | 6351 |
| Prostaglandin-endoperoxide synthase 2 | PTGS2 | 4.626 up | 0.000255 | 5743 |
| PreB-cell colony-enhancing factor 1 | PBEF1 | 4.528 up | 0.00393 | 10135 |
| Jun B proto-oncogene | JUNB | 4.509 up | 0.00017 | 3726 |
| Stanniocalcin 1 | STC1 | 4.340 up | 0.00632 | 6781 |
| Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta | NFKBIZ | 4.189 up | 0.00148 | 64332 |
| Basic helix-loop-helix domain containing, class B, 2 | BHLHB2 | 4.074 up | 0.00000437 | 8553 |
| Dual-specificity phosphatase 1 | DUSP1 | 3.964 up | 0.000822 | 1843 |
| Cysteine-rich, angiogenic inducer, 61 | CYR61 | 3.933 up | 0.0000911 | 3491 |
| Growth arrest and DNA damage-inducible, beta | GADD45B | 3.888 up | 0.00126 | 4616 |
| Hypothetical LOC387763 | LOC387763 | 3.878 up | 0.000147 | 387763 |
| v-Myc myelocytomatosis viral oncogene homolog (avian) | MYC | 3.856 up | 0.000234 | 4609 |
| Nuclear receptor subfamily 4, group A, member 1 | NR4A1 | 3.713 up | 0.000038 | 3164 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 3.655 up | 0.00014 | 6347 |
| Interleukin 6 (interferon, beta 2) | IL6 | 3.611 up | 0.000594 | 3569 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 1 | ADAMTS1 | 3.558 up | 0.00136 | 9510 |
| Serum/glucocorticoid-regulated kinase 1 | SGK1 | 3.443 up | 0.000306 | 6446 |
| OTU domain-containing 1 | OTUD1 | 3.326 up | 0.000515 | 220213 |
| Hairy and enhancer of split 1, (Drosophila) | HES1 | 3.285 up | 0.000000689 | 3280 |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 | B3GNT5 | 3.193 up | 0.00834 | 84002 |
| Chemokine (C-X-C motif) receptor 4 | CXCR4 | 3.187 up | 0.00028 | 7852 |
| CCAAT/enhancer binding protein (C/EBP), delta | CEBPD | 3.006 up | 0.00112 | 1052 |
| Phorbol-12-myristate-13-acetate-induced protein 1 | PMAIP1 | 2.951 up | 0.000119 | 5366 |
| BCL2-related protein A1 | BCL2A1 | 2.787 up | 0.00108 | 597 |
| RAS, dexamethasone-induced 1 | RASD1 | 2.746 up | 0.000181 | 51655 |
| Epstein-Barr virus-induced gene 2 | EBI2 | 2.682 up | 0.0000197 | 1880 |
| SRY (sex determining region Y)-box 17 | SOX17 | 2.638 up | 0.00422 | 64321 |
| S100 calcium-binding protein A9 | S100A9 | 2.600 up | 0.000662 | 6280 |
| Complement component 5a receptor 1 | C5AR1 | 2.564 up | 0.00329 | 728 |
| Immediate early response 2 | IER2 | 2.564 up | 0.00000262 | 9592 |
| Regulator of G protein signaling 1 | RGS1 | 2.562 up | 0.00757 | 5996 |
| Jun oncogene | JUN | 2.561 up | 0.00355 | 3725 |
| Dual-specificity phosphatase 5 | DUSP5 | 2.530 up | 0.000213 | 1847 |
| CD83 molecule | CD83 | 2.521 up | 0.000011 | 9308 |
| CD69 molecule | CD69 | 2.393 up | 0.000213 | 969 |
| KIAA0146 | KIAA0146 | 2.268 up | 0.000752 | 23514 |
| Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | CDKN1A | 2.233 up | 0.000957 | 1026 |
| Interleukin 1 receptor antagonist | IL1RN | 2.219 up | 0.0000786 | 3557 |
| Zinc finger protein 331 | ZNF331 | 2.218 up | 0.00000985 | 55422 |
| Neural precursor cell expressed, developmentally downregulated 9 | NEDD9 | 2.169 up | 0.0052 | 4739 |
| Interleukin 1 receptor, type II | IL1R2 | 2.158 up | 0.00359 | 7850 |
| SERTA domain-containing 1 | SERTAD1 | 2.086 up | 0.0000892 | 29950 |
| Cyclin L1 | CCNL1 | 2.059 up | 0.00213 | 57018 |
| S100 calcium-binding protein P | S100P | 2.033 up | 0.00022 | 6286 |
Genes exhibiting 2-fold or greater change in expression are shown.
Table 3.
Genes differentially expressed in postoperative vs. preoperative left ventricular samples of off-pump patients
| Gene Title | Symbol | Fold | P Value | Gene ID |
|---|---|---|---|---|
| v-fos FBJ murine osteosarcoma viral oncogene homolog | FOS | 9.654 up | 0.000117 | 2353 |
| Early growth response 1 | EGR1 | 7.698 up | 0.0000341 | 1958 |
| Activating transcription factor 3 | ATF3 | 4.914 up | 0.000107 | 467 |
| Early growth response 3 | EGR3 | 4.714 up | 0.000123 | 1960 |
| Interleukin 8 | IL8 | 4.450 up | 0.000186 | 3576 |
| Zinc finger protein 36, C3H type, homolog (mouse) | ZFP36 | 4.082 up | 0.0000034 | 7538 |
| BTG family, member 2 | BTG2 | 3.396 up | 0.0003 | 7832 |
| SNF1-like kinase | SNF1LK | 3.326 up | 0.0000204 | 150094 |
| FBJ murine osteosarcoma viral oncogene homolog B | FOSB | 3.292 up | 0.000772 | 2354 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 3.082 up | 0.00014 | 6347 |
| Cysteine-rich, angiogenic inducer, 61 | CYR61 | 3.026 up | 0.000089 | 3491 |
| Early growth response 2 (Krox-20 homolog, Drosophila) | EGR2 | 3.000 up | 0.0000196 | 1959 |
| Hypothetical LOC387763 | LOC387763 | 2.873 up | 0.000147 | 387763 |
| v-Myc myelocytomatosis viral oncogene homolog (avian) | MYC | 2.570 up | 0.000234 | 4609 |
| Growth arrest and DNA damage-inducible, beta | GADD45B | 2.378 up | 0.00126 | 4616 |
| OTU domain-containing 1 | OTUD1 | 2.316 up | 0.000515 | 220213 |
| Dual-specificity phosphatase 1 | DUSP1 | 2.198 up | 0.000822 | 1843 |
Genes exhibiting twofold or greater change in expression are shown.
Fig. 1.
A: Venn diagram of the significantly regulated genes >2-fold of on-pump post- vs. preoperative and off-pump post- vs. preoperative comparisons groups. B: Venn diagram of the significantly regulated genes >2-fold of on-pump post- vs. preoperative and on-pump postoperative vs. off-pump postoperative comparisons groups. C: bar chart of Gene Ontology (GO) annotations for the category “biologic process.” The chart shows the number of transcripts in the categories separately for on-pump and off-pump comparison.
To uncover the genes implicated in CPB and ischemic-CA response, we compared the post-operative gene expression profiles of on-pump and off-pump patients. Of the genes examined, 38 genes were identified as differentially expressed in the on-pump vs. off-pump patients (P < 0.01; ≥2-fold), with all of them upregulated (Table 4). Importantly 12 genes were exclusive to on-off-pump comparison and 26 were shared with on-pump comparison (Fig. 1B).
Table 4.
Genes differentially expressed in postoperative on-pump vs. postoperative off-pump left ventricular samples
| Gene Title | Symbol | Fold | P Value | Gene ID |
|---|---|---|---|---|
| Chemokine (C-C motif) ligand 3 | CCL3 | 11.328 up | 9.4E-10 | 414062 |
| Nuclear receptor subfamily 4, group A, member 2 | NR4A2 | 6.077 up | 0.0000158 | 4929 |
| Hairy and enhancer of split 1, (Drosophila) | HES1 | 4.922 up | 0.000000689 | 3280 |
| Cholesterol 25-hydroxylase | CH25H | 4.890 up | 0.000106 | 9023 |
| Chemokine (C-C motif) ligand 8 | CCL8 | 4.871 up | 0.000174 | 6355 |
| SNF1-like kinase | SNF1LK | 4.469 up | 0.0000204 | 150094 |
| Solute carrier family 2 (facilitated glucose transporter), member 3 | SLC2A3 | 4.235 up | 0.0000242 | 6515 |
| Chemokine (C-X-C motif) receptor 4 | CXCR4 | 3.628 up | 0.00028 | 7852 |
| S100 calcium-binding protein A12 | S100A12 | 3.553 up | 0.00008 | 6283 |
| Phorbol-12-myristate-13-acetate-induced protein 1 | PMAIP1 | 3.550 up | 0.000119 | 5366 |
| Early growth response 2 (Krox-20 homolog, Drosophila) | EGR2 | 3.548 up | 0.0000196 | 1959 |
| Chemokine (C-C motif) ligand 4 | CCL4 | 3.547 up | 0.000000159 | 6351 |
| Collagen, type I, alpha 1 | COL1A1 | 3.280 up | 0.00571 | 1277 |
| Kruppel-like factor 4 (gut) | KLF4 | 3.185 up | 0.000137 | 9314 |
| SRY (sex determining region Y)-box 17 | SOX17 | 3.034 up | 0.00422 | 64321 |
| Basic helix-loop-helix domain-containing, class B, 2 | BHLHB2 | 3.027 up | 0.00000437 | 8553 |
| Jun B proto-oncogene | JUNB | 2.760 up | 0.00017 | 3726 |
| SAM domain, SH3 domain and nuclear localization signals 1 | SAMSN1 | 2.682 up | 0.00419 | 64092 |
| Immediate early response 2 | IER2 | 2.589 up | 0.00000262 | 9592 |
| AXIN1 upregulated 1 | AXUD1 | 2.566 up | 0.0000355 | 64651 |
| Collagen, type VI, alpha 2 | COL6A2 | 2.528 up | 0.00516 | 1292 |
| Serum/glucocorticoid-regulated kinase 1 | SGK1 | 2.513 up | 0.000306 | 6446 |
| Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | ID3 | 2.439 up | 0.00019 | 3399 |
| S100 calcium-binding protein A9 | S100A9 | 2.435 up | 0.000662 | 6280 |
| Fibulin 1 | FBLN1 | 2.379 up | 0.00336 | 2192 |
| SRY (sex-determining region Y)-box 18 | SOX18 | 2.375 up | 0.00903 | 54345 |
| SRY (sex-determining region Y)-box 7 | SOX7 | 2.299 up | 0.00274 | 83595 |
| CD69 molecule | CD69 | 2.237 up | 0.000213 | 969 |
| Nuclear receptor subfamily 4, group A, member 1 | NR4A1 | 2.223 up | 0.000038 | 3164 |
| SERTA domain containing 1 | SERTAD1 | 2.220 up | 0.0000892 | 29950 |
| Epstein-Barr virus-induced gene 2 | EBI2 | 2.182 up | 0.0000197 | 1880 |
| Tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | TIE1 | 2.152 up | 0.00771 | 7075 |
| CD83 molecule | CD83 | 2.139 up | 0.000011 | 9308 |
| Zinc finger protein 331 | ZNF331 | 2.068 up | 0.00000985 | 55422 |
| Matrix remodeling-associated 5 | MXRA5 | 2.055 up | 0.00244 | 25878 |
| Vascular cell adhesion molecule 1 | VCAM1 | 2.045 up | 0.00762 | 7412 |
| Latent transforming growth factor beta-binding protein 3 | LTBP3 | 2.036 up | 0.000746 | 4054 |
| Cytochrome P450, family 4, subfamily V, polypeptide 2 | CYP4V2 | 2.000 up | 0.00157 | 285440 |
Genes exhibiting 2-fold or greater change in expression are shown.
Gene Ontology biologic process annotations of gene differentially expressed in on- and off pump pre- vs. postoperative.
The DAVID (Database for Annotation, Visualization, and Integrated Discovery) resource (9) was used to obtain Gene Ontology (GO) annotations for the category “biologic process.” The biologic process categories: inflammatory response, apoptosis, cell proliferation, cell development, transcription regulation, cell communication, and angiogenesis were identified. The bar chart (Fig. 1C) shows the number of genes in the categories separately for on-pump and off-pump comparison transcripts. Transcripts in all categories, including inflammatory response and apoptosis, dramatically decreased in off-pump comparison (Fig. 1C). Most strikingly, transcripts in the category angiogenesis are exclusive to on-pump comparison (Fig. 1C). Collectively, these ontological changes indicate genetic remodeling of the myocardium subjected to CPB and ischemic-CA. This genetic alteration seems to be ameliorated by off-pump surgery.
Pathway analysis.
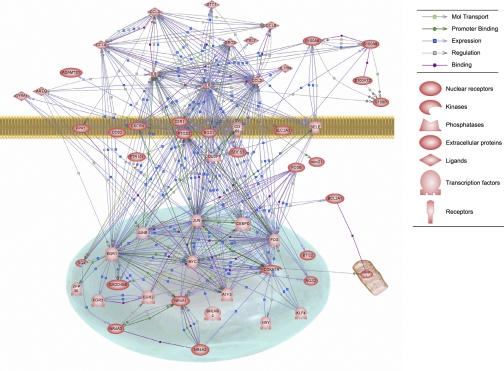
Using Pathway studio (Ariadne Genomics, Rockville, MD), we further analyzed the differentially expressed genes in on-pump post-op vs. pre-op. The analysis looked for the direct pathways connecting genes. The resulting biological association network is presented in Fig. 2. The inflammatory response factors IL-6, IL-8, CCL2, CCL3, and CCL4 showed a high number of direct connections to each other as well as to the transcription factors JUN, FOS, and early growth response (EGR)1. In the inflammation class, IL-6 exhibited the biggest node connecting most of the differentially expressed genes followed by CCL2 and IL-8. In the transcription category, JUN and FOS presented the biggest nodes.
Fig. 2.
Biological association network of the significantly regulated genes >2-fold in on-pump post- vs. preoperative comparison created using Pathway Studio Software.
Validation of microarray with real-time PCR.
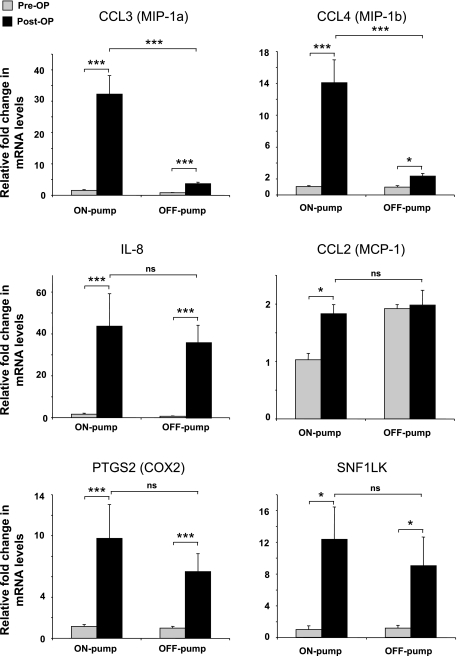
Microarray results were confirmed by using real-time quantitative PCR on six selected genes that demonstrated differential gene expression. The changes in expression levels of CCL2, CCL3, CCL4, IL8, PTGS2, and SNF1LK showed a high degree of correlation with the microarray data (Fig. 3).
Fig. 3.
Confirmation of the results from the microarray analysis of changed genes pre- and postoperatively in on- and off-pump surgery. Changes in mRNA expression of CCL3 (MIP1α), CCL4 (MIP1β), CCL2 (MCP1), IL-8, PTGS2 (COX2), and SNF1LK was verified in all 10 patients by quantitative real-time RT-PCR. Results are shown as means (± SE) fold-change. *P < 0.05; ***P < 0.001; ns, not significant.
ELISA protein analysis.
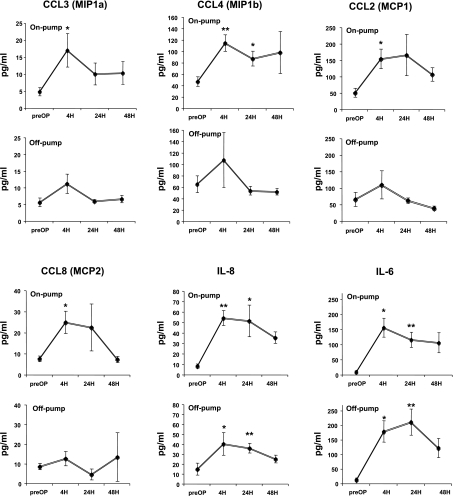
The microarray analysis showed mRNA level increase of a panel of cytokines and chemokines in heart biopsies following on-pump surgery. Assessment of plasma concentrations for these cytokines and chemokines preoperative and 4, 24, and 48 h postoperative, revealed similar profile to that obtained at the transcriptomic level: IL-6 and IL-8 were induced in response to either on-pump or off-pump surgery and this increase was detectable following surgery (Fig. 4). However, MCP1 (CCL2), MCP2 (CCL8), MIP1α (CCL3), and MIP1β (CCL4) circulating levels increased by on-pump surgery but did not change by off-pump surgery (Fig. 4).
Fig. 4.
Time course of plasma levels of IL6, IL8, MCP1, MCP2, MIP1α, and MIP1β. Dots connected by solid line represent the time course for median protein concentration in plasma samples (in pg/ml). For MCP1, MCP2, MIP1α, and MIP1β, significantly enhanced protein expression was confirmed after 4 h of on-pump surgery, whereas no changes were observed following off-pump surgery. For IL-6 and IL-8, significantly enhanced protein expression was confirmed after 4 h of on-pump and off-pump surgeries. Results are shown as means (± SE). *P < 0.05 vs. preoperative, **P < 0.01 vs. preoperative.
DISCUSSION
The development of off-pump surgery, which avoids cardiopulmonary bypass and cardioplegic arrest used in the on-pump procedure, allows for the first time a direct comparison of two groups of patients with the same pathology undergoing coronary revascularization. This is the first genomic study comparing the effect of both interventions on transcriptomic changes in left ventricle biopsies using gene expression profiling technology. The main finding of the study is that off-pump surgery is associated with fewer alterations in gene expression linked to inflammation and apoptosis. This work completes earlier studies (3, 10, 11, 13, 14, 16, 23) that have focused on on-pump surgery with CPB and ischemia-CA.
The categories of genes found to be upregulated in on-pump subjects were early transcription factors such as FOS, EGR1, and EGR2, mediators of the inflammatory response such as IL8 and CCL3 and apoptosis genes such as NR4A1. Performing the surgery off-pump was associated with dramatically fewer number of upregulated genes implicated in transcription, inflammatory response, and apoptosis. A likely explanation for the difference between the two techniques is that on-pump surgery involves an inflammatory response due to CPB and myocardial ischemic stress due to CA.
In the category inflammatory response, several cytokine and chemokine genes were upregulated in on-pump patients. IL-8 showed an important gene expression change and was highly upregulated (14:1 and 4:1 ratios, respectively) in both on-pump and off-pump patients following surgery. IL-6 showed 3.6- and 1.9-fold increase by surgery in on-pump and off-pump subjects, respectively. The plasma protein level of IL-6 and IL-8 reflected the transcriptomic changes observed. It has previously been shown that IL-8 and IL-6 are produced and released by the ischemic myocardium and cardiac myocytes exposed to ischemia-reperfusion (8, 15, 20). Here we provide evidence that the induction of IL-6 and IL-8 is not specific to on- or off-pump surgery and that these two cytokines are part of a general inflammatory response associated with the surgery itself. Our observations are similar to that of an investigation in which the induction of IL-6 and IL-8 in the myocardium was demonstrated following acute ischemia (10). Additionally the global responses in our study were quite similar to this previous investigation (10).
The upregulation of IL-6, CCL2 (MCP1), and CCL4 (MIP1β) gene and protein expressions is consistent with a recent study looking at peripheral blood mononuclear cells transcriptomic and plasma proteomic responses to on-pump and off-pump surgery (22). Indeed Tomic et al. (22) demonstrated an increase in plasma protein level of these three genes after surgery. However, they did not notice any significant alteration of the corresponding mRNA transcripts in the peripheral blood mononuclear cells, concluding that the latter are not the primary source of plasma-enhanced level of these cytokine and chemokines following on-pump surgery. Although the trigger for cytokines and chemokines' upregulation is likely to be systemic (CPB) and cardiac (CA), the question remains of whether an increase in cardiac gene expression is translated into protein production. There is experimental evidence that isolated hearts exposed to ischemia and reperfusion have increased levels of cytokines (8, 20). Our data suggest that ventricular heart cells may contribute to plasma cytokine and chemokine alteration observed. However, these elevations could also be caused by release from other organs and or posttranscriptional changes induced by CPB and ischemic-CA.
Furthermore, here we provide evidence that the myocardial mRNA and plasma protein levels of CCL3 (MIP1α) and CCL4 (MIP1β) were associated with CPB and ischemic-CA. Indeed, performing the surgery off-pump reduced significantly the levels of these two chemokines at both the mRNA and plasma levels. Unlike CCL2, which appears to reflect a more generalized stress response, CCL3 and CCL4 seem to be specifically associated with initiation of CPB and ischemic-CA, suggesting potential role for these factors different from that of CCL2.
In addition to the above cytokine and chemokines, we validated the differential expression of two more genes, SNF1LK (also known as MSK and SIK) and PTGS2, also known as cyclooxygenase 2 (COX2). SNF1LK, upregulated by 11-fold in the on-pump group and 3-fold in the off-pump group, is a serine/threonine kinase previously shown to be expressed specifically in myocardial cells of the developing heart (17). Additionally, the region encompassing SNF1LK locus has been implicated in congenital heart defects often observed in patients with Down syndrome (19). However, no SNF1LK expression has been documented in the normal or ischemic adult heart. The upregulation of SNF1LK mRNA by surgery stress suggests a new role for this kinase in the adult heart. PTGS2 (COX2), upregulated by fivefold in on-pump patients, encodes a key enzyme in prostaglandin biosynthesis. PTGS2 expression is induced in cardiomyocytes during myocardial infarction and thought to play a role in mediating inflammatory reaction (25). Additionally new evidence suggests a potential cause-effect link between PTGS2 expression and enhanced myocardial apoptosis (1). In concert with these previous studies, our findings indicate a role for PTGS2 in myocardial injury following CPB and ischemic-CA.
When comparing on-pump vs. off-pump at the end of surgery, our data suggest that CPB and ischemic-CA upregulates an injury-related response as demonstrated by the upregulation of apoptosis and remodeling markers (NR4A1, FBLN1, COL1A1, COL1A2, MXRA5), whereas surgery stress alone in off-pump surgery upregulates mainly a cytoprotective genetic program (ATF3, EGR1). However, this does not answer the specific CPB and ischemic-CA contribution to the gene expression changes observed. Furthermore, CPB and ischemic-CA elicit both cardioprotective (ATF3, EGR1) and injury-related (NR4A1, PMAIP1) transcriptional changes. This finding is similar to previous investigation showing the activation of both proapoptotic and antiapoptotic mediators after CPB and ischemic-CA (14).
Finally and in support to our findings, 15 of the upregulated genes in our on-pump ventricular biopsies were found to be upregulated by CPB and ischemic-CA in on-pump atrial biopsies in previous study (16). Despite the apparent differences between atrial and ventricular myocardium, both tissues respond in a similar way to CPB and ischemic-CA (16). Interestingly, our data show mainly an upregulation of gene expression in ventricular biopsies following CPB and ischemic-CA in contrast to atrial biopsies showing up- and downregulation of gene expression in response to the same stimulus (16, 23).
Limitation of the study.
In this study we showed that a number of cytokine and chemokine genes were upregulated postoperatively in on-pump patients at the mRNA level in the myocardium and at the protein level in the blood. It is therefore reasonable to suggest that ventricular heart cells contribute to plasma cytokine and chemokine alteration observed in patients' blood. However, because the blood samples used were not transcardiac, we cannot be certain as to whether the increased circulating levels of cytokines and chemokines are from the heart and/or other organs. Furthermore, our study cannot distinguish the individual contribution of CPB and ischemic-CA to the transcriptome changes seen in the on-pump patients.
Conclusion.
Off-pump surgery reduced the alteration in gene expression associated with inflammation, apoptosis, hypertrophy, and remodeling seen after on-pump surgery with cardiopulmonary bypass and ischemic-cardioplegic arrest. This study lends weight to mechanistic understanding of the previously demonstrated advantages of off-pump over on-pump surgery in terms of early functional changes, myocardial protection and clinical outcomes (2, 5). Since on-pump procedures are unavoidable in some patient subgroups, our study also identifies multiple possible targets for intervention to reduce its impact on the ventricular myocardium.
GRANTS
This work was funded by the British Heart Foundation, the Garfield Weston Trust, and the National Institute of Health Research Bristol Biomedical Research Unit in Cardiovascular Medicine.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Professors D. Murphy, A. Newby and S. Suleiman for reading this manuscript and valuable comments.
REFERENCES
- 1.Abbate A, Santini D, Biondi-Zoccai GG, Scarpa S, Vasaturo F, Liuzzo G, Bussani R, Silvestri F, Baldi F, Crea F, Biasucci LM, Baldi A. Cyclo-oxygenase-2 (COX-2) expression at the site of recent myocardial infarction: friend or foe? Heart 90: 440–443, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelini GD, Taylor FC, Reeves BC, Ascione R. Early and midterm outcome after off-pump and on-pump surgery in Beating Heart Against Cardioplegic Arrest Studies (BHACAS 1 and 2): a pooled analysis of two randomized controlled trials. Lancet 359: 1194–1199, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Arab S, Konstantinov IE, Boscarino C, Cukerman E, Mori A, Li J, Liu PP, Redington AN, Coles JG. Early gene expression profiles during intraoperative myocardial ischemia-reperfusion in cardiac surgery. J Thorac Cardiovasc Surg 134: 74–81, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Ascione R, Angelini GD. OPCAB surgery: a voyage of discovery back to the future. Off-pump coronary artery bypass. Eur Heart J 24: 121–124, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Ascione R, Lloyd CT, Underwood MJ, Lotto AA, Pitsis AA, Angelini GD. Inflammatory response after coronary revascularization with or without cardiopulmonary bypass. Ann Thoracic Surg 69: 1198–1204, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Calafiore AM, Teodori G, Mezzetti A, Bosco G, Verna AM, Di Giammarco G, Lapenna D. Intermittent antegrade warm blood cardioplegia. Ann Thoracic Surg 59: 398–402, 1995 [DOI] [PubMed] [Google Scholar]
- 7.Caputo M, Bryan AJ, Calafiore AM, Suleiman MS, Angelini GD. Intermittent antegrade hyperkalaemic warm blood cardioplegia supplemented with magnesium prevents myocardial substrate derangement in patients undergoing coronary artery bypass surgery. Eur J Cardiothorac Surg 14: 596–601, 1998 [DOI] [PubMed] [Google Scholar]
- 8.Chandrasekar B, Mitchell DH, Colston JT, Freeman GL. Regulation of CCAAT/enhancer binding protein, interleukin-6, interleukin-6 receptor, and gp130 expression during myocardial ischemia/reperfusion. Circulation 99: 427–433, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3, 2003 [PubMed] [Google Scholar]
- 10.Gabrielsen A, Lawler PR, Yongzhong W, Steinbruchel D, Blagoja D, Paulsson-Berne G, Kastrup J, Hansson GK. Gene expression signals involved in ischemic injury, extracellular matrix composition and fibrosis defined by global mRNA profiling of the human left ventricular myocardium. J Mol Cell Cardiol 42: 870–883, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Konstantinov IE, Coles JG, Boscarino C, Takahashi M, Goncalves J, Ritter J, Van Arsdell GS. Gene expression profiles in children undergoing cardiac surgery for right heart obstructive lesions. J Thorac Cardiovasc Surg 127: 746–754, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 29: e45, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podgoreanu MV, Michelotti GA, Sato Y, Smith MP, Lin S, Morris RW, Grocott HP, Mathew JP, Schwinn DA. Differential cardiac gene expression during cardiopulmonary bypass: ischemia-independent upregulation of proinflammatory genes. J Thorac Cardiovasc Surg 130: 330–339, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Ramlawi B, Feng J, Mieno S, Szabo C, Zsengeller Z, Clements R, Sodha N, Boodhwani M, Bianchi C, Sellke FW. Indices of apoptosis activation after blood cardioplegia and cardiopulmonary bypass. Circulation 114: I257–I263, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Ren G, Dewald O, Frangogiannis NG. Inflammatory mechanisms in myocardial infarction. Curr Drug Targets Inflamm Allergy 2: 242–256, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Ruel M, Bianchi C, Khan TA, Xu S, Liddicoat JR, Voisine P, Araujo E, Lyon H, Kohane IS, Libermann TA, Sellke FW. Gene expression profile after cardiopulmonary bypass and cardioplegic arrest. J Thorac Cardiovasc Surg 126: 1521–1530, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Ruiz JC, Conlon FL, Robertson EJ. Identification of novel protein kinases expressed in the myocardium of the developing mouse heart. Mech Dev 48: 153–164, 1994 [DOI] [PubMed] [Google Scholar]
- 18.Sehl PD, Tai JT, Hillan KJ, Brown LA, Goddard A, Yang R, Jin H, Lowe DG. Application of cDNA microarrays in determining molecular phenotype in cardiac growth, development, and response to injury. Circulation 101: 1990–1999, 2000 [DOI] [PubMed] [Google Scholar]
- 19.Stephenson A, Huang GY, Nguyen NT, Reuter S, McBride JL, Ruiz JC. snf1lk encodes a protein kinase that may function in cell cycle regulation. Genomics 83: 1105–1115, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Suleiman MS, Ascione R, Chase A, Harrington R, Angelini GL. Cardiac and systemic inflammatory response during open-heart surgery with or without cardiopulmonary bypass. Appl Cardiopulm Pathophysiol 11: 47–55, 2007 [Google Scholar]
- 21.Tarnok A, Hambsch J, Emmrich F, Sack U, van Son J, Bellinghausen W, Borte M, Schneider P. Complement activation, cytokines, and adhesion molecules in children undergoing cardiac surgery with or without cardiopulmonary bypass. Pediatr Cardiol 20: 113–125, 1999 [DOI] [PubMed] [Google Scholar]
- 22.Tomic V, Russwurm S, Moller E, Claus RA, Blaess M, Brunkhorst F, Bruegel M, Bode K, Bloos F, Wippermann J, Wahlers T, Deigner HP, Thiery J, Reinhart K, Bauer M. Transcriptomic and proteomic patterns of systemic inflammation in on-pump and off-pump coronary artery bypass grafting. Circulation 112: 2912–2920, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Voisine P, Ruel M, Khan TA, Bianchi C, Xu SH, Kohane I, Libermann TA, Otu H, Saltiel AR, Sellke FW. Differences in gene expression profiles of diabetic and nondiabetic patients undergoing cardiopulmonary bypass and cardioplegic arrest. Circulation 110: II280–II286, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Watters MP, Ascione R, Ryder IG, Ciulli F, Pitsis AA, Angelini GD. Haemodynamic changes during beating heart coronary surgery with the ‘Bristol Technique’. Eur J Cardiothorac Surg 19: 34–40, 2001 [DOI] [PubMed] [Google Scholar]
- 25.Zidar N, Dolenc-Strazar Z, Jeruc J, Jerse M, Balazic J, Gartner U, Jermol U, Zupanc T, Stajer D. Expression of cyclooxygenase-1 and cyclooxygenase-2 in the normal human heart and in myocardial infarction. Cardiovasc Pathol 16: 300–304, 2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.