Abstract
Although the Escherichia coli fatty acid synthesis (FAS) pathway is the best studied type II fatty acid synthesis system, a major experimental limitation has been the inability to feed intermediates into the pathway in vivo because exogenously-supplied free fatty acids are not efficiently converted to the acyl-acyl carrier protein (ACP) thioesters required by the pathway. We report that expression of Vibrio harveyi acyl-ACP synthetase (AasS), a soluble cytosolic enzyme that ligates free fatty acids to ACP to form acyl-ACPs, allows exogenous fatty acids to enter the E. coli fatty acid synthesis pathway. The free fatty acids are incorporated intact and can be elongated or directly incorporated into complex lipids by acyltransferases specific for acyl-ACPs. Moreover, expression of AasS strains and supplementation with the appropriate fatty acid restored growth to E. coli mutant strains that lack essential fatty acid synthesis enzymes. Thus, this strategy provides a new tool for circumventing the loss of enzymes essential for FAS function.
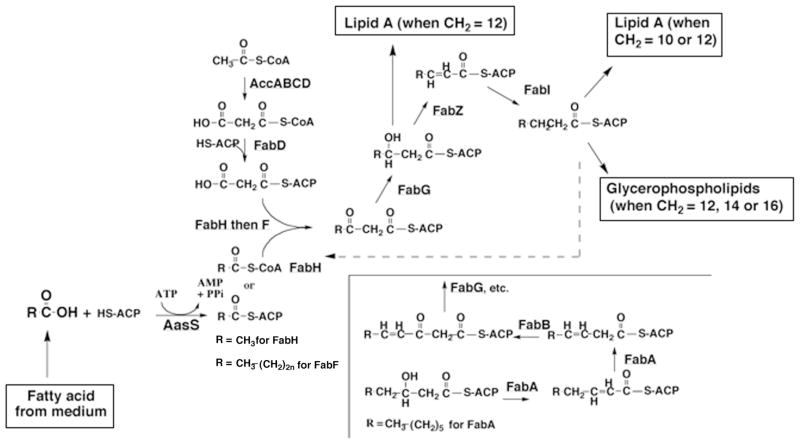
The Escherichia coli fatty acid biosynthetic pathway (Fig. 1) is the archetypical type II fatty acid synthesis (FAS) system (1). Although genetic, enzymological and structural analyses have rendered E. coli FAS by far the best understood type II system, it has one serious disadvantage; the inability of exogenous fatty acids to efficiently enter the fatty acid synthetic pathway. Fatty acids added to cultures of E. coli are readily transported into the cell where they are converted to CoA thioesters and degraded to acetyl-CoA by the β-oxidation system (2). However, if the acyl chains are sufficiently long (>C12), the acyl-CoAs can be used directly as acyl donors by the acyltransferases of glycerophospholipid synthesis (1). However, there is little or no elongation of the fatty acids indicating they can not enter the fatty acid synthetic pathway (3). For example, labeled fourteen carbon fatty acids are incorporated into glycerophospholipids only as fourteen carbon fatty acids, not as a sixteen or eighteen carbon fatty acid (4). This indicates that E. coli lacks the means to efficiently convert either free fatty acids (FFAs) or acyl-CoA thioesters to the acyl-acyl carrier protein (ACP) thioester substrates required by the fatty acid synthesis system. This inability to form acyl-ACPs from exogenous fatty acids has precluded feeding fatty acid synthetic intermediates (or analogues thereof) to address mechanistic and genetic questions in vivo. We report that expression of Vibrio harveyi acyl-ACP synthetase (AasS), a soluble cytosolic enzyme that ligates FFAs to ACP to form acyl-ACPs (5–7) allows exogenously supplemented FFAs to enter the E. coli fatty acid synthesis pathway and become elongated. Moreover, AasS expression allowed growth of fabA, fabB and fabH null mutant strains on fatty acids that do not support growth in the absence of the synthetase. Finally, AasS expression allowed labeling of the primary and secondary acyl chains of the lipid A component of the outer membrane with exogenous dodecanoic and tetradecanoic acids. We also report that the very low background incorporation seen in the absence of AasS is dependent on conversion of exogenous acids to their acyl-CoA thioesters.
Fig. 1.
The fatty acid synthesis pathway of E. coli, its intersection with V. harveyi AasS and incorporation of acyl chains into complex lipids. Unsaturated fatty acid synthesis is shown below the bracket at the lower right of the figure. Unsaturated fatty acids are not found in lipid A unless the cells are grown at low temperatures (30, 31).
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids and Growth Conditions
Bacterial strains and plasmids are listed in Table 1. All E. coli strains are K-12 derivatives. The aasS coding sequence was PCR amplified from pYFJ64 (7) with primers AAS NcoI (5′-CCATGGACCAGTATGTAAATG AT-3′) and AAS HindIII (5′-AAGCTT TTACAGATGAAGTTT ACG CAG T-3′), followed by cloning of the PCR product into pCR2.1-TOPO, resulting in pYFJ82. Plasmid pYFJ82 was digested with NcoI and HindIII and the aasS-containing fragment was then ligated into the same sites of pBAD24 (8) to give pYFJ85. Plasmids pYFJ85 and pBAD24 were transformed into strains K19 (fadE62), K27 (fadD88) and JWC255 (cfa::kan, fadE62). The last strain was constructed by transduction of strain K19 with a phage P1 lysate gown on strain YYC1273 (9). Strain K19 and K27 were obtained from the Coli Genetic Stock Center (Yale University).
Table 1.
E. coli K-12 strains and plasmids used in this work.
| Strain or Plasmid | Relevant Characteristics | Source or Reference |
|---|---|---|
| Strains | ||
| UB1005 | metB1 relA1 spoT1 gyrA216 lr l | Lab collection |
| CL81 | UB1005, attHK022::(plsX’fabH, Spcr Strr) | (10) |
| CL111 | UB1005, attHK022::(plsX’fabH, Spcr Strr), fabH::Kan | (10) |
| YFJ248 | CL111/pBAD24 | This work |
| YFJ249 | CL111/pYFJ85 | This work |
| CAG12094 | MG1655, zcb-3059::Tn10 | (28) |
| CAG18466 | MG1655, zcb-282::Tn10 | (28) |
| YFJ260 | fabH::Kan, zcb-3059::Tn10, Spcs Strs/pYFJ85 | This work |
| YFJ262 | fabH::Kan, zcb-282::Tn10, Spcs Strs/pYFJ85 | This work |
| YFJ293 | UB1005/pBAD24 | This work |
| YFJ294 | UB1005/pYFJ85 | This work |
| YFJ303 | YFJ294 fabH::Kan | This work |
| YFJ304 | YFJ293 fabH::Kan | This work |
| YFJ305 | UB1005 fabH::Kan | This work |
| GRT26 | MC1061 cfa::Cm | Lab collection |
| YFJ315 | YFJ303 cfa::Cm | This work |
| YFJ316 | YFJ304 cfa::Cm | This work |
| YFJ317 | YFJ305 cfa::Cm | This work |
| CHC41 | UB1005, cfa::kan | This work |
| K19 | fadE62 | (23) |
| K27 | fad88 | (23) |
| CAG18483 | fadL771::Tn10 | (28) |
| CAG18496 | fadA 101::Tn10 | (28) |
| JWC255 | fadE62 cfa::kan | This work |
| YYC1273 | cfa::kan | (9) |
| Plasmids | ||
| pBAD24 | Expression vector with pBAD promoter | (8) |
| pCR2.1-TOPO | TOPO TA cloning vector | Invitrogen |
| pYFJ82 | aasS PCR amplified and inserted into pCR2.1-TOPO | This work |
| pYFJ85 | aasS fragment of pYFJ82 inserted in to the NcoI/HindIII sites of pBAD24 | This work |
| pMON5839 | Expression vector | (29) |
| pBB277 | Saccharomyces cerevisiae FAA1 gene in pMON5839 | (29) |
The ΔfabH strains were derived by elimination of the functional copy of fabH from strain CL111 by phage P1 transduction with lysates grown on strains CAG12094 and CAG18466 as previously described (10) except that the strain carried the AasS encoding plasmid pYFJ85 or the vector plasmid, pBAD24 to give strains YFJ249 and YFJ248, respectively. The transduction mixtures were plated on LB supplemented with 1 mM octanoate, 0.2% oleate, 0.02% arabinose, 15 μg/ml tetracycline, 50 μg/ml kanamycin and 100 μg/ml ampicillin. The plates were incubated at 37°C for 48 h to allow growth of small colonies. Both the large and small colonies were screened for sensitivity to a mixture of spectinomycin and streptomycin. Colonies sensitive to the mixture (17.5 μg/ml of each) of spectinomycin and streptomycin (the fabH bypass mutants) were only seen among the transductants of strain YFJ249 and all were small colonies. The bypass strains were designated as YFJ260 (P1 grown on CAG12094) and YFJ262 (P1 grown on CAG18466).
To construct strains YFJ303, YFJ304 and YFJ305 a P1 lysate grown on CL111 was used to transduce strains YFJ294 (UB1005/pYFJ85), YFJ293 (UB1005/pBAD24) and UB10005, respectively. The transductants were plated on LB plates supplemented with 1 mM octanoate, 0.2% oleate, 0.02% arabinose, 50 μg/ml kanamycin and 50 μg/ml neomycin and 100 μg/ml ampicillin (for recipient strains YFJ294 and YFJ293 only). Although addition of octanoate, oleate and arabinose were not expected to aid growth of YFJ293 and UB1005 transductants, these supplements were routinely added to the plating medium. The transduction plates were again incubated at 37°C for 2–3 days to allow growth of small colonies. Strain YFJ303 was essentially a fabH bypass strain similar to YFJ262 except for the absence of a Tn10 element. Strains YFJ304 and YFJ305 were expected to show the fabH deletion mutant phenotype in the absence of AasS expression. The cfa derivatives of YFJ303, YFJ304 and YFJ 305 were obtained by P1 transduction of a cfa::Cm locus from strain GRT26 into the corresponding recipient strains to give strains YFJ315, YFJ316 and YFJ317. To confirm that the fabH locus of YFJ305 had the expected insertion mutation (that of the donor strain CL111), PCR analysis was performed using primers 5′-GAGTCACTCGCAAAGCGAACG-3′ and 5′-CGTTACTGAGTACGCAGCC-3′ (which flank the chromosomal fabH region) followed by restriction mapping of the PCR products in wild type (UB1005), YFJ305 and CL111 strains (Fig. 1S). The fabH bypass and deletion mutant strains were routinely grown on LB supplemented with 1 mM octanoate, 0.2% oleate, 0.02% arabinose and the appropriate antibiotics. For testing the growth phenotypes of these strains on various fatty acids, 1 mM (final concentration) of each fatty acid was added to the growth media.
Radioactive Labeling, Lipid Extraction and Fatty Acid Analysis
For the fabH deletion strains the cfa derivatives of YFJ315 (carrying the aasS plasmid pYFJ85), YFJ316 (carrying the vector pBAD24) and YFJ317 (no plasmid) together with the corresponding wild type strain CHC41 (UB1005 cfa::Kan) were grown at 37oC either in LB or in LB supplemented with 0.5 mM octanoic acid and 0.2% arabinose. Derivatives of strains K19, K27 or JWC255 carrying either pYFJ85 or pBAD24 were grown overnight in LB containing 15 μM [1-14C]-labeled fatty acid (50–55 mCi/mmol), ampicillin (100 μg/ml) and various concentrations of arabinose. Phospholipids were extracted by the method of Bligh and Dyer (11) whereas lipid A was obtained by the following procedure. Labeled cells were recovered by filtration using solvent-resistant 0.2 μm syringe filters (Chromafil Xtra PET-20–25 from Macherey-Nagel) and washed several times with LB to remove unincorporated fatty acids. The filters were then washed 3 times with 10 ml of chloroform-methanol (1/2, v/v) to remove phospholipids. Forcing air through them dried the filters. Each filter was then connected to a fresh syringe that was used to draw 1 M KOH into the filter and the syringe plunger was partially withdrawn to seal the top of the filter and impede drainage of the KOH by creation of a vacuum. The filters were then incubated at 42oC for three days to hydrolyze the ester-linked fatty acids. The KOH was then expelled from the filter, acidified and the fatty acids recovered by Bligh-Dyer extraction. The fatty acids were analyzed by reverse phase chromatography on Partisil KC18 thin layer plates of octadecyl-modified silica gel 60 (Whatman) using a solvent system of acetonitrile-acetic acid-acetone (7:1:1, by volume) followed by autoradiography. Note that only the ester-linked lipid A acyl groups were analyzed since amide-linked 3-hydroxytetradecanoate moieties are not released by base treatment. The phospholipid acyl chains were analyzed as their methyl esters which were obtained by base catalyzed transesterification (12).
The methyl esters were analyzed by reverse phase chromatography as above, except that the solvent was acetonitrile-methanol-water (65/35/0.5) by volume or by argentation thin layer chromatography on 20% AgNO3 (Analtech Silica Gel GHL) plates developed twice in toluene at −20oC followed by autoradiography. The chromatograms were dried and exposed to Kodak BioMax XAR film. Mass spectral analyses of the fatty acid compositions of the membrane phospholipids was done as described previously (13) on phospholipid extracts from cultures grown overnight with the fatty acid to be tested at 0.1 mM final concentration.
RESULTS
AasS Expression Allows Efficient Incorporation and Elongation of Exogenous Short and Medium Chain Fatty Acids into Phospholipids and Lipid A
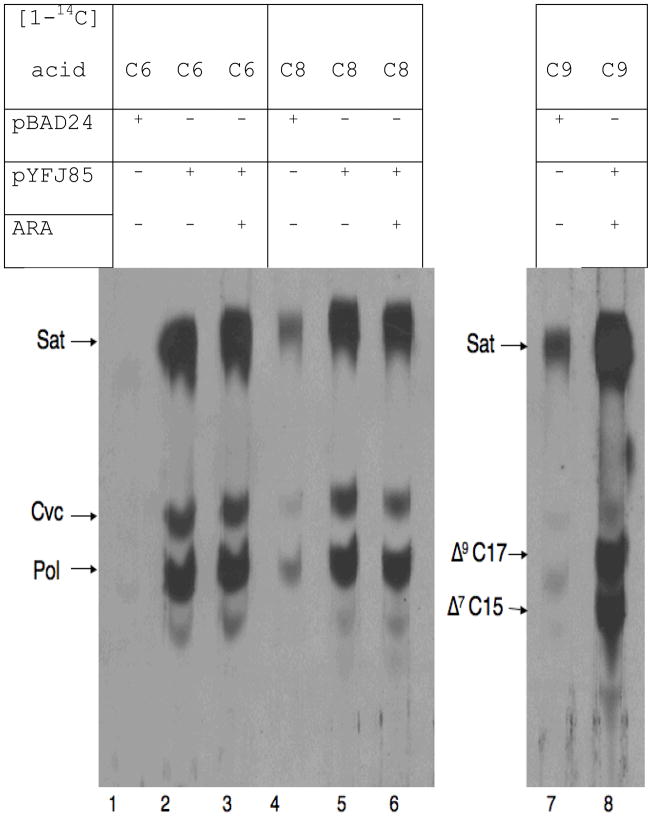
We first tested the ability of strains expressing AasS to elongate exogenous short chain fatty acids to chain lengths of sufficient length to be accepted by the glycerophospholipid acyltransferases for incorporation into membrane phospholipids. In our first experiments the exogenous acids were labeled in the carboxyl group with 14C and both even and odd chain length acids were tested in the presence or absence of AasS expression (Fig. 2). The concentration used was 15 μM since this concentration is more than twice the Michaelis constants of the C8, C12 and C14 acids (5). The most striking result was observed with [1-14C]hexanoate. In the absence of AasS expression (the empty vector control) no detectable label was incorporated into phospholipid acyl chains whereas either induced or basal expression of AasS resulted in high levels of incorporation into both saturated and unsaturated acyl chains (the strains used with the short chain acids were unable to convert unsaturated phospholipid acyl chains to their cyclopropane derivatives due to inactivation of the cfa gene). The lack of labeling of the empty vector control culture indicates that labeling is not due to β-oxidation of the labeled fatty acid followed by incorporation of labeled acetyl-CoA. Moreover, identical labeling was seen in wild type and β-oxidation negative (fadE) strains (data not shown). When the supplements were [1-14C]-labeled with C8 or C9 fatty acids some incorporation was seen in the vector control cultures in both wild type and β-oxidation negative (fadE) strains. As expected the C6 and C8 acids were incorporated into both saturated and unsaturated acyl chains (due to inactivation of the cfa gene the strains used were unable to convert unsaturated phospholipid acyl chains to their cyclopropane derivatives which chromatograph as saturated fatty acids). Unsaturated fatty acids were also synthesized from the C9 acid indicating that the unsaturated fatty acid synthetic enzymes, FabA and FabB, can function (albeit poorly) with a C11 substrate in place of the usual C10 substrate. This is consistent with early enzymatic studies of FabA (14). The C9 acid would be elongated to 3-ketoundecanoyl-ACP which would be reduced to 3-hydroxyundecanoyl-ACP and then dehydrated and isomerized by FabA. The products expected would be cis-7-pentadecenoic and cis-9-heptadecenoic acids. Analysis of both the argentation chromatography of the radioactive products (Fig. 2) and mass spectra of the unlabeled products (see below) provides further evidence for synthesis of these fatty acids.
Fig. 2.
Strain YYC1273 (cfa::kan) carrying either the vector plasmid pBAD24 or the AasS encoding plasmid pYFJ85 was grown overnight at 37oC on each of the [1-14C]-labeled short chain fatty acids shown at 15 μM. The acyl chains were analyzed as their methyl esters by argentation chromatography. When added arabinose was present at 0.02%. Sat, saturated fatty acid esters; Cvc, cis-vaccenic acid (cis-11-octadecenoic acid); Pol, palmitoleic acid (cis-9-hexadecenoic acid, Δ9-C17, cis-9-heptadecenoic acid and Δ7-C15, cis-9-pentadecenoic acid. Identification of the fatty acids were done using authentic standards as described previously (19) excepting the odd chain unsaturated acids which were identified by reference to the prior literature (32, 33).
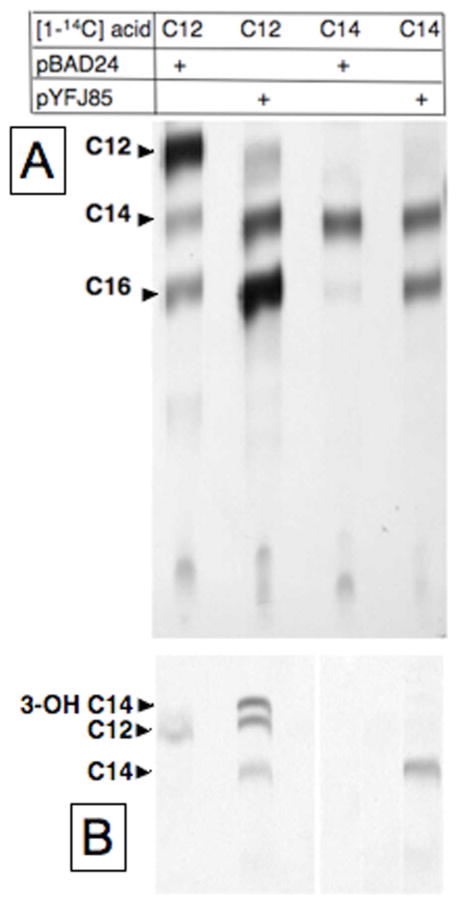
Encouraged by these results we asked if the dodecanoate and tetradecanoate secondary acyl groups of lipid A could be labeled when these fatty acids were provided exogenously in the presence of AasS. The lipid A acyltransferases are considered specific for ACP thioesters (15), although it was recently reported that LpxL has some activity with dodecanoyl-CoA (16). In this case the exogenous acids are sufficiently long to induce the β-oxidation pathway which could result in randomization of the label, hence we used a fadE62 strain (lacking acyl-CoA dehydrogenase). We first examined incorporation into glycerophospholipids. Supplementation of an AasS expressing strain with [1-14C]dodecanoic acid resulted in incorporation of the label into C12, C14 and C16 glycerophospholipid acyl chains indicating efficient elongation of the exogenous acid (Fig. 3A). As expected no label was found in unsaturated acids (data not shown). In a similar manner the [1-14C]tetradecanoic acid was efficiently elongated to the C16 species. In both cases some incorporation of the labeled acids was seen in cultures lacking AasS (Fig. 3A). This seemed primarily due to their incorporation as acyl-CoA thioesters since the chains were incorporated without elongation in the case of the C14 acid and with little elongation in the case of the C12 acid. When the lipid A fractions of these cultures were examined [1-14C]dodecanoic acid labeled both the C12:0 and C14:0 secondary acyl groups as well as the primary 3-hydroxymyristate acyl groups indicating that the labeled fatty acid had entered the elongation cycle. In contrast, supplementation with [1-14C]tetradecanoic acid labeled only the C14:0 secondary acyl group as expected (Fig. 3B). The lack of longer acyl chains showed that the lipid A preparations were largely free of glycerophospholipids. In the case of the C12 acid the strain lacking AasS showed some labeling consistent with the recent report that the LpxL acyltransferase has low activity with dodecanoyl-CoA (16). It should be noted that labeling with the C12 acid was significantly more efficient than labeling with the C14 acid. This can probably be attributed to the 8-fold lower Michaelis constant of AasS for the shorter substrate (5).
Fig. 3.
Incorporation of [1-14C]dodecanoic acid or [1-14C]tetradecanoic acid into the phospholipid (Panel A) or lipid A (Panel B) acyl chains of strain K19 (fadE62) carrying either the vector plasmid pBAD24 or the AasS encoding plasmid pYFJ85 assayed by reverse phase chromatography. The phospholipid acyl chains were analyzed as their methyl esters whereas the lipid A acyl chains were analyzed by reverse phase chromatography as free fatty acids. Note than the right hand two lanes of panel B were exposed three times as long as the two left hand lanes (two exposures of the same plate were done). Arabinose was present at 0.1%. C12, C14 and C16 denote dodecanoic, tetradecanoic and hexadecanoic acids, respectively, whereas 3-OH C14 denotes 3-hydroxytetradecanoic acid. The fatty acids were identified using authentic standards as described previously (19).
The Defective Growth of E. coli fabH Null Mutant Strains is Rescued by Expression of V. harveyi Acyl-ACP Synthetase Plus Exogenous Fatty Acids
Most of the fatty acid synthetic genes are involved in the elongation cycle and therefore are needed throughout the synthetic process. An exception is FabH, the short chain 3-ketoacyl-ACP synthetase (KAS III) thought to be required for only the first elongation, that of an acetate unit to acetoacetyl-ACP, but not for the subsequent elongations required to make long chain length acyl-ACPs. Hence, FabH is thought to provide the “primer” of fatty acid synthesis and to be used only once per acyl chain synthesized (1). E. coli fabH null mutants grow very slowly indicating that the enzyme plays an essential role in the normal fatty acid synthetic pathway (10). However, the slow growth of E. coli fabH null mutants indicates that another inefficient mechanism to generate the primer is present. Indeed, fabH null mutants of Lactococcus lactis were found to synthesize fatty acids at about 10% of the normal rate (10). If the role of FabH is to make the primer, it seemed possible that loss of the enzyme should be compensated by the combination of AasS expression plus supplementation with an exogenous short chain fatty acid such as octanoate.
To test this hypothesis we used strain CL111 previously constructed in this laboratory (10). This strain was constructed by insertion of a second copy of fabH (that of Salmonella enterica) into the attHK022 attachment site of strain UB1005 followed by the deletion of the chromosomal fabH copy. A control strain, CL81, that contained both the S. enterica and E. coli fabH genes, was also constructed. It was then shown that the copy of S. enterica fabH could be deleted in strain CL81 (where a second fabH copy was present), but not in CL111 (which lacked the second copy). These data indicated that a functional FabH is essential for normal cell growth of E. coli (10).
We first introduced the AasS expression plasmid pYFJ85 into strain CL111. The aasS gene was under the control of the arabinose inducible pBAD promoter in the high copy number vector, pBAD24. To remove the functional (S. enterica) copy of fabH strain YFJ249 was then transduced with phage P1 stocks grown on two strains that carried different transposon Tn10 insertions closely linked to the attHK022 site into which the S. enterica fabH gene (marked with a spectinomycin/streptomycin resistance element) had been inserted (10). The transductants were selected on LB supplemented with octanoate plus oleate and arabinose (the inducer of AasS expression) with selection for tetracycline resistance. The tetracycline resistant colonies were then screened for sensitivity to a mixture of spectinomycin and streptomycin. The colonies sensitive to spectinomycin/streptomycin were fabH null mutant strains formed by homologous recombination with the transduced DNA segment that removed the S. enterica fabH gene together with the spectinomycin/streptomycin resistant cassette. Indeed, after 48 h incubation at 37 °C, both large and small colonies formed. All of the small colonies (about 86% of the total tetracycline resistant colonies) were sensitive to spectinomycin/streptomycin whereas all of the large colonies were spectinomycin/streptomycin resistant. In contrast in a parallel experiment, a strain carrying the empty vector plasmid (YFJ248) in place of the aasS plasmid gave no spectinomycin/streptomycin sensitive colonies when exposed to the same protocol.
Given these results we asked if a fabH null strain could be constructed by direct transduction of the fabH::kan null allele from strain CL111 into strains carrying a single functional copy of fabH. We used the wild type strain UB1005 and two different derivative of this strain as transduction recipients. Strain YFJ294 carried pYFJ85 and expressed AasS whereas strain YFJ293 carried the vector plasmid pBAD24. The transduction mixtures were plated on LB medium supplemented with octanoate, oleate and arabinose and incubated at 37°C for two days to permit the strains lacking AasS activity to form colonies.
All three strains gave kanamycin resistant colonies and a colony from each strain background was chosen for further analysis. The strains were designated as strains YFJ303 (derived from YFJ294, the pAasS strain), YFJ304 (derived from YFJ293, the vector plasmid strain) and YFJ305 (derived from the wild type strain, UB1005). The latter two strains YFJ304 and YFJ305 formed very small colonies irrespective of fatty acid supplementation (data not shown). In contrast the growth of the AasS-containing construct, YFJ303, was essentially identical to that of strain YFJ262 (Fig. 4) (strains YFJ303 and YFJ262 were interchangeable except when the cfa mutation of the former strain was required). In addition we confirmed by PCR amplification of the chromosomal fabH region followed by restriction mapping of the PCR products (Fig 1S) that the insertion of strain YFJ305 (10) was as expected. However, strains YFJ304 and YFJ305 could not be used for physiological experiments because significantly larger colonies often appeared on plates after prolonged incubation in the presence or absence of a fatty acid supplement (Fig. 2S). When these larger colonies were picked and purified, they gave uniformly large colonies comparable in size to those of the wild type strain after overnight incubation at 37°C (Fig. 2S). Since the strains retained the ΔfabH::kan marker in which the kanamycin resistance determinant replaced the FabH active site region, they must be extragenic suppressors. Since such fast growing cells would readily take over liquid cultures of the authentic ΔfabH::kan strains, we used strains in which the ΔfabH phenotype was bypassed by the presence of AasS plus octanoate supplementation and engendered the ΔfabH phenotype by removal of the fatty acid supplement or by shutting down AasS expression (or both). However, this bypass approach did not result in cells that grew as rapidly as the extragenic suppressor strains and thus the possibility of take over of liquid cultures by suppressor strains remained. We are currently in the process of identifying the extragenic suppressor mutation(s) that allows growth of ΔfabH strains. It should be noted that in their systematic deletion analysis of the E. coli chromosome (the Keio collection), Baba and coworkers (17) reported that fabH is not essential for normal growth. However, we find (Smith, A. and Cronan, J.E., unpublished data) that the fabH721::kan strain, JW1077-1, of the Keio collection obtained from the Coli Genetic Stock Center (Yale University) carries a suppressor mutation as well as the fabH deletion allele. Phage P1 transduction of the JW1077-1 fabH allele into the wild type strain MG1655 with selection for kanamycin resistance resulted in the same syndrome (very small colonies that give rise to large colonies) that we observed in our constructions. (The selection for the suppressor mutation in the Keio strain is probably due to the natural tendency to select large colonies and discount small colonies as abortive transformants).
Fig. 4.
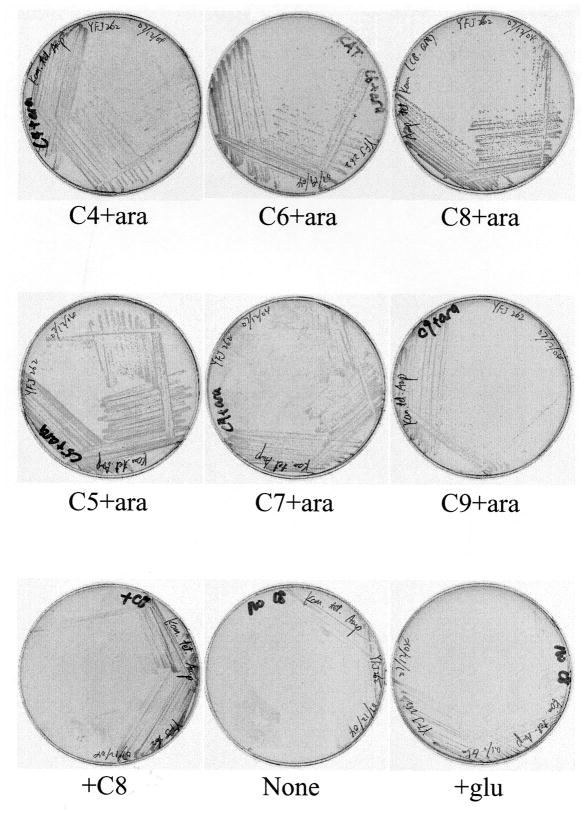
Growth of the ΔfabH pYFJ85(AasS) containing strain YFJ262 streaked on LB plates containing ampicillin and a fatty acid (1 mM) and/or arabinose (0.02%) as shown. The control plates (bottom row) were: +C8, plates containing octanoate but no arabinose (the residual growth can be attributed to basal expression from the pBAD promoter); None, plates with no fatty acid or arabinose supplements and +glu, plates containing 0.1% glucose (which represses basal expression of the pBAD promoter) and None, no fatty acid. The plates were incubated at 37oC for 24 h.
Our original hypothesis was that growth of fabH null mutant strains would require both AasS induction and exogenous fatty acid supplementation. This was only partially the case (Fig. 4). First, the strains grew, albeit very slowly, on media lacking fatty acid supplementation. This very slow growth was also seen when chemically defined medium was used in place of LB medium and when basal level expression of AasS from the pBAD promoter was suppressed by addition of glucose (Fig. 4). These findings suggested that the fabH null mutant retained some fatty acid synthetic activity. (Prior work (10) did not mention the very slow growing colonies formed by the ΔfabH strains presumably because the plates were scored before these colonies appeared). Upon supplementation with a fatty acid plus induction of AasS expression by addition of arabinose, much more rapid growth was obtained (Fig. 4). Note that during our construction of the original mutant strains two fatty acid supplements were present, oleate (to alleviate the need for unsaturated fatty acid synthesis) and octanoate. Octanoate was chosen because it is a good AasS substrate (5, 7, 18) and because octanoyl-ACP enters fatty acid synthesis immediately before the branch point where fatty acid synthesis splits into saturated and unsaturated pathways. Hence, octanoate should provide a primer for synthesis of both saturated and unsaturated fatty acids. This was the case, rapid growth did not require oleate. Further studies showed that butyrate and hexanoate were less effective supplements whereas decanoate (not shown) was growth inhibitory (Fig. 4). The odd chain length fatty acids, pentanoic, heptanoic and nonanoic acids, also supported growth with pentanoic acid being the best of these supplements, although the growth on these acids was less robust than seen on the even chain acids (Fig. 4) perhaps due to decreased synthesis of unsaturated fatty acids.
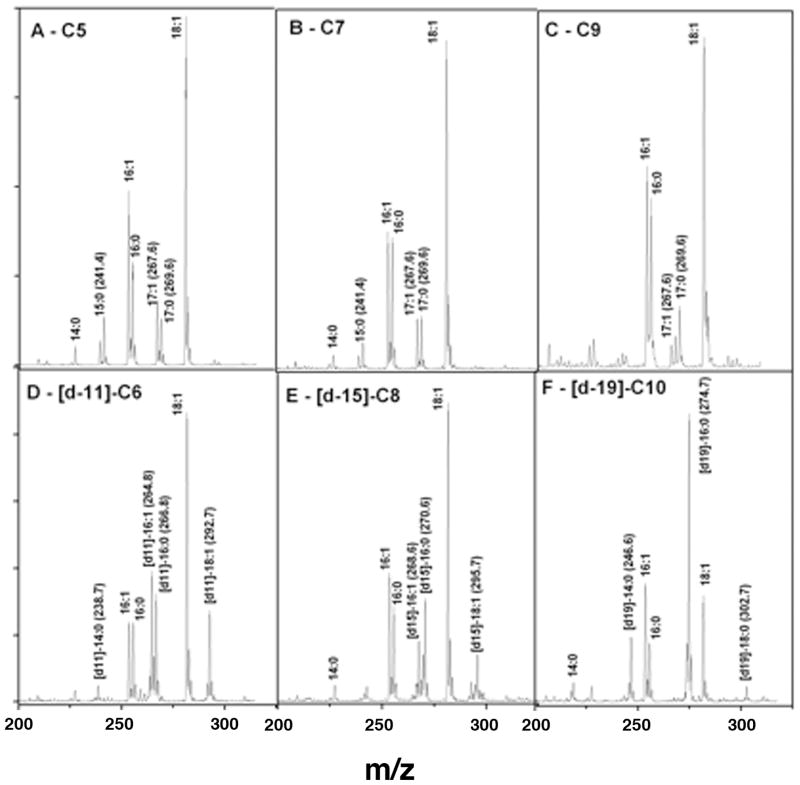
To test if the fabH strain used exogenous short chain fatty acids as primers, we grew cultures of a ΔfabH Δcfa strain supplemented with either a deuterium labeled fatty acid of even chain length ([d-11]-C6:0, [d-15]-C8:0 or [d-19]-C10:0) or with an unlabeled odd chain length fatty acid (C5:0, C7:0 or C9:0). Incorporation was then assayed by electrospray collision-induced mass spectrometry (Fig. 5). Incorporation of these acids would test if significant levels of exogenous fatty acids were elongated and incorporated into glycerophospholipid acyl chains. Moreover, if the labeled acids retained their deuterium contents, this would indicate that they had been incorporated intact. Elongation of each of the fatty acids supplements was observed in the AasS expressing strain YFJ315 (Fig. 5A), whereas as expected from the [14C]-labeled fatty acid incorporation studies, no incorporation was detected in the vector control strain YFJ316 (data not shown). The deuterated C6 and C8 acids were incorporated with full retention of deuterium into long chain saturated fatty acids (C14:0 and C16:0) and unsaturated fatty acids (C16:1 and C18:1). The exception was the third supplement, [d-19]-C10:0. As expected, this precursor gave no labeled unsaturated fatty acids ([d-19]-C16:1 or [d-19]-C18:1) because the chain would enter fatty acid synthesis beyond the point where unsaturated fatty acid synthesis branches from the common (saturated) pathway. A large accumulation of labeled [d-19]-C16:0 and a dramatically reduced peak of de novo synthesized C18:1 were observed. A straightforward explanation for the decreased de novo synthesis of C18:1 would be that the exogenously supplied C10:0 chains would titrate the capacity of the FabF 3-ketoacyl-ACP synthase leading to a deficiency of the longer unsaturated acid (which is the reaction unique to FabF) (1). The growth defect could be due to the decreased fluidity of the cell membrane due to the increased saturated/unsaturated ratio in the glycerophospholipids. Indeed, the defective growth of this strain on decanoate was largely rescued by addition of the unsaturated acid, oleate, to the medium (data not shown). It should be noted that although the strain used was β-oxidation proficient, this was of no consequence since fatty acids shorter than C12 are not degraded by E. coli due to the mechanism that regulates expression of the genes of the pathway (2).
Fig. 5.
Mass spectra (CID ES-MS) of the phospholipid acyl chains of strain YFJ305 ΔfabH cfa::kan carrying the AasS encoding plasmid, pYFJ85. The cultures were grown overnight in the presence of 100 μM fatty acid plus 0.02% arabinose. The spectra shown are those of experiment 2 of Table 1S.
The cfa mutation was essential for analysis of products of the odd chain length fatty acid supplements because the cyclopropane fatty acids of E. coli have the same masses as the unsaturated elongation products (C15:1 and C17:1) that would result from elongation of the odd chain length acid fatty acids. Although E. coli does not normally synthesize odd chain length fatty acids, both C15:0 and C17:0 saturated fatty acids and a C17:1 unsaturated fatty acid were detected in the phospholipids of the cells grown with any of the three odd chain length fatty acid supplements (C5, C7 and C9) (Fig. 5). These data indicated that E. coli fatty acid biosynthetic machinery tolerated the odd chain length acyl-ACPs. As expected from the [1-14C]nonanoic acid labeling studies (Fig. 2), the C9 supplement like the C5 and C7 supplements resulted in a C17:1 unsaturated fatty acid, indicating that FabA can use 3-hydroxyundecanoyl-ACP as a substrate. It was also noteworthy that no C19:1 acid was formed in any of the odd chain fatty acid supplementation experiments indicating that FabF (3-ketoacyl-ACP synthase II), the enzyme responsible for elongation of the C16:1 acid in E. coli was unable to elongate C17:1-ACP.
AasS Expression Allows Supplementation of fabA and fabB Strains with cis-5-Dodecenoic Acid
Mutants of E. coli with lesions in either the fabA or fabB genes are defective in unsaturated fatty acid biosynthesis and require supplementation with an appropriate unsaturated fatty acid (e.g., oleate) for growth (1). FabA is a bifunctional enzyme that introduces the double bond of these acids by first dehydrating 3-hydroxdecanoyl-ACP to trans-2-decenoyl-ACP, the double bond of which is then isomerized to cis-3-decenoyl-ACP (Fig. 1). Several cycles of elongation then result in the unsaturated acyl chains of the membrane lipids (1). Recent in vivo labeling studies have shown that FabB is required for the first of these elongations, that of cis-3-decenoyl-ACP to cis-5-dodecenoyl-ACP (19). These data argued that if cis-5-dodecenoic acid was a suitable AasS substrate, then growth of a fabB mutant strain should result in the presence of cis-5-dodecenoic acid upon expression of AasS. This was the case (Fig. 6). The combination of cis-5-dodecenoic acid supplementation and AasS expression allowed growth of starins carrying null mutations in either fabB or fabA, thereby confirming the in vivo labeling results. However, growth was slow and high levels of AasS expression and high concentrations of the supplement were required. This was expected since unsaturated fatty acids are known to be poor substrates for AasS (5). Indeed, we expected to be able to supplement growth of a fabA strain with cis-3-decenoic acid, but this failed. Subsequent in vitro assays showed that AasS was unable to couple this acid to ACP (data not shown). It should be noted that 0.2% arabinose was needed for growth of the fabA and fabB strains whereas lower concentrations (or no arabinose) sufficed in other experiments. This is presumably due to the relative activities of AasS with the various fatty acid substrates. Note also, that arabinose at 0.2% results in modest growth inhibition of wild type strains carrying plasmid pYFJ85, perhaps due to competition between AasS and malonyl-CoA:ACP transacylase (FabD) for ACP.
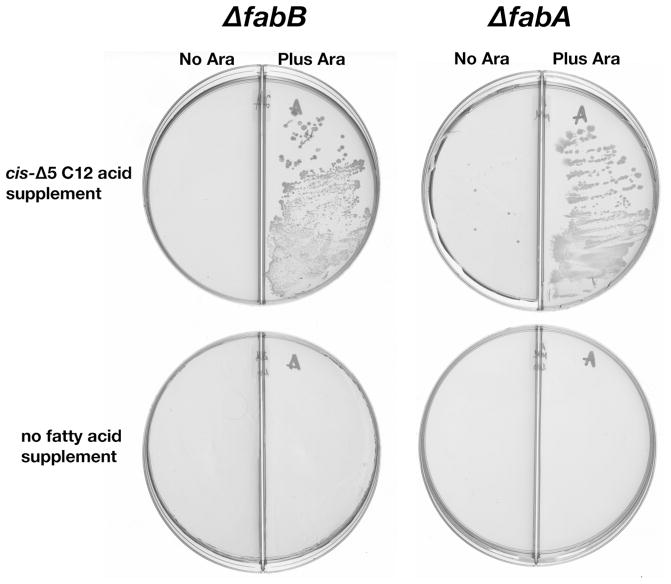
Fig. 6.
Growth of strains JWC280 (ΔfabA ΔfadA::cml ΔfadIJ::kan) and GRT23 (ΔfabB Δcfa::kan) carrying the AasS encoding plasmid pYFJ85 in the presence or absence of cis-5-dodecenoic acid (0.5 mg/ml from Aldrich) and in the presence or absence of arabinose (0.2%). The LB plates were incubated for three days at 37oC. The plates are divided into halves by a wall. Both strains are derivatives of strain BW25111 (34) and were constructed by phage λ Red-mediated recombination and transduction (34). None of these media supported detectable growth of the same strains carrying the vector plasmid pBAD24 in place of pYFJ85 (data not shown
DISCUSSION
Expression of AasS in E. coli clearly allows exogenous fatty acids to access both the fatty acid and lipid A synthetic pathways of E. coli. The acids are efficiently activated to their ACP thioesters and are elongated or incorporated directly into phospholipids or lipid A. The fact that the radioactive short chain fatty acids labeled in the carboxyl group were converted to normal length glycerophospholipid acyl chains argues that at least a portion of the acids had been incorporated intact since the first cycle of β-oxidation would have removed the label from the chain. Subsequent mass spectral studies with deuterated and odd chain length acids showed that the acids were incorporated intact. These findings indicate that mechanistic questions of fatty acid synthesis and subsequent distribution of the acyl chains can now be approached in vivo as well as in vitro. It should be noted that prior work with V. harveyi, the source of AasS, had shown that exogenous fatty acids were efficiently taken up, elongated and incorporated into the complex lipids of this bacterium (20). However, the routes of incorporation remain unclear (21). Recent work showed that strains of V. harveyi with an inactive aasS gene retained the ability to incorporate exogenous fatty acids, although the ΔaasS strains were somewhat defective in incorporation at low exogenous acid concentrations (7).
The AasS-engendered uptake and elongation of short and medium chain fatty acids also allowed growth of unsaturated fatty acid auxotrophs and of strains lacking the fabH-encoded 3-ketoacyl-ACP synthase III, the enzyme responsible for priming fatty acid synthesis. However, the mass spectral data of Fig. 5 show that an appreciable level of fatty acid synthesis occurred in the ΔfabH strain without AasS-facilitated incorporation of the exogenous acid (the unlabeled acids in the deuterated acid experiments and the even numbered chains in the odd chain supplemetation experiments). Moreover, the extent of incorporation of the exogenous acid varied markedly in Table 1S. We believe this is due to extragenic suppressor mutations that allow growth of ΔfabH strains through bypass of the enzyme defect (Fig. 2S); the inactivated fabH allele remained intact. These suppressor mutations arose spontaneously and the suppressed strains grew much more rapidly than ΔfabH strains and more rapidly than ΔfabH strains expressing AasS in the presence of fatty acid supplementation (Fig. 2S). The faster growth rates allowed the suppressor strains to rapidly take over liquid cultures which almost certainly accounts for the variability seen in Table 1S. Hence, the mass spectral data should not be taken as an indicator of the residual fatty acid synthetic capacity of ΔfabH strains, but only as an indication that exogenous fatty acids are incorporated intact.
It should be noted that in the absence of AasS expression (in the empty vector control cultures) some incorporation and elongation of short chain fatty acids occurred (Fig. 2). This incorporation was chain length-dependent and favored octanoate and nonanoate over hexanoate (Fig. 2). Elongation of these acids indicated that they somehow had been converted to acyl-ACPs. This was consistent with prior work from this lab showing that low levels of octanoate were attached to the pyruvate and 2-oxoglutarate dehydrogenases as an intermediate in lipoic acid synthesis in a strain in which the only know pathway of octanoate attachment to these proteins was ACP-dependent (22). In both cases only traces of fatty acids are incorporated. For example the amounts of deuterated or odd chain length fatty acids incorporated are insufficient to alter the fatty acid compositions of the phospholipid as assayed by mass spectrometry (Fig. 3S). What is the source of this trace incorporation? Incorporation is almost entirely dependent on the FadD acyl-CoA synthetase (Fig. 4S) indicating that the incorporation proceeds through an acyl-CoA intermediate. The FadD dependence is also consistent with the chain length dependence because octanoate is an appreciably better FadD substrate than hexanoate (23, 24). Moreover, FadD was not unique in this regard; its function could be replaced by the yeast FAA1 acyl-CoA synthetase (Fig. 5S). The only known route for transfer of an acyl chain from CoA to ACP involves the 3-ketoacyl-ACP synthases (25). The acyl chain would be transferred from CoA to the active site thiol of a 3-ketoacyl-ACP synthase and would subsequently be transferred from the active site thiol to ACP to form an acyl-ACP. Although we were unable to demonstrate such an acyl-CoA:ACP transacylase activity with any of the purified 3-ketoacyl-ACP synthases (data not shown), recent work with the 3-ketoacyl-ACP synthases of gram-positive bacteria shows that these enzymes can be efficiently primed with long chain acyl-CoA substrates (26, 27). In any case, the activity seen in absence of AasS is not sufficient to support either lipoic acid synthesis or growth of strains lacking the fatty acid synthetic enzymes FabH, FabA or FabB. Direct incorporation of the longer fatty acids, dodecanoate and tetradecanoate into phospholipids, was expected since in vitro assays with the relevant acyltransferases showed that acyl-CoA esters were competent acyl donors. Indeed, for dodecanoate and tetradecanoate direct incorporation into complex lipids was dominant over elongation whereas the opposite results were seen upon AasS expression (Fig. 3).
Supplementary Material
Acknowledgments
This work was supported by NIH grant AI15650 to JEC.
Abbreviations
- AasS
acyl-ACP synthetase soluble
- ACP
acyl carrier protein
- FFA
free fatty acid
- FabA
3-hydroxydecanoyl-ACP dehydratase:trans-2-decenoyl-ACP isomerase
- FabB
3-ketoacyl-ACP synthase I
- FabF
3-ketoacyl-ACP synthase II
- FabH
3-ketoacyl-ACP synthase III
- cfa
the gene encoding cyclopropane fatty acid synthase
Footnotes
SUPPORTING INFORMATION: Supporting information consisting of analyses of the genetic constructs, mass spectra, a table of mass spectroscopic values and figures of data characterizing the endogenous elongation activity is available at http://pubs.acs.org/doi/suppl.
References
- 1.Cronan J, Rock C. Biosynthesis of Membrane Lipids. In: Bock A, Curtiss R III, Kaper J, Karp P, Neidhardt F, Nystrom T, Slauch J, Squires C, Ussery D, editors. EcoSal Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, DC: 2008. http://www.ecosal.org. [Google Scholar]
- 2.Clark D, Cronan J. Two-Carbon Compounds and Fatty Acids as Carbon Sources. In: Bock A, Curtiss R III, Kaper J, Karp P, Neidhardt F, Nystrom T, Slauch J, Squires C, Ussery D, editors. EcoSal Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, DC: 2005. http://www.ecosal.org. [Google Scholar]
- 3.Silbert DF, Ruch F, Vagelos PR. Fatty acid replacements in a fatty acid auxotroph of Escherichia coli. J Bacteriol. 1968;95:1658–1665. doi: 10.1128/jb.95.5.1658-1665.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pluschke G, Hirota Y, Overath P. Function of phospholipids in Escherichia coli. Characterization of a mutant deficient in cardiolipin synthesis. J Biol Chem. 1978;253:5048–5055. [PubMed] [Google Scholar]
- 5.Byers DM, Holmes CG. A soluble fatty acyl-acyl carrier protein synthetase from the bioluminescent bacterium Vibrio harveyi. Biochem Cell Biol. 1990;68:1045–1051. doi: 10.1139/o90-154. [DOI] [PubMed] [Google Scholar]
- 6.Fice D, Shen Z, Byers DM. Purification and characterization of fatty acyl-acyl carrier protein synthetase from Vibrio harveyi. J Bacteriol. 1993;175:1865–1870. doi: 10.1128/jb.175.7.1865-1870.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y, Chan CH, Cronan JE. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry. 2006;45:10008–10019. doi: 10.1021/bi060842w. [DOI] [PubMed] [Google Scholar]
- 8.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YY, Cronan JE., Jr Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol Microbiol. 1999;33:249–259. doi: 10.1046/j.1365-2958.1999.01456.x. [DOI] [PubMed] [Google Scholar]
- 10.Lai CY, Cronan JE. β-ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J Biol Chem. 2003;278:51494–51503. doi: 10.1074/jbc.M308638200. [DOI] [PubMed] [Google Scholar]
- 11.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 12.Christie WW. Lipid analysis: Isolation, separation, identification and structural analysis of lipids. 3. Vol. 15. The Oily Press; Bridgewater, United Kingdom: 2003. [Google Scholar]
- 13.Morgan-Kiss RM, Cronan JE. The Lactococcus lactis FabF fatty acid synthetic enzyme can functionally replace both the FabB and FabF proteins of Escherichia coli and the FabH protein of Lactococcus lactis. Arch Microbiol. 2008;190:427–437. doi: 10.1007/s00203-008-0390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloch K. s-hydroxydecanoyl thioester dehydrase. In: Boyer PD, editor. The Enzymes. 3. Academic Press; New York: 1971. pp. 441–464. [Google Scholar]
- 15.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Six DA, Carty SM, Guan Z, Raetz CR. Purification and mutagenesis of LpxL, the lauroyltransferase of Escherichia coli lipid A biosynthesis. Biochemistry. 2008;47:8623–8637. doi: 10.1021/bi800873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Z, Fice D, Byers DM. Preparation of fatty-acylated derivatives of acyl carrier protein using Vibrio harveyi acyl-ACP synthetase. Anal Biochem. 1992;204:343–349. doi: 10.1016/0003-2697(92)90135-t. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, Cronan JE. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J Biol Chem. 2009;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byers DM. Elongation of exogenous fatty acids by the bioluminescent bacterium Vibrio harveyi. J Bacteriol. 1989;171:59–64. doi: 10.1128/jb.171.1.59-64.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Z, Byers DM. Exogenous myristic acid can be partially degraded prior to activation to form acyl-acyl carrier protein intermediates and lipid A in Vibrio harveyi. J Bacteriol. 1994;176:77–83. doi: 10.1128/jb.176.1.77-83.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris TW, Reed KE, Cronan JE., Jr Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J Biol Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- 23.Klein K, Steinberg R, Fiethen B, Overath P. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur J Biochem. 1971;19:442–450. doi: 10.1111/j.1432-1033.1971.tb01334.x. [DOI] [PubMed] [Google Scholar]
- 24.Kameda K, Nunn WD. Purification and characterization of acyl coenzyme A synthetase from Escherichia coli. J Biol Chem. 1981;256:5702–5707. [PubMed] [Google Scholar]
- 25.Alberts AW, Bell RM, Vagelos PR. Acyl carrier protein. XV. Studies of β-ketoacyl-acyl carrier protein synthetase. J Biol Chem. 1972;247:3190–3198. [PubMed] [Google Scholar]
- 26.Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, Colletti SL, Herath K, Cummings R, Salazar O, Gonzalez I, Basilio A, Vicente F, Genilloud O, Pelaez F, Jayasuriya H, Young K, Cully DF, Singh SB. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc Natl Acad Sci U S A. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 28.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoll LJ, Gordon JI. Use of Escherichia coli strains containing fad mutations plus a triple plasmid expression system to study the import of myristate, its activation by Saccharomyces cerevisiae acyl-CoA synthetase, and its utilization by S. cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J Biol Chem. 1993;268:4281–4290. [PubMed] [Google Scholar]
- 30.Carty SM, Sreekumar KR, Raetz CR. Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction At 12 degrees C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J Biol Chem. 1999;274:9677–9685. doi: 10.1074/jbc.274.14.9677. [DOI] [PubMed] [Google Scholar]
- 31.Vorachek-Warren MK, Carty SM, Lin S, Cotter RJ, Raetz CR. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 degrees C. J Biol Chem. 2002;277:14186–14193. doi: 10.1074/jbc.M200408200. [DOI] [PubMed] [Google Scholar]
- 32.Morris LJ, Wharry DM. Chromatographic behaviour of isomeric long-chain aliphatic compounds. I. Thin-layer chromatography of some oxygenated fatty acid derivatives. J Chromatogr. 1965;20:27–37. doi: 10.1016/s0021-9673(01)97362-5. [DOI] [PubMed] [Google Scholar]
- 33.Gunstone FD, Ismail IA. Fatty acids. 16. Thin layer and gas-liquid chromatographic properties of the cis and trans methyl octadecenoates and of some acetylenic esters. Chem Phys Lipids. 1967;1:376–385. [Google Scholar]
- 34.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.