Abstract
Metazoan akirin genes regulate innate immunity, myogenesis, and carcinogenesis. Invertebrates typically have one family member, while most tetrapod and teleost vertebrates have one to three. We demonstrate an expanded repertoire of eight family members in genomes of four salmonid fishes, owing to paralog preservation after three tetraploidization events. Retention of paralogs secondarily lost in other teleosts may be related to functional diversification and posttranslational regulation. We hypothesized that salmonid akirins would be transcriptionally regulated in fast-twitch skeletal muscle during activation of conserved pathways governing catabolism and growth. The in vivo nutritional state of Arctic charr (Salvelinus alpinus L.) was experimentally manipulated, and transcript levels for akirin family members and 26 other genes were measured by quantitative real-time PCR (qPCR), allowing the establishment of a similarity network of expression profiles. In fasted muscle, a class of akirins was upregulated, with one family member showing high coexpression with catabolic genes coding the NF-κB p65 subunit, E2 ubiquitin-conjugating enzymes, E3 ubiquitin ligases, and IGF-I receptors. Another class of akirin was upregulated with subsequent feeding, coexpressed with 14-3-3 protein genes. There was no similarity between expression profiles of akirins with IGF hormones or binding protein genes. The level of phylogenetic relatedness of akirin family members was not a strong predictor of transcriptional responses to nutritional state, or differences in transcript abundance levels, indicating a complex pattern of regulatory evolution. The salmonid akirins epitomize the complexity linking the genome to physiological phenotypes of vertebrates with a history of tetraploidization.
Keywords: Salmonidae family, phylogenetic analysis, phosphorylation, hierarchical clustering
akirins are ∼20- to 25-kDa nuclear localized proteins that regulate gene expression in many physiological processes, including the innate immune response of insects and mammals (25), metazoan myogenesis (24, 40, 47), mammalian carcinogenesis (33), and insect growth/reproduction (17, 18). They cannot bind DNA (25) but interact with cofactors to promote or repress mRNA transcription including 14-3-3 proteins (33) and the basic helix-loop-helix transcription factor Twist (24). The importance of akirin genes to animal development is evident from the lethal embryonic phenotype of mice knockouts (25) as well as the lethal or reduced growth phenotypes demonstrated by targeted knockdown in Drosophila (25), arachnid ticks (17), and the nematode Caenorhabditis elegans (39).
Although akirin genes arose before the origin of plant, fungi, and animal lineages, they are only prevalent in metazoan genomes (38). Many invertebrates have a single family member (akirin), coorthologous to two paralogs (akirin1 and 2) generated by a genomic duplication event in the vertebrate stem of chordates (38). Mammals and amphibians retain both paralogs, whereas avians lost akirin1 (38). Teleosts of the Acanthopterygii lineage have two akirin2 paralogs, called akirin2(1) and 2(2) (38), that likely arose during a tetraploidization event at the base of teleost evolution (30, 37). However, one paralog was lost in the Ostariophysi lineage, which includes the model zebrafish Danio rerio (38). There was also good evidence that akirin1 duplicated in a common teleost ancestor, with both Ostariophysi and Acanthopterygii lineages subsequently losing a paralog (38). Therefore, vertebrates examined to date, including all the main genetic models, have a maximum of three akirin genes. One lineage unstudied in terms of its repertoire of akirin genes is the Salmonidae family of teleosts, an established vertebrate physiological model and rising model for genomics and evolution (50), with genome sequencing projects under way in Atlantic salmon (by cGRASP; http://web.uvic.ca/grasp/) and rainbow trout (by INRA; http://www.international.inra.fr/). The salmonid genome has gone through a further tetraploidization event compared with most other teleosts (3) and, while in the process of reverting to a diploid state, is thought to retain 50–75% of genes as duplicates (5; see, e.g., Ref. 37). Therefore, studies of salmonid gene families can provide important insights into the link between tetraploidization and physiological phenotypes.
The developmentally essential mouse akirin2 gene functions at the end point of cytokine-induced innate immune response pathways as a mediator of the nuclear factor-κ light chain enhancer of activated B cells (NF-κB) transcription factor complex (25). Classical NF-κB is formed of two subunits called p65 and p50 and is predominantly located in the cytoplasm, because of its association with proteins from the inhibitor of κB (IκB) family (34). Upon the binding of various cytokines (e.g., TNF-α, IL-6, IL-1β) to one of several receptors, IκB becomes phosphorylated by a large protein complex called inhibitor of IκB kinase (IKK) and then targeted to the proteosome, allowing NF-κB to enter the nucleus (34). Nuclear localized Akirin2 was essential for the transcriptional activation of a subset of NF-κB target genes at the end of this pathway (25). The classical NF-κB pathway also potently inhibits muscle growth and triggers/maintains muscle atrophy (29). For example, skeletal myogenesis is strongly promoted in mice lacking the p65 subunit (6), whereas the transgenic overexpression of an NF-κB activating protein induced a profound atrophic phenotype (11). Furthermore, NF-κB activates the expression of components of the ubiquitination pathway required for muscle atrophy, including an E2 ubiquitin-conjugating enzyme in response to TNF-α stimulation (35) and the E3 ubiquitin-ligase Murf1, independent of TNF-α (11). While no role for akirin2 in myogenesis has yet been established, it is feasible that this gene can regulate NF-κB target genes at the end point of conserved signaling pathways that mediate both the innate immune response and muscle catabolism.
Unlike its paralog, Akirin1 is not an essential component of the NF-κB-mediated innate immune response of mice (25). However, this protein does function as part of the phosphoinositide 3-kinase (PI3K)-insulin-like growth factor (IGF)-V-akt murine thymoma viral oncogene homolog (Akt) signaling pathway (40, 47), which has a direct opposite role to NF-κB signaling in muscle, where it promotes protein synthesis and cell growth while inhibiting muscle atrophy by negative regulation of components of the ubiquitination pathway such as E3 ubiquitin-ligases (reviewed in Ref. 23). Specifically, in C2C12 cells, Akirin1 overexpression promoted myogenic differentiation and hypertrophy, upregulated IGF-II transcript/protein expression, and increased the expression of phosphorylated Akt (40).
In this research, our first aim was to experimentally obtain and evolutionarily characterize the complete repertoire of akirins conserved in salmonid genomes including Arctic charr, Atlantic salmon (Salmo salar L.), brown trout (S. trutta L.), and rainbow trout (Oncorhynchus mykiss Richardson). Our second aim was to test the hypothesis that the salmonid akirin family has physiological roles in regulating cellular growth and catabolism. Accordingly, we developed quantitative real-time PCR (qPCR) assays to measure transcript abundance levels of each family member, along with a wide assortment of genes that regulate IGF signaling or muscle catabolism though NF-κB-dependent and -independent mechanisms. We also examined whether there was coexpression between akirin and 14-3-3 protein gene families of salmonids, since in rodents 14-3-3 proteins bind Akirin2 during carcinogenesis (33). Expression profiles of 34 genes were measured in Arctic charr skeletal muscle after experimental manipulation of catabolic or anabolic pathways, providing insights into the transcriptional networks in which the expanded salmonid akirin family might function and the complexity of its regulatory evolution in relation to physiological systems.
EXPERIMENTAL PROCEDURES
Bioinformatics.
With human (Homo sapiens) Akirin1 and 2 amino acid sequences as in silico probes, directed tBLASTn searches were performed against National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) nucleotide and expressed sequence tag (EST) databases for the following taxa: Salmonidae, Paracanthopterygii, Ostariophysii, and Aves. Other Akirin sequences were obtained from Ensembl (http://www.Ensembl.org) as described elsewhere (38). Nuclear localization signals (NLSs) for each salmonid Akirin family member were predicted with PSORT II (42) and phosphorylation sites with NetPhos2.0 (7).
Phylogenetic analyses.
Phylogenetic analysis was performed on 61 high-quality complete or near-complete Akirin amino acid sequences from a range of vertebrates as well as outgroups from two chordate and one nonchordate deuterostome species (details in Supplemental Table S1).1 Sequence alignment was performed with Mafft v.6 employing the G-INS-I strategy and the Mafft-homologs feature (31). A 356-site output was manually checked and submitted to Gblocks (13), where the “less stringent” block selection option was used to reduce ambiguous and/or saturated sites. A final alignment of 142 sites (available on request to D. J. Macqueen) was used for phylogenetic analysis with maximum likelihood (ML) and Bayesian inference (BI). The alignment was submitted to ProtTest (1) to determine the best fitting of 112 models of substitution according to Akaike's information criterion statistics. The best-supported model was JTT+G (Jones-Taylor-Thornton substitution model with estimation of the gamma distribution parameter of among-site rate variation). ML was performed with PhyML (28) employing JTT+G with 4 substitution rate categories and 1,000 parametric bootstrap replicates to obtain branch confidence values. BI was performed with MrBayes v.3.12 (45) again with JTT+G. Two runs were used, each of a single chain of 10 million generations, sampled every 10,000 generations. Convergence was assessed by comparing the standard deviation of split frequencies between runs. Visual assessment with Tracer v1.4 (19) indicated that a suitable mixing of Markov chains was obtained. Five hundred trees were excluded, and the independence of the remaining samples was confirmed by the lack of significant increase in autocorrelation in postburnin tree log-likelihood values assessed with the ACF function of Minitab 13.2 (Minitab). A neighbor joining analysis was also performed in Mega 4.0 (49) with 22 akirin1 nucleotide sequences. Maximum composite likelihood was employed, with uniform among-site rate variation, considering all codon positions and both transversions/transitions and using 1,000 bootstrap replicates to obtain branch confidence values.
Comparing functional constraints of akirin family member paralogs.
A set of amino acid sequences similar to that used for the phylogenetic analysis (minus the invertebrate Akirins) was aligned by an equivalent approach. The alignment was manually checked, and a polyalanine tract specific to Akirin2 of mammals and avians was removed, leaving 198 sites in total (available on request to D. J. Macqueen). Aligned salmonid Akirin family members were then submitted to the rate-shift analysis server (www.daimi.au.dk/∼compbio/LRTs) to identity type II sites between Akirin family members at various levels of paralogy, as inferred from the phylogenetic analysis. When comparing two related proteins, type II sites are functionally constrained in both sequences but code distinct residues (27, 32) (type II sites are described further in results and discussion). The method described in Reference 32 employs likelihood ratio tests to identify statistically significant type II sites under a given phylogeny. For each of the four salmonid species, Akirin1(1a) and 1(1b) were together compared with Akirin1(2a) and 1(2b), Akirin2(1a) and 2(1b) were together compared with Akirin2(2a) and Akirin2(2b), Akirin1(1a) was compared with Akirin1(1b), Akirin1(2a) was compared with Akirin1(2b), Akirin2(1a) was compared with Akirin2(1b), and Akirin2(2a) was compared with Akirin2(2b).
Animals and tissue dissections used for akirin cloning experiments.
Arctic charr were reared at the Sauôárkrókur research station of Hólar University College and represented a laboratory-maintained wild anadromous population ranging from ∼3 to 15 g in weight. Atlantic salmon were obtained from Landcatch (Lochgilphead, UK) and represented an aquaculture population ranging from ∼40 to 70 g in weight. Brown trout and rainbow trout represented aquaculture populations purchased from College Mill trout farm (Almondbank, Perth, UK) and ranged in body size from ∼430 to 560 g in weight. For each species, three or four individuals representing both males and females were sampled after being fed to satiation. Pure dissections of nine tissues were obtained, including skeletal muscle (pool of fast- and slow-twitch fiber types), heart ventricle, liver, brain, gill, eye, skin, and ovary. Tissues were flash frozen in liquid N2 and stored at −80°C.
Experiment manipulating nutritional state of Arctic charr skeletal muscle.
The Animal Welfare Committee of Hólar University College approved all experiments with Arctic charr. Approximately four hundred individuals representing 15 populations from several distinct geographical locations were placed in two replicate tanks and then maintained under identical temperature (∼8°C) and photoperiod regimes (12 h dark:12 h light). The different populations were distinguished by fin clipping. Fish were fed to satiation with standard aquaculture feed (EWOS) supplemented thrice daily with bloodworms. Fish were acclimated to laboratory conditions for several weeks and then sampled before being fasted for a period of 21 days and sampled again. Subsequently, fish were fed to satiation as described above and sampled after a period of 2, 4, 6, 9, and 14 days. At each sampling point, six fish from the same population as used for akirin cloning experiments were randomly sampled from each tank and killed humanely by a sharp blow to the head and severing of the spinal cord. Pure samples of fast-twitch skeletal muscle were dissected from a common portion of the dorsal epaxial myotome and flash frozen in liquid N2.
Cloning and sequencing of complete amino acid coding cDNA sequences of akirin in four salmonid species.
For each salmonid species, total RNA was extracted from ∼100 mg of each tissue per individual with Tri reagent (Sigma) according to the manufacturer's instructions. Total RNA was separated by gel electrophoresis, and intact 18 and 28S RNA bands were used to infer integrity of the mRNA. RNA purity was also examined with a NanoDrop spectrophotometer (Thermo Scientific), where 230-to-260-nm ratios exceeded 2.2. One microgram of pooled total RNA (equally representing each tissue RNA from each species) was then used as a template for first-strand cDNA synthesis with QuantiTect Reverse Transcriptase (Qiagen) according to the manufacturer's instructions, including a genomic DNA (gDNA) removal step. Each cDNA sample was diluted 25-fold in nuclease-free water. Standard reverse transcription-polymerase chain replication reactions were performed for each species, containing a first-strand cDNA template (40 ng of reverse-transcribed total RNA), 1.25 U of Taq polymerase (Bioline), and each primer pair at 200 nM (details in Table 1). Cycling conditions included 1 step of 95°C for 10 min, followed by 35 cycles of 30 s at 95°C, 30 s of a primer-dependent temperature (Table 1), and 1 min at 72°C, followed by 1 cycle of 72°C for 10 min. Products were separated by gel electrophoresis, purified with a QIAquick Gel Extraction Kit (Qiagen), ligated into a pCR4-TOPO T/A vector (Invitrogen), and transformed into competent Escherichia coli (Invitrogen). Ten transformed clones of each product were sequenced in a single direction with BigDye Terminator v3.1 Ready Reaction Mix (Applied Biosystems) and were read with an Applied Biosystems 3730 DNA sequencer (Geneservice, Oxford, UK).
Table 1.
Details of primer pairs used for cloning and qPCR experiments
| Gene | Accession No. | Sense Primer (5′-3′) | Tm, °C | Antisense Primer (5′-3′) | Tm, °C | Spanning Exon-Junction | Annealing Temp, °C | Product Size, bp |
|---|---|---|---|---|---|---|---|---|
| akirin1(1a) | GQ247756 | TTCGTTGTCTCTTTTGAGCAGG | 60 | CAGTAGAAGAGCAGCCATCTTGTG | 61 | N/A | 58 | 642 |
| GTCTCTTTTGAGCAGGCACCAT | 61 | CGGGGGAGACATCGAGTGTGATG | 68 | Neither | 62 | 197 | ||
| akirin1(1b) | GQ247757 | TACATCTTCGTTGTCTATTTTGCTCAG | 62 | TCTAGTGGAGGAAGGGGCAGTAG | 62 | N/A | 59 | 664 |
| CATCTTCGTTGTCTATTTTGCTCAGG | 63 | GTCGGGGGAGACATCGAGTGCG | 71 | Neither | 62 | 206 | ||
| akirin1(2a) | GQ247758 | CTACATTTTGACCGACACCCC | 59 | TGGAGATAGGCGAGCAGCAGT | 62 | N/A | 59 | 675 |
| CTCAAGGACCATGACGAGAACATCC | 66 | GACATAGCTGGAGGGTCTGGCG | 66 | Antisense | 62 | 141 | ||
| akirin1(2b) | GQ247759 | CTAGCATGGCGTGCGGAG | 61 | GCAGGTAGCTTACTGTTAAGAGTTCA | 59 | N/A | 59 | 574 |
| GAGAACATAGACTTACCCCAGAGCAGA | 64 | GGTCCCTCTTCATAGACACTCCTGATG | 66 | Both | 64 | 185 | ||
| akirin2(1a) | GQ247760 | ATGGCTTGTGGGGCTACTTTG | 62 | TCAGGAAACATAGCTAGCAGGTTG | 61 | N/A | 59 | 555 |
| TCGTCCAGACTCACAACGGAGCAA | 69 | GATGGGGAGATGGCACCAGAAGATGTT | 71 | Both | 64 | 191 | ||
| akirin2(1b) | GQ247761 | TCAGGAAACATAGCTAGCAGGTTG | 62 | TCAGGAAACATAGCTAGCAGGTTG | 61 | N/A | 59 | 555 |
| GCCCCAATGTCTCCAGCCGCTTAC | 71 | TATTGTGCAGGATTTGCTCGGTTGTG | 69 | Antisense | 64 | 115 | ||
| akirin2(2a) | GQ247762 | ATCATGGCGTGTGGAGCGAC | 64 | TCAGGAAACATAGCTAGCAGGTTG | 61 | N/A | 59 | 546 |
| CGGTGTCCCCATCATCATCCT | 64 | TCTCCTCATATTCCTCCCGCACTTTG | 69 | Neither | 64 | 354 | ||
| akirin2(2b) | GQ247763 | ATCATGGCGTGTGGAGCGAC | 64 | TCAGGAAACATAGCTAGCAGGTTG | 61 | N/A | 59 | 549 |
| GTTTCCCCATCATCCTCCTCATCC | 67 | GGTCTCCTCATATTCCTCCCGAACTTTA | 68 | Neither | 65 | 242 | ||
| IGF-I | GU933431 | GTGATGTCTTCAAGAGTGCGATGTG | 64 | CGCCCCTGTTGCCGCCGAA | 74 | Sense | 61 | 99 |
| IGFBP5.2 | GU933428 | GCAGCCTGAGTTTGGACCCT | 61 | CGTGATGGTTTACACTGCTTGTG | 61 | Sense | 60 | 151 |
| IGFBP6 | GU933435 | CGCATAGTGGTGAAATAGAAAAGGC | 64 | CCCTTGGAGGAACGACACTGCTT | 67 | Both | 60 | 160 |
| pappa | GU933427 | ACCGCAGACAGCGTACTCAAAG | 62 | TTGTCTCCCATAAAGGGCTCAC | 60 | Antisense | 60 | 140 |
| stat3 | GU933429 | CCTTAAAAGTCAGGGTGAGTTGTCC | 63 | CAGTCAGCCAGCTCGTCATCGG | 68 | Sense | 60 | 201 |
| mstn1a | NM_001123549 | GGGGGCAGGCACATAGGT | 60 | TCCATGAAGGGTTGCAGTC | 56 | Antisense | 60 | 211 |
| UBE2H(1) | NM_001165354 | ACAGCCCTTTACGATCTCACCAAT | 58 | TTCTGGATGTACTCTTTGATTTTCTG | 59 | Neither | 59 | 155 |
| UBE2H(2) | BX080911 | AAAGGAGCAAGAGGAGGGAGGTG | 65 | GAAATGATAGTCCGTGTGGGTGAG | 63 | Neither | 60 | 136 |
| NF-κB p65 | EL548185 | AGCATCCCTGGAGAGAAGAGCAA | 64 | ATGCCCAGGTTTTGAAAACTGTG | 63 | Antisense | 60 | 217 |
| 14-3-3γ1 | AY370885 | AGCCATGAAATCGGTGACAGAGT | 61 | CGCTGTAGGACTTCTCAGAGGACT | 63 | Sense | 61 | 391 |
| 14-3-3γ2 | AY370886 | GCTGCCATGAAATCGGTAACAG | 62 | TTGTAAGCCTTCTCTGATGATTCG | 61 | Sense | 61 | 391 |
| 14-3-3β2 | NM_001124469 | CTTCTATGAAATCCTCAACAACCCA | 62 | CCGATGTCCACAGAGTCAGGTGTC | 64 | Antisense | 61 | 160 |
| rps13 | BT059859 | CCCTCTCAGATCGGTGTGATCC | 63 | TCCTTGTCCTTTCTGTTCCTCTCC | 63 | Both | 60 | 193 |
| rps29 | NM_001139600 | GGGTCATCAGCAGCTCTATTGG | 61 | AGTCCAGCTTAACAAAGCCGATG | 63 | Antisense | 60 | 167 |
| rpl4 | BT057966 | CCTTCAGAAACATCCCTGGTATCAC | 63 | GGGCAGATTGTAGTCTACCTTGAGAG | 62 | Both | 62 | 182 |
| rpl13 | BT060300 | CGCTCCAAGCTCATCCTCTTCCC | 68 | CCATCTTGAGTTCCTCCTCAGTGC | 64 | Antisense | 62 | 79 |
Primer pairs in italic type were used for cloning; primer pairs in roman type were used for quantitative real-time PCR (qPCR). Accession no, NCBI GenBank accession number related to the sequence; Tm, primer melting temperature, calculated with NetPrimer (http://www.premierbiosoft.com), stated to the nearest 0.5°C; product size, product size for Arctic charr (Salvelinus alpinus). Spanning Exon-Junction column indicates whether the sense, antisense, both, or neither primer is predicted to span an exon-exon junction, based on conservation in teleost, bird, and mammal orthologs; N/A, not applicable. Details of primer sequences for IGF-II, IGFBP2a, IGFBP2b, IGFBP4, IGF-IR1a, IGF-IR1b, β-actin and RNA Pol-II were published in Ref. 8. Details of primer sequences for murf1 and mafbx were published in Ref. 9.
Primer design and validation for qPCR assays.
Many of the salmonid akirin family members share very high nucleotide sequence similarity. Thus special care was required to ensure that qPCR assays were specific to the targeted sequence. This was achieved by constructing a nucleotide alignment of 32 distinct akirin cDNA sequences (8 family members per species) with Mafft v.6 (31). Subsequently, regions were identified with the highest dissimilarity between different family members, and primers (Table 1) were designed to span these regions. Importantly, the nucleotides distinguishing family members were fixed in each of the four salmonid species. To test each assay's specificity, qPCR products amplified with the given cycling conditions were cloned and 25 separate transformants were sequenced as described above. For each optimized assay, no products representing undesired akirin family members were sequenced by this approach, demonstrating that cross-binding of primers between similar paralogs was avoided. For four of eight akirin family members, at least one primer in a pair was designed to span a conserved exon-exon junction (see Table 1). For other family members, this was not possible because of constraints in the number of distinguishing regions (see Table 1). However, these primers were still positioned in different exons.
Primers used for 26 other qPCR assays are listed in Table 1 along with accession numbers of sequences used for primer design. All primers either spanned an exon-exon boundary or were positioned in different exons (detailed in Table 1). The specificity of each assay was confirmed by sequencing several transformants of each qPCR product with the approach described above.
qPCR protocol.
For each sampling point in the experiment manipulating nutritional state, total RNA was extracted as described above from muscle samples of six individuals (3 per tank) as closely matched in body size as possible (mean weight = 8.2 g, SD = 2.3 g). cDNA was synthesized from 1 μg of total RNA as described above and diluted 100-fold in nuclease-free water. The qPCR protocol conformed to recent “minimum information for publication of quantitative real-time PCR experiments” (MIQE) guidelines (10). Each 15-μl qPCR reaction contained 5 μl of first-strand cDNA as a template (2.5 ng of reverse-transcribed total RNA), 7.5 μl of Brilliant SYBR Green qPCR master mix (Stratagene), and 400 nM sense and antisense primers (34 primer pairs in total; Table 1). Reactions were performed in duplicate with a Mx30005P qPCR thermocycler (Stratagene) with 1 cycle of 10 min at 95°C and 40 cycles of 30 s at 95°C, 30s at a primer-dependent temperature (Table 1), and 30s at 72°C, followed by an amplicon dissociation analysis from 55 to 95°C, where a single peak was observed in optimized assays. SYBR Green fluorescence was recorded during the extension phase of cycling, and each plate contained all samples. Negative controls were included in duplicate and contained either all components of the reverse transcription mixture (using a pool of all total RNAs as a template), excepting reverse transcriptase (no-reverse transcriptase control), or just water instead of first-strand cDNA (no-template control).
Raw data were analyzed with Mx30005P qPCR software (Stratagene), and the threshold fluorescence of dRn values was adjusted to 0.5 for all assays, which was in the linear phase of amplification. The efficiency of each qPCR assay was calculated with LinRegPCR v.11, as recommended by the authors (46). Specifically, each assay's efficiency was taken as the mean of reaction efficiencies calculated for every sample, by fitting a regression equation to data points in the log-linear phase of amplification while correcting for baseline heterogeneity (46). Cycle threshold (Cq) values were manually exported into GenEx v.4.4.2 (MultiD Analyses) and corrected for differences in amplification efficiencies. The Normfinder application (4) was used to examine the suitability of six reference genes, β-actin, RNA Pol-II, rps13, rps29, rpl13, and rpl4 (Table 1). According to the Normfinder results, Cq values for experimental genes were normalized to rps13, rps29, rpl13, and rpl4 Cq values, which were each highly stably expressed across the samples. Normalizing with various combinations of these four reference genes had little effect on final transcript abundance values of any experimental gene.
Statistical analysis of qPCR data.
All statistics on qPCR transcript abundance data were performed in Minitab v.13.2 (Minitab). There was no significant difference in gene expression between tanks (not shown), so data were combined. Statistical differences in mean transcript abundance values of individual akirin family members at different sampling days of the nutritional state experiment were initially determined. The Anderson-Darling test was used to examine significant departures from a normal distribution and Levene's test to assess the equality of variance between sampling days. Data for akirin1(1a), 1(1b), 1(2a), 1(2b), 2(1a), and 2(1b) followed a normal distribution and showed equality in variance and were therefore considered suitable for parametric statistics. akirin2(2a) data were made to better approximate a normal distribution with a fourth-root transformation using the equation y = x0.25, where y is the transformed transcript abundance value and x is the untransformed value. Equality in variance was maintained in the transformed data. Data for akirin2(2b) were made to better approximate a normal distribution with a logarithmic power transformation using the equation y = logx(2), where y is the transformed transcript abundance value and represents the exponent to which the untransformed value x must be raised to obtain 2. Equality in variance was maintained in the transformed data. One-way ANOVA was used on each akirin data set with Fisher's individual error rate test (error rate of 0.01) to establish statistical differences at the P < 0.01 level.
Statistical differences in mean transcript abundance values for a subset of other genes were then determined. 14-3-3β2 data were normally distributed and showed equality in variance. 14-3-3γ1 and 14-3-3γ2 data were made to better approximate a normal distribution with the same logarithmic power transformation as for akirin2(2b). NF-κB p65 data were made to better approximate a normal distribution with a Box-Cox transformation. UBE2H(1) and IGF-IR(1a) data were made to better approximate a normal distribution with the fourth-root transformation described above. UBE2H(1), mafbx, and murf1 data were made to better approximate a normal distribution with a power transformation with the equation y = x0.02, where y is the transformed transcript abundance value and x is the untransformed value. All transformed values for these genes showed equality in variance, and one-way ANOVA was used on each data set as described above.
Statistical differences in mean relative transcript abundances of different akirin family members at either satiation or the point of maximal fasting were then determined. Both data sets were right skewed and did not show equal variances. Thus a fourth-root transformation was used as described above, which brought the data closer to a normal distribution and made data variance equal. One-way ANOVA was used on each data set as described above.
Pearson's correlation coefficient (R) was used to analyze the strength and direction of linear relationships in relative transcript abundances of the 42 individual samples in all pairwise combinations of the 28 nonreference genes assayed across the experimental model of nutritional state, using log2-transformed relative transcript abundance values. A cutoff value of R = 0.7 (P < 0.0001) was used as evidence for gene coregulation.
Hierarchical clustering of qPCR data.
Relationships among relative transcript abundance profiles of the 28 nonreference genes assayed across the model of nutritional state were examined with PermutMatrix (12). Clustering and seriation were based on Pearson's correlation coefficient of z-score normalized relative transcript abundance values (scaled from 0 to 1). The multiple-fragment heuristic seriation method was used with WPGMA as the aggregation criterion for hierarchical clustering.
RESULTS AND DISCUSSION
Many more distinct akirin sequences are present in salmonids than other vertebrates.
We identified numerous distinct akirin sequences in salmonid genomes and then experimentally sequenced each, plus several closely related sequences, using a PCR-cloning-sequencing strategy in Arctic charr, Atlantic salmon, rainbow trout, and brown trout. Eight unique full-coding akirin sequences were obtained in each species and deposited in GenBank (accession numbers in Supplemental Table S1). We also identified two distinct akirin2 sequences in the Atlantic cod (Gadus morhua) representing the Paracanthopterygii lineage (Supplemental Table S1). Furthermore, in previously unexamined species of the Ostariophysi lineage, a single akirin2 sequence was identified in two catfish species and a single akirin1 and 2 sequence in the common roach (Rutilus rutilus) (Supplemental Table S1). No distinct akirin sequences to those previously discovered (38) were identified in teleost genetic model species from Acanthopterygii and Ostariophysi lineages, or in mammals and amphibians. However, as shown before for birds (38), we observed that the reptile Anolis carolinensis has only akirin2 in its genome sequence (not shown). Therefore, salmonid genomes contain between five and seven additional unique akirin sequences compared with a wide range of other vertebrate lineages.
Phylogenetic analysis of salmonid Akirin proteins.
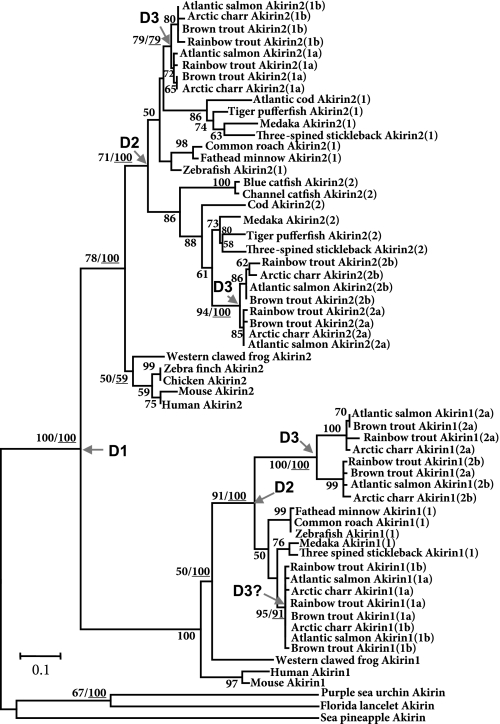
Amino acid translations of each salmonid akirin family member were used for ML and BI phylogenetic analyses, employing the best-fitting available models of substitution and incorporating Akirin1 and 2 orthologs/coorthologs from a wide range of vertebrate lineages (detailed in Supplemental Table S1). Rooted to the single Akirin protein of invertebrate deuterostomes, trees initially branched into strongly supported monophyletic vertebrate clades for Akirin1 and 2 (Fig. 1). In the Akirin2 clade, teleost samples branched from tetrapod sequences and immediately split into two subclades, each representing species from all the major lineages, with nodes closely following expected taxonomic relationships (Fig. 1). Within both subclades, salmonid sequences were further split into two monophyletic groups, each containing a sequence common to the four included species (Fig. 1). These results can be explained if the akirin2 gene duplicated in a common teleost ancestor, leading to paralogs that we previously named akirin2(1) and 2(2) (38), which each duplicated again in a common salmonid ancestor, leading to two respective sets of paralogs that we hereby name akirin2(1a)/2(1b) and akirin2(2a)/2(2b) (Fig. 1, Supplemental Table S1).
Fig. 1.
Phylogenetic tree constructed with maximum likelihood (ML), demonstrating the evolutionary relationships of salmonid Akirin proteins in relation to family members from a broader range of vertebrates. A Bayesian inference (BI) analysis produced a very similar topology. All ML bootstrap confidence values >50% are shown, as well as BI posterior probabilities >50% at important nodes (underlined numbers). Three putative genomic duplication events are marked with arrows, the first in a common vertebrate ancestor [duplication 1 (D1)], the second in a common ancestor to teleosts (D2), and the third in a common ancestor to salmonids (D3). D3? indicates that D2-level salmonid Akirin1(1) sequences did not spilt into 2 monophyletic clades as observed elsewhere in the tree (discussed in text; see Supplemental Fig. S1). A consensus nomenclature proposal accounting for the evolutionary relationships of vertebrate akirins is clearly demonstrated. Vertebrate sequences were rooted to the single Akirin ortholog of 3 deuterostome invertebrates. Scale bar shows number of substitutions per site.
Within the Akirin1 clade, teleost sequences branched from tetrapods and split into two clades, one represented by all major lineages and the other containing only salmonid sequences (Fig. 1). This topology suggests a duplication event in a common teleost ancestor, leading to gene paralogs we hereby name akirin1(1) and akirin1(2), followed by the secondary loss of akirin1(2) genes in lineages outside the Salmonidae. The Akirin1(2) clade further splits into two monophyletic groups of sequences represented once in each salmonid species (Fig. 1). We hereby name the genes coding these putative paralogs as akirin1(2a) and 1(2b) (Fig. 1, Supplemental Table S1). In the teleost Akirin1(1) clade, eight salmonid sequences form a cluster, which, unlike elsewhere in the tree, was not resolved into two monophyletic groups represented by the four salmonid species (Fig. 1). A closer examination of fixed substitutions and indels in the corresponding nucleotide sequences suggests that they do represent two separate sequences commonly inherited from a salmonid ancestor (Supplemental Fig. S1A). Furthermore, phylogenetic analysis employing nucleotide data with exclusion of akirin2 sequences retrieved monophyletic akirin1(1a) and 1(1b) clades with 99% confidence (Supplemental Fig. S1B). We hereby name these putative akirin1(1) paralogs as akirin1(1a) and 1(1b), in line with our proposed nomenclature system (Fig. 1, Supplemental Table S1).
These results are compatible with no less than three genomic duplication events leading to eight akirin genes, the first in an ancestor to vertebrates [duplication 1 (D1)], the second in an ancestor to teleosts (D2), and the third in a common salmonid ancestor (D3). We use these terms here when describing combinations of family member paralogs. Previous phylogenetic analysis of gene families proximal to akirin1 and 2 suggested that a duplication event occurred in a vertebrate ancestor affecting a large chromosomal region encompassing an akirin protogene (38). Additionally, comparisons of shared synteny between teleosts and Sarcopterygians indicated that large chromosomal regions containing akirin1 and 2 duplicated again in a teleost ancestor (38). Considering these results and the well-supported proposals of tetraploidization events at the base of vertebrates (43), teleosts (30), and salmonids (3), we suggest that the current repertoire of akirin sequences in salmonid genomes was attained by successive rounds of tetraploidization, with full retention of generated paralogs. It is essential that a common nomenclature accounting for the evolutionary relationships of vertebrate akirins be utilized by the research community (outlined in Fig. 1 and Supplemental Table S1). The forthcoming Atlantic salmon and rainbow trout genome sequences will be useful in providing comparative genomic evidence (e.g., shared synteny), to confidently establish the scale of the duplication events underlying the structure of salmonid gene families like the akirins.
Comparison of salmonid Akirin family member proteins related to physiological functions.
We next compared salmonid Akirin protein sequences, testing a hypothesis that the families' expansion through paralog retention was related to functional diversification. Mammalian and insect Akirins are nuclear localized, dependent on a single NH2-terminal motif (25). Furthermore, they function within signaling pathways governed by kinase-mediated phosphorylation cascades, including both NF-κB (25) and IGF-PI3K-Akt (40, 47) pathways, and were suggested to be themselves phosphorylated (18). Accordingly, we were interested in identifying putative phosphorylation motifs and NLSs in the eight salmonid Akirin family members. We were also interested in identifying “type II” sites, which are positions functionally constrained to conserve distinct residues in related proteins representing distinct phylogenetic clades, e.g., gene family member paralogs (27, 32). Type II sites identified between two proteins likely reflect that a shift in evolutionary rates occurred at those positions in one or both proteins shortly after their origin (e.g., by gene duplication), leading to distinct amino acid replacements being fixed in each protein that subsequently became subject to strong purifying selection in ongoing lineages. In cases where type II sites are fixed as residues with radically distinct biochemical properties, it is possible that selection is acting to maintain functional specificities of the proteins under comparison (27, 32). We focus on the unique evolution of the gene family in salmonids, comparing paralog groups generated by D2 and D3 events.
All vertebrate Akirins, including the eight salmonid family members, have a similar NH2-terminal NLS (Fig. 2, Fig. 3), although this motif was predicted to include three more residues in salmonid Akirin2(2) than Akirin2(1) family members (Fig. 3). Similarly, an additional conserved NLS at positions 88–91 was present in all Akirin2 proteins (Fig. 3). These results suggest that all the salmonid Akirin family members are constrained to functionally retain the ability to localize to the nucleus.
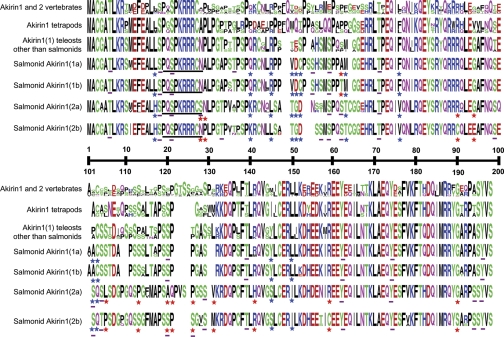
Fig. 2.
Sequence logo alignment showing the amino acid sequence of individual salmonid Akirin1 proteins (4 species) compared with nonsalmonid teleost Akirin1(1) proteins (5 species), Akirin1 of tetrapods (3 species), and Akirin1 and 2 across vertebrate lineages (58 species). Amino acids are color coded by biochemical property and are vertically scaled to represent their conservation at that site. Blue and red stars indicate type II sites in salmonid Akirin family members at the D2 and D3 levels, respectively. Residues underlined with black lines indicate putative nuclear localization signals (NLSs). Residues underlined with purple lines indicate putative phosphorylated sites conserved across the 4 salmonid species. A scale bar is shown on the black line splitting the first and second halves of the alignment.
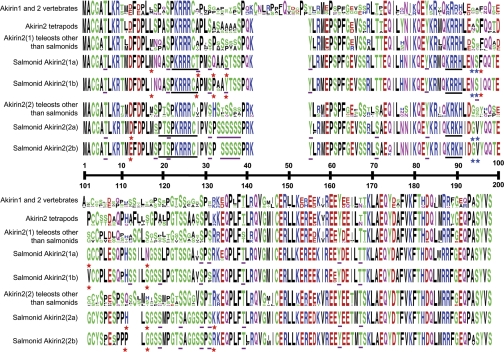
Fig. 3.
Sequence logo alignment showing the amino acid sequence of individual salmonid Akirin2 family members (4 species per family member) compared with nonsalmonid teleost Akirin2(1) and Akirin2(2) (7 and 5 species, respectively), Akirin1 of tetrapods (5 species), and Akirin1 and 2 across vertebrates (58 species). Other details are as described in Fig. 2.
Twelve of 198 (6.1%) of all positions between salmonid Akirin1(1) and 1(2) proteins were type II sites, of which 9 were functionally constrained to retain radically distinct residues (Fig. 2). At the D2 level, the majority of residues at type II sites of salmonid Akirin1(1) proteins are also conserved in teleost Akirin1(1) proteins (Fig. 2). For 5 of 12 of these sites, salmonid Akirin1(2) residues are also more biochemically dissimilar to tetrapod Akirin1 than Akirin1(1) family members (Fig. 2). Therefore, salmonid Akirin1(2) proteins, which were secondarily lost in other teleost lineages (Fig. 1), have a set of residues under a radically distinct set of biochemical constraints to Akirin1(1) proteins, which supports a hypothesis that functional divergence contributed to the retention of the gene coding the ancestral Akirin1(2) protein, prior to D3.
The single nonsynonymous change fixed between Akirin1(1a) and 1(1b) family members was a type II site (position 62) (Fig. 2). Comparing Akirin1(2a) and 1(2b), 13 of 198 (6.6%) positions were type II sites, of which 9 were constrained to conserve radically distinct amino acids (Fig. 2). For many such sites, one paralog retains a residue identical or at least biochemically similar to other teleost Akirin1(1) proteins/Akirin1 of tetrapods (Fig. 2). There was no clear bias as to which D3 paralog retained a similar or distinct residue compared with other Akirin1 proteins (Fig. 2). Thus, at the D3 level, salmonid Akirin1(2) paralogs have many more type II sites than Akirin1(1) paralogs. This is also reflected in the overall rate of nonsynonymous substitution among D3-level Akirin1 paralogs: for any salmonid species, Akirin1(1) paralogs retain ∼97% sequence identity, compared with ∼85% in Akirin1(2) paralogs (not shown; evident in branch lengths in Fig. 1). Thus Akirin1(2) paralogs have been freer to change in protein sequence during evolution than Akirin1(1) family members, including in terms of rate constraints leading to type II sites. This suggests that Akirin1(1) family members are more likely to fulfill functions conserved from the pre-D2 state.
The total number of predicted phosphorylation sites in Akirin1(1a), 1(1b), 1(2a), and 1(2b) were 12, 11, 13, and 19, respectively. At the D2 level, some phosphorylated residues predicted in Akirin1(2) but absent in Akirin1(1) family members either were type II sites (Fig. 2, position 102) or were located proximally to type II sites (positions 63, 105). A phosphorylated serine (position 28) present in Akirin1(2a) but not Akirin1(2b) or Akirin1(1) family members was a type II site (Fig. 2). Similarly, most of the additional phosphorylated residues predicted in Akirin1(2b) compared with Akirin1(2a) and Akirin1(1) family members either were type II sites (Fig. 2, positions 120 and 126) or were located proximally to type II sites (Fig. 2, positions 33, 99, and 139). Thus at both D2 and D3 levels, some type II sites fixed for phosphorylated residues in Akirin1(2) but not Akirin1(1) proteins could underlie differences in posttranslational regulation of these family members. Since the specificity of kinases to their substrates is strongly dependent on the residues proximal to the phosphorylated site (48), the same argument applies for those type II sites flanking paralog-specific phosphorylation sites.
The total number of predicted phosphorylation sites in Akirin2(1a), 2(1b), 2(2a), and 2(2b) were 12, 14, 18, and 19, respectively (Fig. 3). The additional phosphorylation sites in Akirin2(2) compared with 2(1) proteins were mainly accounted for by a unique tract of phosphorylated serines (Fig. 3, positions 31–38). Comparing Akirin2(1) and 2(2), only 2 of 198 (∼1%) positions were type II sites (positions 94–95) and these were not located proximally to any predicted phosphorylated residues (Fig. 3). These two residues have radically distinct biochemical properties and are directly proximal in the polypeptide sequence residues, just COOH-terminal to the Akirin-2 specific NLS (Fig. 3). Thus, for salmonid Akirin2 family members, there is less evidence for type II functional divergence at the D2 level compared with Akirin1 proteins.
Comparing Akirin2(1a) and 2(1b), 7 of 198 (∼3.5%) of all positions were type II sites, of which 4 coded radically different residues (Fig. 3). Many of these type II sites were proximal to phosphorylated sites shared by both paralogs and are therefore unlikely to be related to paralog-specific phosphorylation patterns (Fig. 3). However, one type II site was a phosphorylated threonine in Akirin2(1a) but fixed as alanine in Akirin2(1b) (Fig. 3, position 28). Four of 198 (∼2%) positions were type II sites between Akirin2(2a) and 2(2b), of which 2 were fixed as radically different residues and none was related to paralog-specific phosphorylation of adjacent residues (Fig. 3). Thus the level of type II functional divergence observed between both Akirin2(1) and 2(2) D3-level paralogs is lower than observed for Akirin1(2) paralogs and there is less evidence that the identified type II sites act to modify posttranslational regulation by phosphorylation.
D2- and D3-level type II sites were often directly proximal (e.g., D2-level Akirin1 positions 50–51-52 or 102–103; D2-level Akirin2 positions 94–95; D3-level Akirin1 positions 28–29 and 120–121) or clustered (e.g., D2-level Akirin1 positions 40–53; D3-level Akirin1 positions 105–131) (Fig. 2 and Fig. 3). Considering that Akirin proteins interact with a multitude of proteins (reviewed in Ref. 38), we suggest that such clustering of type II sites may be related to conserving protein-protein interfaces, or interactions specific to D2- or D3-level family members.
Distinguishing expression patterns of salmonid akirin family members.
Our next aim was to measure each salmonid akirin family member's transcript expression with qPCR. Since all D3 paralogs other than akirin1(2) paralogs are >95% similar in the coding sequence (not shown), it was important to design primer sequences spanning the most highly distinguishing available regions. We were also careful to ensure that primer-binding regions were fixed across the four salmonid species. These optimized qPCR assays specifically amplified each akirin family member in both Arctic charr (this study) and Atlantic salmon (not shown) and performed with the given parameters (Table 1) should perform equally well in rainbow trout, brown trout, or other salmonid species.
Distinct transcriptional responses of arctic charr akirins to altered nutritional state.
We measured how in vivo transcript abundance of each akirin family member varied with respect to nutritional state in fast-twitch skeletal muscle of Arctic charr using qPCR, with a robust normalization strategy, exploring the suitability of six reference genes. Target products were not amplified in any negative controls (not shown). The experiment included a “prefasted” sampling point, at which fish had been fed to satiation for an extended period, followed by a “maximal fasting” sampling point after 3 wk without feeding. Subsequently, five samples were taken during a 14-day period of recovery refeeding. At the point of maximal fasting, there was no material visible in the stomach or intestines, the gallbladder was distended with bile, and a loss of fat reserves was evident around the gut compared with the prefasted state (not shown). After 2 days of refeeding and in subsequent samples, the stomach and gut were distended with food (not shown). Fat reserves became evident around the gut after 4 days of refeeding and increased in extent for the remaining period (not shown). Of 42 fish (n = 6 per sampling point), 3 fish were objectively removed from this analysis, since they displayed a clear signature of fasting during feeding phases, including marked upregulation of E3 ubiquitin-ligases (described below), which we have found to be invariably upregulated after feed restriction in salmonids (see, e.g., Ref. 9) and zebrafish (I. G. P. Amaral and I. A. Johnston, unpublished result).
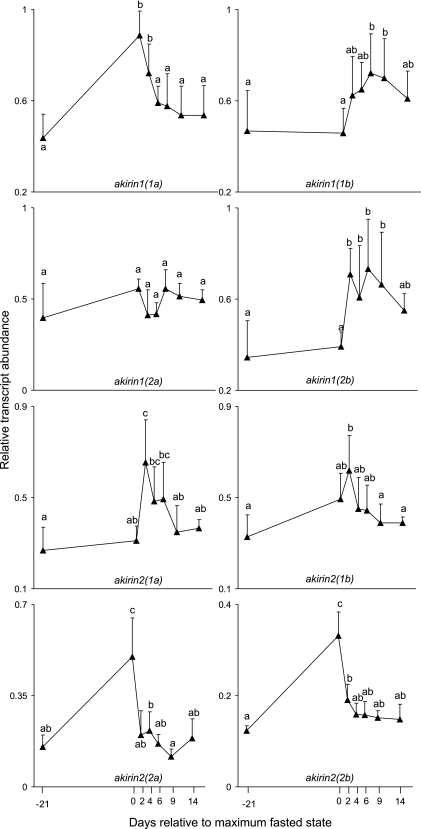
Two principal responsive patterns were evident across the model considering statistical differences at the P < 0.01 level. The first, observed for akirin1(1a), 2(2a), and 2(2b), saw greatest relative transcript abundances at maximal fasting (between ∼2- and 3.3-fold elevated from the prefasted state), followed by a gradual or immediate return to near the prefasted level during the refeeding period (Fig. 4). The second responsive pattern involved either no change [akirin1(1b), 1(2b), and 2(1)] or a notable but nonsignificant increase [akirin2(1b)] in transcript levels between satiation feeding and maximal fasting states, with highest transcript levels at refeeding, either after 2 days for akirin1(2b), 2(1a), and 2(1b) (between ∼1.9- and 2.4-fold elevated from the prefasted state) or more gradually for akirin1(1b), which peaked after 6 days of refeeding (∼1.6-fold elevated from the prefasted state) (Fig. 4). After their maximum expression level, the transcript abundance of these genes returned to levels close to the prefasted state (Fig. 4). akirin1(2a) showed no significant differences in relative transcript abundance with respect to nutritional state (Fig. 4). There was coexpression between akirin1(2b) and akirin2(1a), akirin1(1b) and akirin1(2b), and akirin1(1b) and akirin2(1a), with respective Pearson's R values of 0.85, 0.72, and 0.75 (Supplemental Fig. S2).
Fig. 4.
Relative transcript abundance levels of Arctic charr akirin family members in fast-twitch skeletal muscle at different nutritional states. Values are means + SD; n = 6 for all days except days −21 and 14 (n = 5 and n = 4, respectively), where data points were removed by an objective criterion (discussed in text). Details of the normalization strategy can be found in experimental procedures. Transcript abundance values for each gene are on a scale ranging from 0 to 1, relative to the lowest expression value for that gene; thus no between-family member comparisons are provided in this figure. Statistical differences between sampling day means at the P < 0.01 level are indicated by different letter groupings: if 2 means are grouped with distinct letters (e.g., a vs. b, ab vs. c, bc vs. a) they are statistically different or if grouped with identical letters (e.g., a vs. a, a vs. ab, bc vs. c) they are statistically equivalent.
These results demonstrate that the majority of Arctic charr akirin family members are transcriptionally responsive to altering nutritional state of skeletal muscle. The evolutionary relatedness of family members was not closely reflected in correlation values between transcript levels, with no D3 paralogs receiving R values >0.7. In fact, the most closely coexpressed family members have been evolving independently since D1 and D2 events, which occurred at the base of vertebrates and teleosts, respectively. Considering known tetraploidization events, D1 and D2 paralogs potentially separated ∼500 (43) and ∼350 (30) million years ago, respectively. If D1 and D2 events were tetraploidizations, the resulting paralogs would be located on different chromosomes, certainly initially. Thus these examples of coexpression between D1- and D2-level akirin paralogs suggest that selection has acted over a vast evolutionary time to independently maintain or introduce nutritionally responsive regulatory elements leading to similar transcriptional responses in genes with markedly different protein products (e.g., 40–50% sequence identity at the D1 level). Forthcoming salmonid genome sequences will facilitate in delineating the controlling factors governing the cumulative expression patterns of family member paralogs, for example, through study of conserved transcription factor binding sites in proximal promoters and enhancers.
Comparative transcript abundance of different akirin family members.
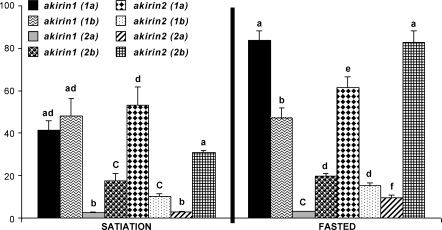
We compared differences in relative transcript abundances of Arctic charr akirin family members at satiation and maximal fasting (Fig. 5). At both nutritional states, there were large differences in transcript levels among all family members, up to ∼24-fold with satiation feeding and ∼28-fold at maximal fasting (Fig. 5). At the D2 level, both akirin1(1) paralogs were significantly more abundant than either akirin1(2a) or akirin1(2b), respectively, by ∼19.0- to 27.6-fold or ∼2.0- to 4.0-fold, depending on nutritional state (Fig. 5). Conversely, considering both nutritional states, akirin2 family members did not show a pattern in which both genes in a pair of D2-level paralogs were more transcriptionally abundant than both genes in the other paralog pair (Fig. 5). Since Akirin1(1) family members have diverged less at the protein level from the ancestral state shared with Akirin1(2) proteins (Fig. 2 and previous section), it is feasible that their higher mRNA abundance in muscle at both nutritional states (Fig. 5) simply reflects that more units of protein are required to fulfill certain fundamental cellular functions absent in akirin1(2) family members.
Fig. 5.
Differences in relative transcript abundance levels among akirin family members in Arctic charr fast-twitch skeletal muscle after feeding to satiation and at maximal fasting. Values are scaled between family members relative to the lowest expression value among all genes. Other details are as described in Fig. 4.
For three D3-level paralogs, skeletal muscle transcript levels were higher in one paralog independent of nutritional state (Fig. 5). At satiation and the point of maximal fasting, akirin1(2a) was ∼8- and 6.5-fold less abundant than akirin1(2b), respectively, akirin2(1b) was ∼5.2- and 4.0-fold less abundant than akirin2(1a), respectively, and akirin2(2a) was ∼10.5- and 8.7-fold less abundant than akirin2(2b), respectively (Fig. 5). In these cases, it is possible that enhancers or other regulatory elements present in one, but not both D3-level akirin paralogs account for a difference in basal skeletal muscle transcription rates. Conversely, akirin1(1a) and 1(1b) paralogs, which showed distinct transcriptional responses to nutrition (Fig. 4), had comparable transcript levels at satiation and were only 1.8-fold different upon maximum fasting (Fig. 5).
Positioning Arctic charr akirin expression data within a network of genes that regulate catabolism and growth in skeletal muscle.
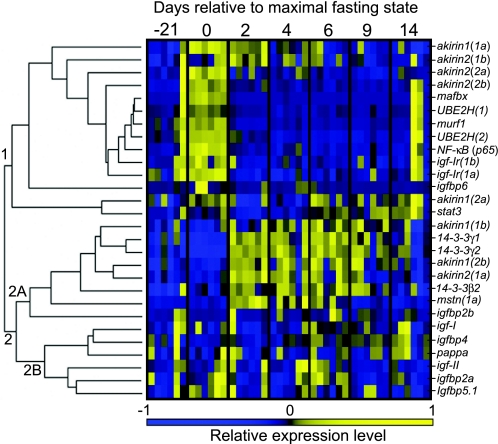
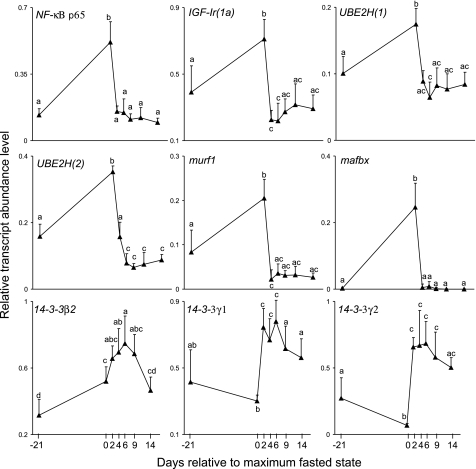
Hierarchical clustering of qPCR data revealed two major clusters of gene expression in Arctic charr skeletal muscle at various nutritional states (Fig. 6). In cluster 1, a clade of closely regulated genes is evident that were most transcriptionally abundant at maximal fasting, including NF-κB p65, two genes coding E3 ubiquitin-ligases, (mafbx and murf1), two E2 conjugating enzyme paralogs [UBE2H(1) and (2)], and two IGF-I receptor paralogs [igf-Ir(1a) and igf-Ir(1b)] (Fig. 6). These genes were upregulated by fasting by ∼3.8-, 99.8-, 2.5-, 1.7-, 2.2-, 1.8-, and 1.6-fold, respectively, and then downregulated by 2 days of subsequent feeding by ∼3.4-, 51.8-, 9.2-, 2.0-, 2.2-, 1.9-, and 1.9-fold, respectively, remaining low thereafter (Fig. 7). All combinations of these genes share R values between 0.74 and 0.91 (Supplemental Fig. S2). The next most external branch is akirin2(2b) (Fig. 6), which shares R values of 0.74–0.79 with mafbx, NF-κB p65, and UBE2H(1) and (2) (Supplemental Fig. S2). This is followed by its D3 paralog, akirin2(2a), which was relatively abundant at maximal fasting but was less coexpressed with internal cluster genes during feeding phases (Fig. 6), sharing no R values >0.7 (Supplemental Fig. S2). The next branches are akirin2(1b) and 1(1a), which were relatively highly expressed at day 2 as well as at day 0 (Fig. 6). The most external branches of cluster 1 form a subclade containing akirin1(2a) and the gene coding the Signal transducer and activator of transcription 3 protein (stat3) (Fig. 6), coexpressed with an R value of 0.71 (Supplemental Fig. S2). We note that some individuals presented a clear signature of fasting at days −21 (1 fish) and 14 (2 fish) based on the expression patterns of cluster 1 genes (Fig. 6). This indicates the usefulness of hierarchical clustering for objectively removing outlier points due to nonfeeding individuals in experiments manipulating whole animal nutritional state, as done in this study (Fig. 4 and Fig. 5).
Fig. 6.
Hierarchical clustering and seriation of Arctic charr akirin expression data (i.e., from Fig. 4) among equivalent profiles for 20 further genes involved in regulating the balance between anabolic and catabolic metabolism in fast-twitch skeletal muscle. Data are represented as a heat map showing changes in relative transcript abundances for each gene across the same experimental model (42 samples, n = 6 per sampling day) with an accompanying dendrogram representing among-gene profile similarities. Numbers identify clusters for discussion purposes. Note the strong upregulation of catabolic genes in 2 samples at 14 days of refeeding and in 1 sample at day −21.
Fig. 7.
Expression profiles of a subset of cluster 1 and 2 genes other than akirin family members that were markedly regulated at the transcript level across the experimental model altering nutritional state in Arctic charr skeletal muscle. Other details are as described in Fig. 4.
Considering their coexpression, we suggest that genes clustering from akirin2(2b) inward are under some level of common regulation and form an important component of the molecular response required to reach a catabolic state in Arctic charr skeletal muscle. One supported mechanism is the targeting of muscle proteins for proteosomal breakdown by ubiquitination. For example, NF-κB activation of the mouse ortholog of UBE2H, but not other E2 conjugating enzymes, was essential for ubiquitin conjugation during skeletal muscle atrophy (35). In addition, the mouse ortholog of Murf1 was essential for fasting- or denervation-induced atrophy of thick filaments of skeletal muscle myofibrils (16) and is also transcriptionally regulated by NF-κB (11). Mammalian Mafbx is also an essential component of proteosome-mediated atrophy of skeletal muscle and is activated by p38-MAPK signaling (23), but not by NF-κB (11). The expression of both Mafbx and Murf1 is also regulated by FoxO transcription factors in mammalian skeletal muscle (23), as is the IGF-I receptor in cardiomyocytes (36). Thus it is possible that the coexpression of cluster 1 genes reflects the cumulative activation (during fasting) or repression (during feeding) of more than one pathway mediating catabolism. In any case, our data are limited to identifying coexpression of genes and cannot reveal the regulating factors. Thus we suggest that while the tight coexpression of akirin2(2b) with the NF-κB gene, as well as with components of the ubiquitination pathway that are NF-κB regulated, is consistent with an involvement in skeletal muscle catabolism through classical NF-κB signaling (as might be expected considering previous research; Ref. 25), other pathways could be equally important. This possibility is strengthened by the fact that Akirin2 of rats is involved in the transcriptional regulation of MAPK-phosphatase 1 (33), which regulates p38 MAPK and cytokines including TNF-α and IL-6 (15), which in turn activate the NF-κB pathway (34), including in skeletal muscle (29). Additionally, akirin1 of mice was transcriptionally regulated by p38 MAPK signaling in muscle, as well as by PI3K-Akt signaling (40, 47), which controls FoxO transcription factors (23).
The coexpression of Arctic charr akirin2(2b) and UBE2H genes proteins is worth discussion, since orthologs of these proteins physically interact in Drosophila. The vertebrate UBE2H gene has two putative coorthologs in Drosophila called Ubc-E2H (Flybase ID: FBgn0029996) and CG14739 (FBgn0037987), and the latter was shown by yeast two-hybrid screening to bind to Akirin (22). In certain prokaryotes and yeast (Saccharomyces cerevisiae), a highly significant pattern exists that interacting proteins are more likely to be coexpressed than expected by chance (21, 26) and that there exists strong selective pressure to maintain coexpression of interacting proteins over long timescales (20). It was suggested that this trend reflects the need for interacting proteins to be coordinated spatiotemporally at controlled levels to maintain stoichiometry and thus proper function of protein complexes (20). The coexpression of Arctic charr akirin2(2b) with UBE2H raises the intriguing possibility that a protein-level interaction has been maintained during evolution, with an accompanying selective pressure to preserve coexpression.
Cluster 2 splits into two subclades we called 2A and 2B (Fig. 6). Genes in cluster 2A are generally characterized by lowest relative transcript abundance values at day −21 and maximal fasting with upregulation during recovery feeding and include three akirin family members and all genes coding 14-3-3 proteins (Fig. 6). 14-3-3γ2 was remarkably responsive to nutritional state and was downregulated by 3.3-fold between day −21 and day 0 and upregulated by ∼10-fold after 2 days of refeeding (Fig. 7). The 14-3-3γ2 expression profile was positively correlated to akirin1(1b) (R = 0.77) and to its paralog 14-3-3γ1 (R = 0.89) and highly negatively correlated to profiles for cluster 1A genes [R values from −0.71 to −0.89 with akirin2(2b), IGF-IR1a and 1b, mafbx, murf1, UBE2H(1)/(2), and NF-κB p65] (Supplemental Fig. S2). The 14-3-3β2 expression profile (Fig. 7) was positively correlated to akirin1(2b) (R = 0.77) (Supplemental Fig. S2).
It has been shown previously that TNF-α treatment promoted the recruitment of 14-3-3β/γ proteins to promoters of NF-κB target genes. Furthermore, 14-3-3β/γ physically interact with the p65 subunit of NF-κB (dependent on phosphorylation) and TNF-α treatment enhances this association, while abrogation of 14-3-3 signaling leads to the constitutive association of p65 with chromatin (2). Thus 14-3-3 proteins were suggested to regulate the nuclear activities of p65 by facilitating its nuclear export, ensuring its proper regulation in relation to upstream signaling (2). It was also shown that 14-3-3β/γ bound to IL6 and RANTES promoters (2), which are transcriptional targets of Akirin2/NF-κB (25), and that 14-3-3β binds Akirin2 (33). Furthermore, Akirin2 does not interact with NF-κB (25). Considering that 14-3-3 proteins can serve as phosphorylation-dependent scaffolding proteins between proteins that may otherwise not interact (51), we hypothesize that 14-3-3 facilitates some interaction between Akirin and NF-κB in order to regulate their nuclear location and activities.
In light of the above proposal, it is equally feasible that 14-3-3 proteins regulate the association of NF-κB with promoters of its target genes involved in initiating and maintaining muscle catabolism. While such an event would likely be regulated by phosphorylation, the strikingly opposite transcriptional response of 14-3-3γ and NF-κB target genes as well as NF-κB p65 (Fig. 6, Fig. 7) does indicate strong negative regulation of these genes at the mRNA level during the switch from catabolism to anabolism. We suggest that the relatively high levels of 14-3-3γ genes after 2 days of feeding following fasting reflect a basic requirement for more protein units to enter the nucleus to remove NF-κB, leading to the observed downregulation of its target genes like murf1 and UBE2H. Considering the negative regulation of 14-3-3γ and akirin2(2b) (Supplemental Fig. S2), as well as results discussed in the last paragraph, it is also possible that the nuclear activities of Akirin are regulated by 14-3-3 proteins. This hypothesis warrants further attention in a species with a smaller complement of akirin genes, the zebrafish model being ideal because of the available protein-level resources.
Within cluster 2A, mstn(1a), which codes a paralog of Myostatin (a negative regulator of muscle growth), was not regulated by the physiological transition from satiation feeding to fasting (Fig. 6) but was upregulated by ∼4.5-fold (not shown) by feeding after fasting, before returning to satiation/fasted levels by day 9 (profile evident in Fig. 6). The lack of an mRNA-level response of mstn to fasting has been observed in skeletal muscle of adult tilapia (Oreochromis mossambicus) (44) and rainbow trout (41). However, in striking contrast to our result, it was also shown in rainbow trout that mstn transcript levels were unaffected (14) or decreased (41) by feeding after fasting. Previous work also showed that mouse akirin1 is transcriptionally regulated by myostatin (40). We observed no R values to suggest coexpression between Arctic charr akirin family members and mstn(1a) in response to nutritional state (Supplemental Fig. S2).
Genes in clade 2B code IGF-I and II hormones, several IGF-binding proteins (IGFBPs) (including two IGFBP2 paralogs, IGFBP4, and a paralog of IGFBP5) and Pappa, a protease for IGFBPs (Fig. 6). However, no markedly similar responses to nutritional state (Fig. 6) or R values >0.7 were evident in any of these genes (Supplemental Fig. S2). Thus at the mRNA level there is no evidence supporting that salmonid akirin genes are coexpressed with IGF pathway genes in skeletal muscle at distinct nutrition states (Fig. 6). However, since protein levels were not studied, a role for salmonid Akirins in mediating IGF signaling should not be ruled out without further investigation.
GRANTS
This work was supported by Natural Environment Research Council Grant NE/E015212/1.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
D. J. Macqueen and I. A. Johnston conceived the study. D. J. Macqueen, I. A. Johnston, and B. K. Kristjánsson carried out the nutritional experiment with help from Jennifer King, Rán Sturlaugsdóttir, Camille LeBlanc, and Dr. Vera Vieira-Johnston. D. J. Macqueen designed and executed qPCR and other molecular biology experiments/analyzed subsequent data, performed the phylogenetic and rate-shift analyses, produced all figures, and wrote the manuscript together with I. A. Johnston. We are grateful to Dr. Neil Bower for providing several primer pairs.
Footnotes
The online version of this manuscript contains supplemental material.
REFERENCES
- 1.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Aguilera C, Fernández-Majada V, Inglés-Esteve J, Rodilla V, Bigas A, Espinosa L. Efficient nuclear export of p65-IkappaBalpha complexes requires 14-3-3 proteins. J Cell Sci 119: 3695–3704, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Allendorf FW, Thorgaard GH. Tetraploidy and the evolution of salmonid fishes. In: Evolutionary Genetics of Fishes, edited by Turner BJ. New York: Plenum, 1984 [Google Scholar]
- 4.Andersen CL, Ledet-Jensen J, Ørntoft T. Normalization of real-time quantitative RT-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Bailey GS, Poulter RT, Stockwell PA. Gene duplication in tetraploid fish: model for gene silencing at unlinked duplicated loci. Proc Natl Acad Sci USA 75: 5575–5579, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, Acharyya S, Rudnicki MA, Hollenbach AD, Guttridge DC. IKK/NF-kappaB regulates skeletal myogenesis via a signalling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol 180: 787–802, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blom N, Gammeltoft S, Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294: 1351–1362, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Bower NI, Li X, Taylor R, Johnston IA. Switching to fast growth: the insulin-like growth factor (IGF) system in skeletal muscle of Atlantic salmon. J Exp Biol 211: 3859–3870, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Bower NI, Taylor RG, Johnston IA. Phasing of muscle gene expression with fasting-induced recovery growth in Atlantic salmon. Front Zool 6: 18, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Cai D, Frantz JD, Tawa NEJr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell 119: 285–298, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21: 1280–1281, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Chauvigné F, Gabillard JC, Weil C, Rescan PY. Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. Gen Comp Endocrinol 132: 209–215, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Chi H, Barry SP, Roth RJ, Wu JJ, Jones EA, Bennett AM, Flavell RA. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc Natl Acad Sci USA 103: 2274–2279, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol 185: 1083–1095, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Fuente J, Almazán C, Blas-Machado U, Naranjo V, Mangold AJ, Blouin EF, Gortazar C, Kocan KM. The tick protective antigen, 4D8, is a conserved protein involved in modulation of tick blood ingestion and reproduction. Vaccine 24: 4082–4095, 2006 [DOI] [PubMed] [Google Scholar]
- 18.de la Fuente J, Maritz-Olivier C, Naranjo V, Ayoubi P, Nijhof AM, Almazán C, Canales M, Pérez de la Lastra JM, Galindo RC, Blouin EF, Gortazar C, Jongejan F, Kocan KM. Evidence of the role of tick subolesin in gene expression. BMC Genomics 9: 372, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser HB, Hirsh AE, Wall DP, Eisen MB. Coevolution of gene expression among interacting proteins. Proc Natl Acad Sci USA 101: 9033–9038, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge H, Liu Z, Church GM, Vidal M. Coevolution of gene expression among interacting proteins. Nat Genet 29: 482–48611694880 [Google Scholar]
- 22.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez K, Baylies M. Bhringi: a novel Twist co-regulator. A Dros Res Conf 46: 320B, 2005 [Google Scholar]
- 25.Goto A, Matsushita K, Gesellchen V, El Chamy L, Kuttenkeuler D, Takeuchi O, Hoffmann JA, Akira S, Boutros M, Reichhart JM. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in Drosophila and mice. Nat Immunol 9: 97–104, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigoriev A. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res 29: 3513–3519, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol 23: 1937–1945, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol 287: C834–C843, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biémont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigó R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quétier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Knudsen B, Miyamoto MM, Laipis PJ, Silverman DN. Using evolutionary rates to investigate protein functional divergence and conservation. A case study of the carbonic anhydrases. Genetics 164: 1261–1269, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komiya Y, Kurabe N, Katagiri K, Ogawa M, Sugiyama A, Kawasaki Y, Tashiro F. A novel binding factor of 14-3-3beta functions as a transcriptional repressor and promotes anchorage-independent growth, tumorigenicity, and metastasis. J Biol Chem 283: 18753–18764, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol 2: 725–734, 2002 [DOI] [PubMed] [Google Scholar]
- 35.Li YP, Lecker SH, Chen Y, Waddell ID, Goldberg AL, Reid MB. TNF-alpha increases ubiquitin-conjugating activity in skeletal muscle by up-regulating UbcH2/E220k. FASEB J 17: 1048–1057, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Liu TJ, Lai HC, Ting CT, Wang PH. Bidirectional regulation of upstream IGF-I/insulin receptor signaling and downstream FOXO1 in cardiomyocytes. J Endocrinol 192: 149–158, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Macqueen DJ, Johnston IA. An update on MyoD evolution in teleosts and a proposed consensus nomenclature to accommodate the tetraploidization of different vertebrate genomes. PLoS One 3: e1567, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macqueen DJ, Johnston IA. Evolution of the multifaceted eukaryotic akirin gene family. BMC Evol Biol 9: 34, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol 11: 171–176, 2001 [DOI] [PubMed] [Google Scholar]
- 40.Marshall A, Salerno MS, Thomas M, Davies T, Berry C, Dyer K, Bracegirdle J, Watson T, Dziadek M, Kambadur R, Bower R, Sharma M. Mighty is a novel promyogenic factor in skeletal myogenesis. Exp Cell Res 314: 1013–1029, 2008 [DOI] [PubMed] [Google Scholar]
- 41.Montserrat N, Gabillard JC, Capilla E, Navarro MI, Gutiérrez J. Role of insulin, insulin-like growth factors, and muscle regulatory factors in the compensatory growth of the trout (Oncorhynchus mykiss). Gen Comp Endocrinol 150: 462–472, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36, 1999 [DOI] [PubMed] [Google Scholar]
- 43.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-IT, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071, 2008 [DOI] [PubMed] [Google Scholar]
- 44.Rodgers BD, Weber GM, Kelley KM, Levine MA. Prolonged fasting and cortisol reduce myostatin mRNA levels in tilapia larvae; short-term fasting elevates. Am J Physiol Regul Integr Comp Physiol 284: R1277–R1286, 2003 [DOI] [PubMed] [Google Scholar]
- 45.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574, 2003 [DOI] [PubMed] [Google Scholar]
- 46.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salerno MS, Dyer K, Bracegirdle J, Platt L, Thomas M, Siriett V, Kambadur R, Sharma M. Akirin1 (Mighty), a novel promyogenic factor regulates muscle regeneration and cell chemotaxis. Exp Cell Res 315: 2012–2021, 2009 [DOI] [PubMed] [Google Scholar]
- 48.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol 4: 973–982, 1994 [DOI] [PubMed] [Google Scholar]
- 49.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Thorgaard GH, Bailey GS, Williams D, Buhler DR, Kaattari SL, Ristow SS, Hansen JD, Winton JR, Bartholomew JL, Nagler JJ, Walsh PJ, Vijayan MM, Devlin RH, Hardy RW, Overturf KE, Young WP, Robison BD, Rexroad C, Palti Y. Status and opportunities for genomics research with rainbow trout. Comp Biochem Physiol B Biochem Mol Biol 133: 609–646, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Tzivion G, Shen YH, Zhu J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20: 6331–6338, 2001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.