Abstract
Background
We previously identified a number of genes which were methylated significantly more frequently in the tumor compared to the non-cancerous lung tissues from non-small cell lung cancer (NSCLC) patients. Detection of methylation profiles of genes in NSCLC could provide insight into differential pathways to malignancy and lead to strategies for better treatment of individuals with NSCLC.
Methods
We determined the DNA methylation status of 27 genes using quantitative MethyLight assays in lung tumor samples from 117 clinically well-characterized NSCLC patients.
Results
Hypermethylation was detected in one of more of the genes in 106 (91%) of 117 cases and was detected at high levels (Percentage of Methylation Reference (PMR)≥4%) in 79% of NSCLC cases. Methylation of APC, CCND2, KCNH5 and, RUNX was significantly more frequent in adenocarcinomas compared to squamous cell carcinomas (SCC), while methylation of CDKN2A was more common in SCC. Hypermethylation of KCNH5, KCNH8, and RARB was more frequent in females compared to males. Hypermethylation of APC and CCND2 was inversely associated with proliferation score assessed by Ki-67 level.
Conclusions
Our findings of differential gene hypermethylation frequencies in tumor tissues from patients with adenocarcinoma or squamous cell cancers and in females compared to males suggests that further investigation is warranted in order to more fully understand the potential disparate pathways and/or risk factors for NSCLC associated with histologic type and gender.
Keywords: hypermethylation, lung cancer, gender, histology
Introduction
Primary lung cancer is a major cause of death worldwide (1) and remains the leading cause of cancer death in the United States, with an estimated 215,020 new patients diagnosed and 161,840 deaths expected in 2008 (2). Non-small cell lung cancer (NSCLC) accounts for 80% of these new cases and includes the following histologic types: adenocarcinoma, squamous cell carcinoma, large cell carcinoma and mixed histologies. Approximately 25-33% of NSCLC patients present with stage I or II disease, which permits surgical resection with curative intent. However, despite a complete and presumably curative resection, approximately 40-50% of patients with resected NSCLC die of recurrent disease (3).
In recent years, much attention has focused on whether women and men differ with respect to the epidemiologic and clinicopathologic features of NSCLC. Recent trends in the United States show that while incidence in men has declined during the past decades, lung cancer has continued to increase in women (4). Further, much of the current evidence suggests that women have an increased susceptibility to lung cancer, are younger at age of diagnosis, are more likely to have adenocarcinoma compared to squamous cell carcinomas, and have better response to therapy and improved survival (5-9). Identifying the molecular profiles of these patients could provide insight into differential pathways to malignancy in females compared to males and could lead to strategies for better treatment of individuals with NSCLC.
It has recently become clear that epigenetic alterations play an important role in cancer development and result in changes in gene function that occur without changes in nucleotide sequence (10). One of the most well studied epigenetic changes is DNA methylation, which adds a methyl group to cytosines preceding guanidines (also called CpG dinucleotides). Methylation of CpG islands in the promoter region of the gene leads to gene silencing and inactivation, and it has been proposed that DNA methylation of promoter regions of tumor suppressor genes plays an important role in tumor development (11, 12). A number of genes appear to be aberrantly methylated in tumor as opposed to non-tumor tissues from NSCLC patients (13-19). Multiple studies have attempted to assess the clinical significance of one gene, or a small set of genes, in differentiating the clinicopathological features of these tumors (16, 18, 20-25). However, most of these studies utilized qualitative methylation specific PCR (MSP) in order to detect DNA methylation, a method that can sometimes lead to false positive results and does not distinguish between low and high level methylation (26). Furthermore, results in these studies have been inconsistent due to varying methylation detection protocols, PCR primers, and study populations, and none have comprehensively studied more than 10 genes.
In the present study, we chose to quantify methylation of genes which were potentially important in lung cancer, many of which have been assessed by others but some which have not been previously evaluated (Appendix 1). The selected genes affect apoptosis (DAPK1, RUNX3, TMS1, PTEN, and SOCS3), cell adhesion, invasion and/or metastasis (OPCML, CDH13, BVES, APC, CDH1, IGSF4, KCNH5, and PCSK6), cell cycle control (CCND2, RASSF1, APC, FHIT, CDKN2A, CDKN2B, P14, and PTEN), cell proliferation and/or differentiation (RARB, IGFBP3, KCNH5, KCNH8, SOCS3 and PTGS2), and DNA repair and/or detoxification of DNA adducts (FHIT, MLH1, MGMT, FANCF, and GSTP1). In order to assess more broadly the clinical significance of gene hypermethylation in NSCLC, we determined the DNA methylation status of these 27 genes using quantitative MethyLight assays in lung tumor samples from 117 clinically well-characterized NSCLC patients.
Methods
Study Population
The subjects in the present study are a subset of patients included in a larger prospective study of fluorodeoxyglucose (FDG) PET imaging in NSCLC conducted under University of Washington Human Subjects Division approval (27). Briefly, 208 patients were enrolled into the imaging trial and were followed using the standard NSCLC care algorithm previously described (28). Results of this imaging trial have been recently reported (27). In 117 of the 208 subjects, a paraffin-embedded tumor block of the primary tumor was available for methylation analysis, the results of which are presented here.
Pathology
All biopsy and resection specimens were reviewed by the pathology department of the University of Washington Medical Center or the Veterans Affairs Puget Sound Health Care System to verify non-small cell histology of the lung cancer samples and to determine the histologic subtype. NSCLC histology was classified as adenocarcinoma, squamous, large cell, bronchoalveolar adenocarcinoma (BAC), and NSCLC with NOS/other/mixed histology. Histologic BAC was defined using the WHO definition: a variant of adenocarcinoma characterized by surface growth over alveolar septa and without an invasive component. The tumor proliferation rate was assessed by the Ki-67 score as described previously (28). For each subject, the tissue size and stage had been thoroughly assessed as described previously (29).
DNA isolation from paraffin blocks
Six 20 μm sections were cut from each block and deparaffined by xylene extraction. The resulting tissue pellets were digested with proteinase K at 48°C overnight. The genomic DNA was isolated by phenol/chloroform extraction and ethanol precipitation. Finally the DNA was purified using QIAamp DNA mini-column according to the manufacturer's protocol (Qiagen, Valencia, CA).
Sodium bisulfite conversion
Unmethylated human sperm DNA (U-DNA) and in-vitro fully methylated DNA (M-DNA) were converted with clinical samples as described before (30). Briefly, about 1 μg DNA was modified by 5M sodium bisulfite, desulfonated with NaOH, then purified and resuspended in 80 μl elution buffer (10 mM Tris-HCl, pH 8.0, Qiagen).
DNA Methylation (MethyLight) Analysis
For each gene, the primers and probe were designed specifically for bisulfite-converted fully methylated DNA (Appendix 2). Amplification of bisulfite converted β-Actin (ACTB) was used to normalize for input DNA. Samples that were negative for ACTB were excluded in the methylation analysis. A plasmid containing bisulfite converted ACTB gene of known concentration was diluted and used as the standard curve for quantification. The assay for a given set of samples was considered valid only if the converted U-DNA was not amplified, while the converted M-DNA was amplified. The Percentage Methylated Reference (PMR) for each locus was calculated by dividing the GENE: reference ratio of a sample by the GENE: reference ratio of M-DNA and multiplying by 100 (31). Twenty-seven genes, plus the control gene ACTB, were analyzed in this study.
Statistical Analysis
MethyLight data were dichotomized semi-quantitatively, initially classifying a specific gene positive for hypermethylation using both a PMR>0% cutoff as well as a PMR≥4% cutoff, as we have used in our previous study of lung cancer methylation (19). However, for assessing associations of hypermethylation with behavioral and clinical data, we used a cutoff value of PMR>4% which has been validated in the literature as a standard cutoff and is associated with loss of protein expression (31-34). When multiple cancerous tissue samples were available, we randomly chose one of the cancerous blocks to be used in this analysis. Factors associated with patient and tumor characteristics were assessed univariately with chi-square tests for trend and logistic regression. In the final analysis, multivariable adjustments were made to adjust for the potentially confounding effects of histologic type, gender, and tumor size. Exact logistic regression was utilized to compute risk estimates and confidence intervals when the prevalence of gene methylation was low. In the multivariable analyses, p-values and 95% confidence intervals were adjusted to take into account multiple comparisons by setting the false discovery rate (FDR) equal to 0.05, using PROC MULTTEST (35) and subsequently recalculating confidence intervals (36). A two-sided 0.05 test level determined statistical significance for all analyses. All analyses were conducted using SAS version 9.1 (SAS Institute Inc., Cary, NC).
Results
Demographics
In the present study, we chose genes which were potentially important in lung cancer and determined the DNA methylation status of these 27 genes using quantitative MethyLight assays in lung tumor samples from 117 well-described patients. The mean age of the 117 NSCLC cases at the time of enrollment in the imaging trial was 64.8 years (Table 1).
Table 1. Baseline patient and tumor characteristics of NSCLC cases.
| NSCLC Cases (n=117) |
|
|---|---|
| Age (mean years ± sd) | 64.8 ± 8.9 |
| Female Gender | 45 (38%) |
| Caucasian Race | 105 (90%) |
| Smoking History - Pack Years | |
| 0 | 5 (4%) |
| 1-39 | 28 (24%) |
| ≥40 | 80 (68%) |
| Smoker, amount unknown | 4 (3%) |
| Tumor Size (mean cm ± sd) | 3.0 ± 1.5 |
| <2 | 30 (26%) |
| 2-3 | 37 (32%) |
| 3-5 | 38 (32%) |
| ≥5 | 12 (10%) |
| Histologic Stage | |
| I | 65 (56%) |
| II | 28 (24%) |
| III | 18 (15%) |
| IV | 6 (5%) |
| Histologic Type | |
| Adenocarcinoma | 51 (44%) |
| Bronchoalveolar Cell (BAC) | 8 (7%) |
| Adenocarcinoma (Non-BAC) | 43 (37%) |
| Squamous | 39 (33%) |
| Large Cell | 20 (17%) |
| NSCLC, NOS / mixed | 7 (6%) |
| Tumor Differentiation | |
| Well | 9 (8%) |
| Moderate | 40 (34%) |
| Poor | 68 (58%) |
Somewhat less than half were female (38%), and most were caucasian (90%). All but five patients reported a history of smoking, with 68% reporting at least 40 lifetime pack-years of smoking. The majority of these NSCLC cases had surgical stage I (56%) or II (24%) disease and more than half (57%) had tumors < 3cm. By histology, 44% of the tumors were adenocarcinomas, including 7% bronchioloalveolar carcinomas (BAC) and 37% non-BAC adenocarcinomas, 33% were squamous cell carcinomas, and 17% were large cell carcinomas. Fifty-eight percent had poorly differentiated tumors.
Methylation and Clinical and Tumor Characteristics
The methylation profiles of twenty-seven genes were assessed (Table 2). Hypermethylation at PMR>0% was detected in at least one NSCLC case for all genes except P14 and SOCS3. Three genes (APC, CCND2, and RARB) were had detectable methylation in at least 50% of cases, and nine other genes (BVES, CDH1, CDH13, CDKN2A, DAPK1, IGSF4, KCNH5, RASSF1 and RUNX) were methylated in 20-49% of cases. Hypermethylation at any level was detected in one or more of the genes in 106 (91%) of 117 cases. The frequency of high levels of methylation (PMR≥4%) in NSCLC tissues was at least 20% for seven genes, including APC (38%), RARB (31%), RASSF1 (30%), CCND2 (29%), CDKN2A (26%), BVES (23%), and RUNX (21%). Hypermethylation at PMR≥4% was detected in one or more of the genes in 92 (79%) of 117 cases; 28 (24%) cases were positive for one gene, 16 (14%) were positive for two genes, 26 (22%) were positive for 3 to 5 genes, and 22 (19%) were positive for at least 6 and up to 12 genes. Subsequent analyses are presented only for genes (n=15) with high levels of methylation (PMR≥4%) detected in at least 4% of the NSCLC cases.
Table 2. Promoter hypermethylation in NSCLC tissue (n=117).
| Any methylation PMR>0% | High methylation PMR≥4% | |
|---|---|---|
| APC* | 64 (57%) | 43 (38%) |
| BVES | 47 (40%) | 27 (23%) |
| CCND2* | 63 (56%) | 33 (29%) |
| CDH1 | 48 (41%) | 7 (6%) |
| CDH13 | 29 (25%) | 13 (11%) |
| CDKN2A | 30 (26%) | 30 (26%) |
| CDKN2B | 5 (4%) | 0 (0%) |
| DAPK1 | 24 (21%) | 7 (6%) |
| FANCF | 1 (1%) | 1 (1%) |
| FHIT | 1 (1%) | 0 (0%) |
| GSTP1 | 2 (2%) | 2 (2%) |
| IGFBP3 | 14 (12%) | 2 (2%) |
| IGSF4 | 26 (22%) | 3 (3%) |
| KCNH5 | 39 (33%) | 22 (19%) |
| KCNH8 | 22 (19%) | 12 (10%) |
| MGMT | 12 (10%) | 5 (4%) |
| MLH1 | 1 (1%) | 1 (1%) |
| OPCML | 18 (15%) | 16 (14%) |
| P14 | 0 (0%) | 0 (0%) |
| PCSK6 | 11 (9%) | 3 (3%) |
| PTEN | 1 (1%) | 1 (1%) |
| PTGS2 | 1 (1%) | 0 (0%) |
| RARB | 58 (50%) | 36 (31%) |
| RASSF1 | 35 (30%) | 35 (30%) |
| RUNX | 25 (21%) | 25 (21%) |
| SOCS3 | 0 (0%) | 0 (0%) |
| TMS1 | 15 (13%) | 9 (8%) |
112 LC Blocks for APC and CCND2
Methylation patterns varied substantially by histologic type (Table 3), as the percentage of patients with hypermethylation of a number of genes was significantly different in tumors from those with squamous cell carcinomas compared to non-BAC adenocarcinomas, large cell carcinomas, and bronchioloalveolar cell (BAC) adenocarcinomas. In non-BAC adenocarcinomas (n=43) compared to squamous cell carcinomas (n=36), methylation at PMR>4% was significantly more frequent (p<0.05) for APC (45% vs. 17%, respectively), CCND2 (45% vs. 3%), KCNH5 (26% vs. 0%), KCNH8 (12% vs 0%), OPCML (16% vs. 0%), and RUNX (33% vs. 3%). However, in squamous cell carcinomas, methylation of CDKN2A (46% vs. 14%) and MGMT (10% vs. 0%) was more common than in non-BAC adenocarcinomas. The methylation patterns in large cell carcinomas (n=19) and BACs (n=8) were similar to those of non-BAC adenocarcinomas.
Table 3. Promoter hypermethylation in NSCLC tissues, by histologic type.
| Histology | |||||
|---|---|---|---|---|---|
| All Adenocarcinoma | Squamous (n=39) |
Large Cell (n=20) |
Mixed, NOS, or Other (n=7) |
||
| Non-BAC (n=43) |
BAC (n=8) |
||||
| APC* | 19 (45%) | 6 (75%) | 6 (17%) | 10 (53%) | 2 (29%) |
| BVES | 8 (19%) | 3 (38%) | 7 (18%) | 6 (30%) | 3 (43%) |
| CCND2* | 19 (45%) | 4 (50%) | 1 (3%) | 8 (42%) | 1 (14%) |
| CDH1 | 3 (7%) | 0 (0%) | 2 (5%) | 0 (0%) | 2 (29%) |
| CDH13 | 5 (12%) | 1 (13%) | 4 (10%) | 2 (10%) | 1 (14%) |
| CDKN2A | 6 (14%) | 0 (0%) | 18 (46%) | 5 (25%) | 1 (14%) |
| DAPK1 | 2 (5%) | 0 (0%) | 5 (13%) | 0 (0%) | 0 (0%) |
| KCNH5 | 11 (26%) | 3 (38%) | 0 (0%) | 6 (30%) | 2 (29%) |
| KCNH8 | 5 (12%) | 1 (13%) | 0 (0%) | 6 (30%) | 0 (0%) |
| MGMT | 0 (0%) | 0 (0%) | 4 (10%) | 1 (5%) | 0 (0%) |
| OPCML | 7 (16%) | 1 (13%) | 0 (0%) | 5 (25%) | 3 (43%) |
| RARB | 14 (33%) | 2 (25%) | 12 (31%) | 7 (35%) | 1 (14%) |
| RASSF1 | 14 (33%) | 2 (25%) | 7 (18%) | 9 (45%) | 3 (43%) |
| RUNX | 14 (33%) | 2 (25%) | 1 (3%) | 6 (30%) | 2 (29%) |
| TMS1 | 7 (16%) | 0 (0%) | 2 (5%) | 0 (0%) | 0 (0%) |
Only had 42 Non-BAC, 36 Squamous, and 19 Large Cell tissue samples sufficient for methylation analysis
In univariate analyses, female gender was associated with increased methylation frequency among the 117 NSCLC cases, as RARB, OPCML, KCNH5, and KCNH8 had significantly higher frequencies of methylation among females compared to males (p<0.05, data not shown). However, as 46% of adenocarcinomas but only 26% of squamous cell carcinomas were in female subjects, the relationships between gender and gene methylation are likely confounded by histologic type. In order to assess the independent associations between gender and methylation after taking histology into account, we analyzed methylation frequencies stratified by gender and histology (Table 4). Interestingly, among those with adenocarcinoma, hypermethylation of two (KCNH8 and RARB) of the four genes more frequently hypermethylated overall was significantly more frequent in females compared to males. Conversely, among those with squamous cell carcinoma, hypermethylation of APC, BVES, and CDKN2A were somewhat more frequent in males compared to females, although none of those comparisons were statistically significant. Finally, in the small number (n=20) of subjects with large cell NSCLC, hypermethylation at PMR≥4% of KCNH5, KCNH8, OPCML, and RUNX was common (>50%) in females (n=7) but rare (<20%) in males (n=13).
Table 4. Promoter hypermethylation in NSCLC tissues, by histologic type and gender.
| All Adenocarcinoma | Squamous | Large Cell | ||||
|---|---|---|---|---|---|---|
| Female (n=24) |
Male (n=27) |
Female (n=10) |
Male (n=29) |
Female (n=7) |
Male (n=13) |
|
| APC* | 13 (54%) | 12 (46%) | 0 (0%) | 6 (22%) | 2 (29%) | 8 (67%) |
| BVES | 7 (29%) | 4 (15%) | 1 (10%) | 6 (21%) | 3 (43%) | 3 (23%) |
| CCND2* | 11 (46%) | 12 (46%) | 1 (11%) | 0 (0%) | 4 (57%) | 4 (33%) |
| CDH1 | 1 (4%) | 2 (7%) | 0 (0%) | 2 (7%) | 0 (0%) | 0 (0%) |
| CDH13 | 5 (21%) | 1 (4%) | 1 (10%) | 3 (10%) | 1 (14%) | 1 (8%) |
| CDKN2A | 4 (17%) | 2 (7%) | 3 (30%) | 15 (52%) | 2 (29%) | 3 (23%) |
| DAPK1 | 1 (4%) | 1 (4%) | 2 (20%) | 3 (10%) | 0 (0%) | 0 (0%) |
| KCNH5 | 9 (38%) | 5 (19%) | 0 (0%) | 0 (0%) | 4 (57%) | 2 (15%) |
| KCNH8 | 6 (25%) | 0 (0%) | 0 (0%) | 0 (0%) | 5 (51%) | 1 (8%) |
| MGMT | 0 (0%) | 0 (0%) | 1 (10%) | 3 (10%) | 1 (14%) | 0 (0%) |
| OPCML | 4 (17%) | 4 (15%) | 0 (0%) | 0 (0%) | 4 (57%) | 1 (8%) |
| RARB | 13 (54%) | 3 (11%) | 3 (30%) | 9 (31%) | 4 (57%) | 3 (23%) |
| RASSF1 | 7 (29%) | 9 (33%) | 2 (20%) | 5 (17%) | 4 (57%) | 5 (38%) |
| RUNX | 6 (25%) | 10 (37%) | 0 (0%) | 1 (3%) | 4 (57%) | 2 (15%) |
| TMS1 | 3 (13%) | 4 (15%) | 0 (0%) | 2 (7%) | 0 (0%) | 0 (0%) |
Only had 26 Adenocarcinoma male, 9 Squamous female and 27 Squamous male, and 12 Lage Cell male tissue samples sufficient for methylation analysis
Methylation at PMR≥4% of KCNH8 (OR=0.4, 95% CI=0.2-0.9 per 10 years of age) and RARB (OR=0.6, 95% CI=0.4-0.9 per 10 years of age) was somewhat less frequent in older as opposed to younger NSCLC patients (Table 5). Since only 5 NSCLC patients were never-smokers, we were unable to assess hypermethylation associated with smoking status. However, among smokers the estimated pack-years of lifetime cigarette smoking was available. Methylation of CDH1 (OR=1.2 per 10 pack-years) and CDKN2A (OR=1.1 per 10 pack years) was somewhat associated with increased lifetime quantity of cigarette smoking, although these associations did not reach statistical significance. In further univariate analyses, there was a trend (OR's > 1.0) for most genes to be somewhat more likely to be methylated in larger compared to smaller tumors, although this reached statistical significance only for CDKN2A (OR=1.4, 95% CI=1.0-1.8 per 1 cm increase in tumor size, respectively). Gene hypermethylation was not strongly or systematically associated with tumor stage or nodal tumor involvement (data not shown). In terms of tumor differentiation, APC was more commonly detected in well (56%) or moderately (49%) differentiated tumors compared to poorly differentiated tumors (30%, p=0.04 test for trend). Similar patterns were present for CCND2 and RASSF1, although these trends did not reach statistical significance.
Table 5. Promoter hypermethylation in NSCLC tissues - univariate associations with age, pack-years of smoking, and tumor size. (# OR and 95% CI).
| Age (per 10 years) |
Quantity of smokingˆ (per 10 pack-years) |
Tumor Size (per cm) |
|
|---|---|---|---|
| APC | 0.9 (0.6-1.4) | 1.0 (0.9-1.2) | 1.1 (0.9-1.4) |
| BVES | 1.2 (0.7-2.0) | 1.0 (0.9-1.2) | 1.3 (1.0-1.7) |
| CCND2 | 0.8 (0.5-1.2) | 0.9 (0.8-1.1) | 0.9 (0.7-1.2) |
| CDH1 | 1.1 (0.5-2.6) | 1.2 (1.0-1.4) | 1.2 (0.7-1.9) |
| CDH13 | 0.7 (0.4-1.4) | 0.8 (0.6-1.1) | 1.3 (0.9-1.9) |
| CDKN2A | 1.1 (0.7-1.8) | 1.1 (1.0-1.3) | 1.4 (1.0-1.8)* |
| DAPK1 | 0.7 (0.3-1.8) | 1.0 (0.8-1.3) | 1.0 (0.6-1.6) |
| KCNH5 | 0.9 (0.6-1.6) | 0.9 (0.7-1.1) | 1.2 (0.9-1.7) |
| KCNH8 | 0.4 (0.2-0.9)* | 0.9 (0.7-1.1) | 1.1 (0.7-1.6) |
| MGMT | 0.7 (0.3-2.0) | 0.9 (0.6-1.3) | 1.2 (0.7-2.1) |
| OPCML | 1.0 (0.6-1.8) | 1.0 (0.9-1.2) | 1.3 (0.9-1.8) |
| RARB | 0.6 (0.4-0.9)* | 1.0 (0.8-1.1) | 1.0 (0.8-1.3) |
| RASSF1 | 0.8 (0.5-1.3) | 1.0 (0.8-1.1) | 0.9 (0.7-1.2) |
| RUNX | 0.8 (0.5-1.4) | 0.9 (0.8-1.1) | 1.1 (0.8-1.4) |
| TMS1 | 1.6 (0.7-3.5) | 1.0 (0.8-1.2) | 1.2 (0.8-1.8) |
OR – Odds ratio with 95% confidence interval
among 108 subjects who smoked and had known pack-years of smoking
significant at p<0.05
Hypermethylation at PMR≥4% of APC (p=0.05), CCND2 (p=0.009), and to a lesser degree RUNX (p=0.08) was somewhat inversely associated with proliferation score assessed by Ki-67 level (Figure 1). Conversely, hypermethylation of RARB (p=0.06) is associated with increased proliferation score. Specifically, CCND2 was methylated in 49% of tumors with low Ki-67 (<30%), 23% of tumors with medium Ki-67 (30-70%), and 20% of tumors with high Ki-67 (>70%). Similarly, APC was methylated more frequently in those with low (51%) compared to medium (33%) or high (29%) Ki-67 levels.
Figure 1.
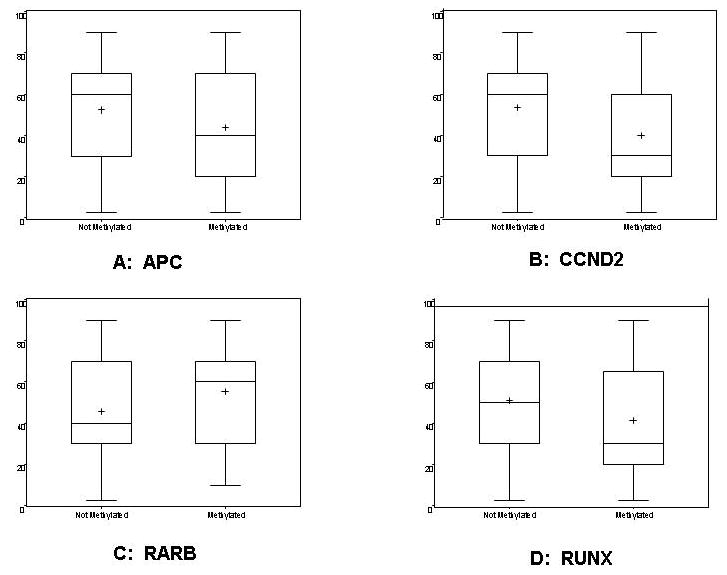
Proliferation score (Ki-67) distribution in tumors with and without hypermethylation of APC (A), CCND2 (B), RARB (C), and RUNX (D).
In order to assess the independent associations between gene hypermethylation and histologic type, tumor size, and gender, we conducted multiple multivariable logistic regressions (Table 6). Since the BAC (n=8) adenocarcinomas showed similar methylation patterns as the non-BAC adenocarcinomas, these two histologic groups were combined for these analyses. In multivariable analyses adjusting for the potential false discovery rate associated with multiple comparisons (15 different models), methylation of a number of genes remained associated with histologic type. Methylation at PMR≥4% of APC (OR=0.2, 95% CI 0.0-0.6), CCND2 (OR=0.0, 95% CI 0.0-0.4), KCNH5 (OR=0.1, 95% CI 0.0-0.5) and RUNX (OR=0.1, 95% CI=0.0-0.6) was significantly less likely in squamous cell carcinomas compared to adenocarcinomas. Conversely, methylation of CDKN2A (OR=5.8, 95% CI=1.6-20.7) was significantly more likely in squamous cell carcinomas compared to adenocarcinomas.
Table 6. Promoter hypermethylation in NSCLC tissues – multivariable models assessing histologic type, tumor size, and gender. (# OR and 95% CI).
| Squamous vs. Adeno* | Tumor Size (per cm) | Female vs. Male | |
|---|---|---|---|
| APC | 0.2 (0.0-0.6) | 1.2 (0.7-2.1) | 0.7 (0.1-4.4) |
| BVES | 0.8 (0.1-4.3) | 1.3 (0.8-2.2) | 1.8 (0.2-13.0) |
| CCND2 | 0.0 (0.0-0.4) | 1.0 (0.6-1.7) | 1.6 (0.2-14.3) |
| CDH1** | 0.6 (0.0-α) | 1.1 (0.4-3.0) | 0.4 (0.0-α) |
| CDH13 | 1.0 (0.0-37.8) | 1.5 (0.7-2.9) | 3.5 (0.5-24.6) |
| CDKN2A | 5.8 (1.6-20.7) | 1.3 (0.8-2.2) | 0.9 (0.0-20.0) |
| DAPK1** | 3.6 (0.2-71.6) | 1.0 (0.3-2.7) | 1.5 (0.0-α) |
| KCNH5** | 0.1 (0.0-0.5) | 1.4 (0.8-2.6) | 4.9 (1.1-22.1) |
| KCNH8** | 0.3 (0.0-5.0) | 1.3 (0.3-6.1) | 33.4 (3.3-340) |
| MGMT** | 7.3 (0.4-121) | 1.1 (0.4-2.9) | 1.8 (0.0-α) |
| OPCML** | 0.1 (0.0-1.5) | 1.3 (0.7-2.6) | 3.1 (0.3-30.9) |
| RARB | 1.2 (0.3-5.2) | 1.1 (0.6-1.8) | 3.2 (1.0-9.7) |
| RASSF1 | 0.5 (0.1-2.2) | 0.9 (0.5-1.6) | 1.1 (0.0-82.3) |
| RUNX | 0.1 (0.0-0.7) | 1.1 (0.6-2.3) | 1.3 (0.0-96.8) |
| TMS1** | 0.3 (0.0-8.9) | 1.3 (0.5-3.4) | 0.8 (0.0-α) |
OR – Odds ratio with 95% confidence interval, adjusted for multiple comparisons
Adenocarcinoma includes BAC and Non-BAC
Exact Logistic Regression
MGMT never methylated in Adenocarcinoma
After adjustment for histologic type, trends of increased methylation in larger tumors and females compared to males remained. Methylation of APC, BVES, CDH13, CDKN2A, KCNH5 and OPCML was somewhat increased with increasing tumor size, with ORs ranging from 1.2 to 1.5, although none of these differences were not statistically significant after adjustment for multiple comparisons. In multivariable analysis, adjusting for histologic type and tumor size, females remained significantly more likely than males to have methylation of KCNH5 (OR=4.9, 95% CI 1.1-22.1), KCNH8 (OR=33.4, 95% CI 3.3-∞), and RARB (OR=3.2, 95% CI 1.1-9.7). These associations were not substantially changed by further adjustments for age or Ki-67 level.
Discussion
In this study of 117 well-characterized NSCLC subjects, we assessed the gene methylation status for 27 genes in tumor tissue blocks with a number of patient and tumor characteristics using quantitative MethyLight assays. Methylation at any level in at least one gene was present in 91% of tumor samples, and a high level of methylation (PMR≥4%) in at least one gene was present in 79% of tumor samples. Conversely, we previously showed that only one of the genes (APC) is methylated at high levels in more than 10% of matched noncancerous tissue from NSCLC patients and that only 5 of the other 26 genes in the current study (RARB, CCND2, CDH1, CDKN2A, and OPCML) were ever methylated (in 2-8% of patients) at high levels in noncancerous tissues (19). In the current study we found that methylation of a number of genes was significantly associated with histologic type; adenocarcinoma was associated with increased methylation of APC, CCND2, KCNH5, and RUNX, and squamous cell carcinoma was associated with increased methylation of CDKN2A. There were trends towards larger tumors being associated with increased methylation frequency. Finally, females with lung cancer were significantly more likely than males to have methylation of a number of genes, including KCNH5, KCNH8, and CDH13.
Our findings of substantial differences in gene methylation patterns depending on histologic typing of NSCLCs have been reported to some degree in the literature. Similar to what we observed in the present study, a number of others (13, 16, 17, 21, 37) have noted that APC is more frequently methylated in adenocarcinomas compared to squamous cell carcinomas, although this has not always been noted (38). Similarly, methylation of RUNX was detected with higher frequency in adenocarcinomas compared to squamous cell carcinomas in previous studies (16, 39, 40). Conversely, methylation of CDKN2A (also known as p16INK4a) has previously been reported to be more common in squamous cell carcinomas compared to adenocarcinomas in most (13, 16, 18, 20, 24, 40) but not all (21, 37) prior studies. However, we believe that our findings that methylation of CCND2 is frequent in adenocarcinomas (46%) but not squamous cell carcinomas (3%) and that methylation of KCNH5 and KCNH8 is less frequent in adenocarcinomas (26% and 12%, respectively) but is specific to this class of tumors, are new to the literature. Aberrant methylation of cyclin D2 (CCND2) has been previously noted as potentially contributing to the pathogenesis of NSCLC (41). However, in that report, potential differences in methylation of CCND2 between squamous and adenocaricomas in NSCLC tumors were not evaluated. Methylation of APC and CCND2, while common in cancer tissues, is also detected frequently in matched non-cancerous lung tissue (19), suggesting that methylation of these two genes may not be a cancer-specific change, but may be the result of environmental exposure, perhaps due to smoking.
We noted a number of important differences in hypermethylation frequency in females compared to males, even after taking into account histologic type. Females with adenocarcinoma were significantly more likely to have methylated CDH13, KCNH5, KCNH8, and RARB compared to males. Interestingly, these differences were not observed in squamous cell carcinomas, although KCNH5 and KCNH8 were not hypermethylated in any squamous cell cancers, regardless of gender. Conversely, males with squamous cell carcinomas were more likely to have hypermethylation of APC, BVES, and CDKN2A (p16INK4a). These substantial differences have not been consistently noted in the literature (16, 18, 20-22, 42), perhaps because few studies have stratified by histologic type when assessing hypermethylation differences by gender. Since females are more likely to have adenocarcinomas compared to males (43), in other studies, differences in hypermethylation frequency due to gender may have been obscured by differences due to histologic type. These substantial differences in hypermethylation by gender suggest the possibility of differential pathways and/or risk factors for NSCLC between genders. Hypermethylation of RARB was similarly common (31%) in males and females with squamous cell carcinoma, but was substantially more common in females with adenocarcinama (54%) or large cell carcinoma (57%) compared to males (11% and 23%, respectively).
Gender differences have historically been noted and debated in the literature with respect to susceptibility and risk factors for NSCLC, especially with regards to the effect of cigarette smoking as well as survival and effectiveness of treatment. The relevance of these variations remains unclear (6-9, 44-47). Epidemiologic studies have generally noted that women with NSCLC are more likely to have developed adenocarcinoma, tend to be younger than men, and are more likely never to have smoked (4, 5, 48, 49). Additionally, for a given stage and treatment regimen, women may have better outcomes than men (7, 45). Hormonal factors may account for differences between men and women with regards to lung cancer (50, 51) although how hormones relate to gene methylation patterns in NSCLC remains unclear. In one study, estrogen receptor (ER) alpha promoter methylation was increased in males with NSCLC compared to females and was associated with poorer prognosis (52). Another proposed explanation is that there is decreased DNA-repair capacity in women compared to men (53), although we did not find evidence that methylation of DNA repair associated genes was differential in males compared to females in the five DNA-repair genes we studied (FHIT, MLH1, MGMT, FANCF, and GSTP1).
To our knowledge, hypermethylation of KCNH5 and KCNH8, two voltage-gated, ether a go-go (Eag 1) family potassium channels, has not been previously studied with regards to human cancers, although KCNH2 has been reported to be downregulated in breast cancer (54). The role of potassium channels in cancer has gained increasing interest in recent years as a possible target for tumor therapy (55, 56). In the present study, hypermethylation of KCNH8 was observed in tumors from six (25%) of 24 females with adenocarcinoma, but was not observed in any males with adenocarcinoma (n=27) or tumors from squamous cell carcinomas from either gender (n=39). Interestingly, hypermethylation of KCNH8 was present in five (71%) of 7 female cases but only one (8%) of 13 male cases of large cell carcinoma. With regards to hypermethylation of KCNH5, we observed similar but less dramatic differences. Importantly, we previously reported that hypermethylation of KCNH5 and KCNH8 is specific to cancerous tissue, as none of 49 matched noncancerous lung tissues from patients with NSCLC were hypermethylated (19).
Our study, and its comparison to other's work, has a number of important limitations. Detection of methylation of the various genes is dependent on the primers and probes used to indicate specific sequence regions, and differences in gene-specific detection rates and as well as associations with clinical features of the tumors may be due to variation in the methodologies used across studies. In the current study, we used quantitative MethyLight, with a cut-off of 4% PMR, to determine associations with methylation. This method is considered sensitive and specific for gene-specific methylation (30, 57). Study findings were similar when a cut-off of 0% PMR was utilized. However, a number of methods, some quantitative and others less sensitive and/or specific qualitative procedures, have been utilized in previous reports, so direct comparisons in study findings across studies may not be entirely possible. In the literature, methylation of FHIT, GSTP1, and MGMT frequently occurs in NSCLC (21, 24, 37, 42, 58) but in the current study, methylation of these genes was rarely detected. The gene-specific discrepancies we observed are likely due to a number of factors, such as assay-specific differences including target sites of CpG island loci, primers or probes, assay conditions including annealing temperatures and the conditions of sodium bisulfaite conversion, differences in tumor characteristics such as type of sample, tumor histology, and level of tumor heterogeneity, and finally, differences in patient populations with regards to age and gender. Primary tumor blocks were unavailable for many of the subjects in the original imaging study, potentially limiting the generalizability of our findings, and sample sizes within histologic subtypes and gender were limited. Further, we were unable to assess the effect of smoking status on methylation in those with NSCLC, as only 5 subjects (4%) were never smokers. Our findings may not be generalizable to populations in which a larger proportion of NSCLC is attributed to cigarette smoking. The distribution of histologic types in the current study was somewhat dissimilar to cases of NSCLC in the United States in general, as squamous and large cell carcinomas were somewhat overrepresented (43). However, the large proportion of non-adenocarcinoma cases and relative heterogeneity of our study tissue samples with respect to histologic subtypes allowed us to identify important differences between adenocarcinomas and squamous cell NSCLCs, comparisons which have rarely been assessed in other studies due to the homogenous nature of their tumor samples. Finally, it is unclear how strongly methylation status is correlated with expression status for the examined genes. Gene expression analyses were not conducted in the current study. However, prior studies using MethyLight have shown that promoter methylation (PMR>4%) of CDKNsA, MLH1, and MGMT was strongly associated with respective loss of protein expression assessed by immunohistochemistry in colorectal cancers (32). In a more recent study in diffuse large B-cell lymphoma, there was good correlation between the presence of MGMT protein expression and unmethylated status using MethyLight (34) and MethyLight performed better than qualitative methylation-specific PCR (MSP). However, in another study, methylation of DAPK (using MSP) was not related with gene silencing in lung cancer cell lines and a number of CpG dinucleotides were often methylated in expression-negative tissues (59). Clearly there may be mechanisms other than promoter hypermethylation contributing to down-regulation of gene expression.
In summary, in the present study of 117 patients with NSCLC, we found that methylation of a number of genes was strongly associated with histologic type; adenocarcinoma was associated with increased methylation of APC, CCND2, and RUNX, squamous cell carcinoma was associated with increased methylation of CDKN2A (p16INK4a), and large cell carcinoma was associated with increased methylation of KCNH8. Among patients with adenocarcinoma, females were significantly more likely to have methylated CDH13, KCNH5, KCNH8, and RARB compared to males. Conversely, males with squamous cell carcinomas were more likely to have hypermethylation of APC, BVES, and CDKN2A (p16INK4a).
The dissimilar pattern of gene hypermethylation in adenocarcinomas compared to other forms of NSCLC implies different pathogenesis of cancer by histologic type. Further, our findings of differential gene hypermethylation frequencies among those with adenocarcinoma or squamous cell cancers in females compared to males suggests that further investigation is warranted in order to more fully understand the potential disparate pathways and risk factors, especially with regard to smoking and hormones, as well as potential differential treatment modalities for NSCLC associated with histology and gender.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institute of Health/National Cancer Institute (CA115559, CA80907, CA107264).
Footnotes
Informed consent was obtained according to procedures approved by the Human Subjects Committee of the University of Washington.
Conflict of interest statement: None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Facts & Figures. Atlanta, Ga: American Cancer Society; 2008. [Google Scholar]
- 3.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111(6):1710–7. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 4.Fu JB, Kau TY, Severson RK, Kalemkerian GP. Lung cancer in women: analysis of the national Surveillance, Epidemiology, and End Results database. Chest. 2005;127(3):768–77. doi: 10.1378/chest.127.3.768. [DOI] [PubMed] [Google Scholar]
- 5.Risch HA, Howe GR, Jain M, Burch JD, Holowaty EJ, Miller AB. Are female smokers at higher risk for lung cancer than male smokers? A case-control analysis by histologic type. Am J Epidemiol. 1993;138(5):281–93. doi: 10.1093/oxfordjournals.aje.a116857. [DOI] [PubMed] [Google Scholar]
- 6.Patel JD, Bach PB, Kris MG. Lung cancer in US women: a contemporary epidemic. JAMA. 2004;291(14):1763–8. doi: 10.1001/jama.291.14.1763. [DOI] [PubMed] [Google Scholar]
- 7.Cerfolio RJ, Bryant AS, Scott E, Sharma M, Robert F, Spencer SA, et al. Women with pathologic stage I, II, and III non-small cell lung cancer have better survival than men. Chest. 2006;130(6):1796–802. doi: 10.1378/chest.130.6.1796. [DOI] [PubMed] [Google Scholar]
- 8.Caldarella A, Crocetti E, Comin CE, Janni A, Pegna AL, Paci E. Gender differences in non-small cell lung cancer: a population-based study. Eur J Surg Oncol. 2007;33(6):763–8. doi: 10.1016/j.ejso.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Freedman ND, Leitzmann MF, Hollenbeck AR, Schatzkin A, Abnet CC. Cigarette smoking and subsequent risk of lung cancer in men and women: analysis of a prospective cohort study. Lancet Oncol. 2008;9(7):649–56. doi: 10.1016/S1470-2045(08)70154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr KM, Galler JS, Hagen JA, Laird PW, Laird-Offringa IA. The role of DNA methylation in the development and progression of lung adenocarcinoma. Dis Markers. 2007;23(1-2):5–30. doi: 10.1155/2007/985474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21(2):163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–9. [PubMed] [Google Scholar]
- 13.Toyooka S, Toyooka KO, Maruyama R, Virmani AK, Girard L, Miyajima K, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther. 2001;1(1):61–7. [PubMed] [Google Scholar]
- 14.Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21(35):5450–61. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- 15.Zochbauer-Muller S, Minna JD, Gazdar AF. Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist. 2002;7(5):451–7. doi: 10.1634/theoncologist.7-5-451. [DOI] [PubMed] [Google Scholar]
- 16.Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94(7):589–92. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9(4):1370–5. [PubMed] [Google Scholar]
- 18.Hanabata T, Tsukuda K, Toyooka S, Yano M, Aoe M, Nagahiro I, et al. DNA methylation of multiple genes and clinicopathological relationship of non-small cell lung cancers. Oncol Rep. 2004;12(1):177–80. [PubMed] [Google Scholar]
- 19.Feng Q, Hawes SE, Stern JE, Wiens L, Lu H, Dong ZM, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17(3):645–54. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakata S, Sugio K, Uramoto H, Oyama T, Hanagiri T, Morita M, et al. The methylation status and protein expression of CDH1, p16(INK4A), and fragile histidine triad in nonsmall cell lung carcinoma: epigenetic silencing, clinical features, and prognostic significance. Cancer. 2006;106(10):2190–9. doi: 10.1002/cncr.21870. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Berman D, Lu C, Wistuba II, Roth JA, Frazier M, et al. Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006;12(24):7329–38. doi: 10.1158/1078-0432.CCR-06-0894. [DOI] [PubMed] [Google Scholar]
- 22.Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007;110(12):2785–92. doi: 10.1002/cncr.23113. [DOI] [PubMed] [Google Scholar]
- 23.Tomizawa Y, Iijima H, Nomoto T, Iwasaki Y, Otani Y, Tsuchiya S, et al. Clinicopathological significance of aberrant methylation of RARbeta2 at 3p24, RASSF1A at 3p21.3, and FHIT at 3p14.2 in patients with non-small cell lung cancer. Lung Cancer. 2004;46(3):305–12. doi: 10.1016/j.lungcan.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61(1):249–55. [PubMed] [Google Scholar]
- 25.Shivapurkar N, Stastny V, Suzuki M, Wistuba II, Li L, Zheng Y, et al. Application of a methylation gene panel by quantitative PCR for lung cancers. Cancer Lett. 2007;247(1):56–71. doi: 10.1016/j.canlet.2006.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shames DS, Minna JD, Gazdar AF. Methods for detecting DNA methylation in tumors: from bench to bedside. Cancer Lett. 2007;251(2):187–98. doi: 10.1016/j.canlet.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Vesselle H, Freeman JD, Wiens L, Stern J, Nguyen HQ, Hawes SE, et al. Fluorodeoxyglucose uptake of primary non-small cell lung cancer at positron emission tomography: new contrary data on prognostic role. Clin Cancer Res. 2007;13(11):3255–63. doi: 10.1158/1078-0432.CCR-06-1128. [DOI] [PubMed] [Google Scholar]
- 28.Vesselle H, Grierson J, Muzi M, Pugsley JM, Schmidt RA, Rabinowitz P, et al. In vivo validation of 3′deoxy-3′-[(18)F]fluorothymidine ([(18)F]FLT) as a proliferation imaging tracer in humans: correlation of [(18)F]FLT uptake by positron emission tomography with Ki-67 immunohistochemistry and flow cytometry in human lung tumors. Clin Cancer Res. 2002;8(11):3315–23. [PubMed] [Google Scholar]
- 29.Vesselle H, Turcotte E, Wiens L, Schmidt R, Takasugi JE, Lalani T, et al. Relationship between non-small cell lung cancer fluorodeoxyglucose uptake at positron emission tomography and surgical stage with relevance to patient prognosis. Clin Cancer Res. 2004;10(14):4709–16. doi: 10.1158/1078-0432.CCR-03-0773. [DOI] [PubMed] [Google Scholar]
- 30.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61(8):3410–8. [PubMed] [Google Scholar]
- 32.Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8(2):209–17. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman WB, Rivenbark AG. Quantitative DNA methylation analysis: the promise of high-throughput epigenomic diagnostic testing in human neoplastic disease. J Mol Diagn. 2006;8(2):152–6. doi: 10.2353/jmoldx.2006.060026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uccella S, Cerutti R, Placidi C, Marchet S, Carnevali I, Bernasconi B, et al. MGMT methylation in diffuse large B-cell lymphoma: validation of quantitative methylation-specific PCR and comparison with MGMT protein expression. J Clin Pathol. 2009;62(8):715–23. doi: 10.1136/jcp.2009.064741. [DOI] [PubMed] [Google Scholar]
- 35.Storey J. A direct approach to false discovery rates. J R Stat Soc, Ser B Stat Methodol. 2002;64:479–98. [Google Scholar]
- 36.Ludbrook J. Multiple inferences using confidence intervals. Clin Exp Pharmacol Physiol. 2000;27(3):212–5. doi: 10.1046/j.1440-1681.2000.03223.x. [DOI] [PubMed] [Google Scholar]
- 37.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103(2):153–60. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 38.Brabender J, Usadel H, Danenberg KD, Metzger R, Schneider PM, Lord RV, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001;20(27):3528–32. doi: 10.1038/sj.onc.1204455. [DOI] [PubMed] [Google Scholar]
- 39.Sato K, Tomizawa Y, Iijima H, Saito R, Ishizuka T, Nakajima T, et al. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep. 2006;15(1):129–35. [PubMed] [Google Scholar]
- 40.Yanagawa N, Tamura G, Oizumi H, Kanauchi N, Endoh M, Sadahiro M, et al. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58(1):131–8. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Virmani A, Rathi A, Heda S, Sugio K, Lewis C, Tonk V, et al. Aberrant methylation of the cyclin D2 promoter in primary small cell, nonsmall cell lung and breast cancers. Int J Cancer. 2003;107(3):341–5. doi: 10.1002/ijc.11393. [DOI] [PubMed] [Google Scholar]
- 42.Vaissiere T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69(1):243–52. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ries L, Melbert D, Krapcho M, Stinchcomb D, Howlader N, Horner M, et al. SEER Cancer Statistics Review, 1975-2005. Bethesda, MD: National Cancer Institute; 2008. [August 15, 2008]. [Google Scholar]
- 44.Blot WJ, McLaughlin JK. Are women more susceptible to lung cancer? J Natl Cancer Inst. 2004;96(11):812–3. doi: 10.1093/jnci/djh180. [DOI] [PubMed] [Google Scholar]
- 45.Wakelee HA, Wang W, Schiller JH, Langer CJ, Sandler AB, Belani CP, et al. Survival differences by sex for patients with advanced non-small cell lung cancer on Eastern Cooperative Oncology Group trial 1594. J Thorac Oncol. 2006;1(5):441–6. [PubMed] [Google Scholar]
- 46.Foegle J, Hedelin G, Lebitasy MP, Purohit A, Velten M, Quoix E. Specific features of non-small cell lung cancer in women: a retrospective study of 1738 cases diagnosed in Bas-Rhin between 1982 and 1997. J Thorac Oncol. 2007;2(6):466–74. doi: 10.1097/01.JTO.0000275340.39960.25. [DOI] [PubMed] [Google Scholar]
- 47.Wakelee HA, Gomez SL, Chang ET. Sex differences in lung-cancer susceptibility: a smoke screen? Lancet Oncol. 2008;9(7):609–10. doi: 10.1016/S1470-2045(08)70162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zang EA, Wynder EL. Differences in lung cancer risk between men and women: examination of the evidence. J Natl Cancer Inst. 1996;88(3-4):183–92. doi: 10.1093/jnci/88.3-4.183. [DOI] [PubMed] [Google Scholar]
- 49.Visbal AL, Williams BA, Nichols FC, 3rd, Marks RS, Jett JR, Aubry MC, et al. Gender differences in non-small-cell lung cancer survival: an analysis of 4,618 patients diagnosed between 1997 and 2002. Ann Thorac Surg. 2004;78(1):209–15. doi: 10.1016/j.athoracsur.2003.11.021. discussion 215. [DOI] [PubMed] [Google Scholar]
- 50.Schabath MB, Wu X, Vassilopoulou-Sellin R, Vaporciyan AA, Spitz MR. Hormone replacement therapy and lung cancer risk: a case-control analysis. Clin Cancer Res. 2004;10(1 Pt 1):113–23. doi: 10.1158/1078-0432.ccr-0911-3. [DOI] [PubMed] [Google Scholar]
- 51.Ganti AK, Sahmoun AE, Panwalkar AW, Tendulkar KK, Potti A. Hormone replacement therapy is associated with decreased survival in women with lung cancer. J Clin Oncol. 2006;24(1):59–63. doi: 10.1200/JCO.2005.02.9827. [DOI] [PubMed] [Google Scholar]
- 52.Lai JC, Cheng YW, Chiou HL, Wu MF, Chen CY, Lee H. Gender difference in estrogen receptor alpha promoter hypermethylation and its prognostic value in non-small cell lung cancer. Int J Cancer. 2005;117(6):974–80. doi: 10.1002/ijc.21278. [DOI] [PubMed] [Google Scholar]
- 53.Wei Q, Cheng L, Amos CI, Wang LE, Guo Z, Hong WK, et al. Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J Natl Cancer Inst. 2000;92(21):1764–72. doi: 10.1093/jnci/92.21.1764. [DOI] [PubMed] [Google Scholar]
- 54.Kuznetsova EB, Kekeeva TV, Larin SS, Zemliakova VV, Babenko OV, Nemtsova MV, et al. Novel methylation and expression markers associated with breast cancer. Mol Biol (Mosk) 2007;41(4):624–33. [PubMed] [Google Scholar]
- 55.Pardo LA, Contreras-Jurado C, Zientkowska M, Alves F, Stuhmer W. Role of voltage-gated potassium channels in cancer. J Membr Biol. 2005;205(3):115–24. doi: 10.1007/s00232-005-0776-1. [DOI] [PubMed] [Google Scholar]
- 56.Hemmerlein B, Weseloh RM, Mello de Queiroz F, Knotgen H, Sanchez A, Rubio ME, et al. Overexpression of Eag1 potassium channels in clinical tumours. Mol Cancer. 2006;5:41. doi: 10.1186/1476-4598-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trinh BN, Long TI, Laird PW. DNA methylation analysis by MethyLight technology. Methods. 2001;25(4):456–62. doi: 10.1006/meth.2001.1268. [DOI] [PubMed] [Google Scholar]
- 58.Kim JS, Kim H, Shim YM, Han J, Park J, Kim DH. Aberrant methylation of the FHIT gene in chronic smokers with early stage squamous cell carcinoma of the lung. Carcinogenesis. 2004;25(11):2165–71. doi: 10.1093/carcin/bgh217. [DOI] [PubMed] [Google Scholar]
- 59.Toyooka S, Toyooka KO, Miyajima K, Reddy JL, Toyota M, Sathyanarayana UG, et al. Epigenetic down-regulation of death-associated protein kinase in lung cancers. Clin Cancer Res. 2003;9(8):3034–41. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


