Abstract
Alcohol and nicotine are coabused, and preclinical and clinical data suggest that common genes may influence responses to both drugs. A gene in a region of mouse chromosome 9 that includes a cluster of three nicotinic acetylcholine receptor (nAChR) subunit genes influences the locomotor stimulant response to ethanol. The current studies first used congenic mice to confirm the influential gene on chromosome 9. Congenic F2 mice were then used to more finely map the location. Gene expression of the three subunit genes was quantified in strains of mice that differ in response to ethanol. Finally, the locomotor response to ethanol was examined in mice heterozygous for a null mutation of the α3 nAChR subunit gene (Chrna3). Congenic data indicate that a gene on chromosome 9, within a 46 cM region that contains the cluster of nAChR subunit genes, accounts for 41% of the genetic variation in the stimulant response to ethanol. Greater expression of Chrna3 was found in whole brain and dissected brain regions relevant to locomotor behavior in mice that were less sensitive to ethanol-induced stimulation compared to mice that were robustly stimulated; the other two nAChR subunit genes in the gene cluster (α5 and β4) were not differentially expressed. Locomotor stimulation was not expressed on the genetic background of Chrna3 heterozygous (+/−) and wild-type (+/+) mice; +/− mice were more sensitive than +/+ mice to the locomotor depressant effects of ethanol. Chrna3 is a candidate gene for the acute locomotor stimulant response to ethanol that deserves further examination.
Keywords: Alcohol, ethanol, locomotor stimulation, nicotinic acetylcholine receptors, quantitative trait locus, mouse
One factor involved in alcohol and nicotine coabuse (Grant et al. 2004; Istvan & Matarazzo 1984; Madden et al. 2000; Talcott et al. 1998) may be common genetic influence. Common genetic factors account for as much as 40% of the covariance between alcohol and cigarette use (Swan et al. 1997), and the genetic correlation between alcohol dependence and nicotine dependence may be as high as 0.68 (Madden & Heath 2002). However, there is some genetic specificity (Volk et al. 2007). Responses to these two drugs in animal models of addiction-related traits also share some genetic codetermination. Mice selectively bred for their heightened acute locomotor response to ethanol were also more stimulated by nicotine compared to mice bred for reduced sensitivity to ethanol stimulation (Bergstrom et al. 2003). Selective breeding of rats and mice for differences in sensitivity to the sedative–hypnotic effects of ethanol also resulted in differences in response to nicotine (Collins et al. 1993; de Fiebre et al. 1987, 1990, 1991, 2002). These genetic correlations suggest that common neural mechanisms associated with common genetic regulation may influence certain responses to alcohol and nicotine.
Individuals who report higher levels of stimulation following alcohol administration in a laboratory setting drink more alcohol outside the laboratory than those that report lower levels of stimulation (Erblich & Earleywine 2003; Holdstock et al. 2000; King et al. 2002; Young et al. 2005). Therefore, magnitude of behavioral stimulation in response to ethanol has been proposed to be an endophenotype for excessive alcohol use (Gabbay 2005), and as such, defining the genetic basis of this response may lead to a greater understanding of alcohol use patterns. Quantitative trait locus (QTL) mapping has been used to locate genes that contribute to variation in traits influenced by multiple genes, including the acute locomotor response to ethanol (Dudek et al. 1991). QTL mapping results indicate that genes on several chromosomes influence this trait, but two laboratories have provided evidence for the influence of at least one gene on mouse chromosome 9 (Cunningham 1995; Palmer et al. 2006). Genes encoding the α5 (Chrna5), α3 (Chrna3) and β4 (Chrnb4) subunits of the nicotinic acetylcholine receptor (nAChR) reside in a gene cluster in the relevant region of chromosome 9. Pretreatment with the non-specific nAChR antagonist mecamylamine attenuated ethanol-induced locomotor stimulation (Blomqvist et al. 1992; Kamens & Phillips 2008; Larsson et al. 2002), but incomplete information exists regarding the involvement of specific nAChR subunits.
To explore the α5, α3 and β4 nAChR subunit genes as candidates for the ethanol stimulation QTL, we combined evidence from several approaches. We studied the ethanol responses of congenic and knockout mice, used a congenic F2 population of mice for finer mapping, and performed quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) gene expression analysis. The ethanol stimulation QTL on mouse chromosome 9 was confirmed, and supporting evidence was provided for one of the subunit genes as the stronger candidate for influencing the acute response to ethanol.
Methods
Subjects
All mice tested in these experiments were produced by breeding pairs at the Portland Veterans Affairs Medical Center. Congenic mice were obtained from J.K.B., whereas C57BL/6J (B6), DBA/2J (D2), B6.129S7-Chrna3tm1Bay/J wild-type (Chrna3 +/+) and B6.129S7-Chrna3tm1Bay/J heterozygous (Chrna3 +/−) mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA).
Congenic mice
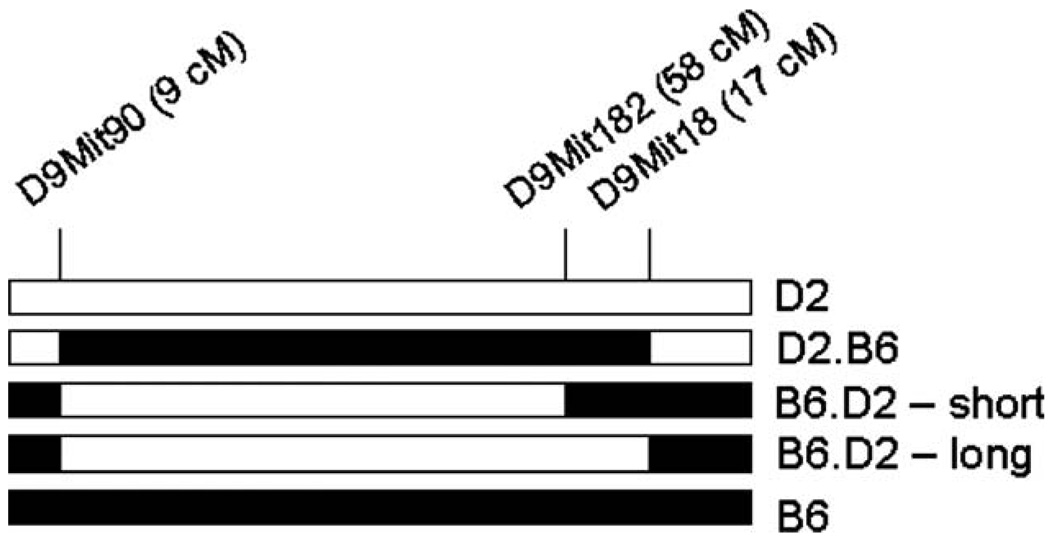
Three chromosome 9 congenic strains were included in these studies: chromosome 9 D2.B6 (D9Mit90,18; 9–71 cM), chromosome 9 B6.D2 – short (D9Mit90,182; 9–58 cM) and chromosome 9 B6.D2 – long (D9Mit90,18; 9–71 cM) congenics (see Fig. 1). The indicated DNA markers (e.g. D9Mit90 at 9 cM and D9Mit18 at 71 cM) define the proximal and distal boundaries of the introgressed regions. The congenic strain notation (e.g. D2.B6) defines the background strain (in this case D2) and the strain that provided the donor region (B6).
Figure 1. A graphical depiction of the chromosome 9 congenic mice.
Black represents B6 alleles and white represents D2 alleles. The markers listed are those that define the ends of the congenic interval.
D2 × chromosome 9 D2.B6 (D9Mit90,18) congenic F2
The congenic F2 mice were derived by crossing male chromosome 9 D2.B6 (D9Mit90, 18) congenic mice to female D2 mice. The F1 offspring from this cross, which were heterozygous for the introgressed region, were then interbred to produce the F2 mice used in the current study.
B6.129S7-Chrna3tm1Bay/J heterozygous (Chrna3 +/−) and wild-type (Chrna3 +/+) mice
Mice deficient in Chrna3 were created using now standard homologous recombination procedures (Xu et al. 1999). Homozygous null mutant animals die shortly after birth; therefore, it was necessary to use only heterozygous (+/−) and wild-type (+/+) mice for this study. Mice used in these experiments had been backcrossed to the B6 strain for 10 generations prior to cryopreservation by the Jackson Laboratory. Wild-type and +/− mice were rederived from frozen embryos and shipped to us. Three types of breeding pairs (+/− × +/−; +/+ × +/−; +/− × +/+) produced the littermates used in these studies.
Animal husbandry
Mice were isosexually housed two to five per cage in shoebox size cages (internal dimensions: 28.5 cm long × 17.5 cm wide × 12 cm high) under standard laboratory temperature and humidity conditions. Mice had ad libitum access to rodent chow (Purina Laboratory Rodent Chow #5001; Purina Mills, St. Louis, MO, USA) and tap water. The lighting in the animal rooms was maintained on a 12-h light/dark cycle (lights on at 0600 h).
At the start of the experiment, all animals were between 51 and 114 days of age, and mice of both sexes were included in these studies. All procedures were approved by the Portland Veterans Affairs Medical Center’s Institutional Animal Care and Use Committee and were in accordance with the National Institutes of Health (1985) principles of laboratory animal care.
Drugs
Ethyl alcohol (200 proof) was obtained from Pharmco (Brookfield, CT, USA) and diluted in physiological saline (Baxter Healthcare Corporation, Deerfield, IL, USA) to a 20% v/v solution. Injection volumes were adjusted for body weight to achieve doses of 1–2.75 g/kg ethanol.
Testing apparatus
AccuScan automated activity monitors (40 × 40 × 30 cm; AccuScan Instruments, Columbus, OH, USA) were used to assess locomotor activity by interruption of eight infrared beams, located 2 cm above the chamber floor. Beam breaks were recorded by a computer and converted into horizontal distance traveled (in cm). The activity monitors were housed in custom-made (Flair Plastics, Portland, OR, USA) black acrylic test chambers designed to separate the monitors from the test room environment. Each chamber was lined with sound attenuating foam, illuminated with an 8W fluorescent light and ventilated with a small fan that also provided background noise. Mice were tested during the light phase of the light/dark cycle, between 0800 and 1600 h, with the fluorescent lights on in the chambers.
Experiment 1
In this experiment, we set out to confirm the presence of a chromosome 9 QTL for ethanol-induced locomotor stimulation using mice congenic for the relevant region of chromosome 9. Chromosome 9 D2.B6 congenic, chromosome 9 B6.D2 – short congenic, B6 and D2 mice were moved from the colony room to the test room about 1 h before behavioral testing. The 3-day testing procedure used in these studies is identical to the procedure that was originally used to map this QTL (Palmer et al. 2006; Phillips et al. 1995). Animals were injected with saline on days 1 and 2 and ethanol (2 g/kg) on day 3. Immediately after the injection on each day, mice were placed into the activity monitors and horizontal distance traveled was measured for 15 min, in 5-min epochs.
The acute locomotor response to ethanol was defined as the day 3 ethanol score minus the day 2 habituated baseline score (day 3 – day 2). Subtracting out the baseline score provides a measure of locomotor response to ethanol that factors out differences among animals in level of baseline locomotor activity. Data from the first 5 min of the test session were used as the primary dependent variable consistent with the original QTL mapping experiment (Phillips et al. 1995). This period during the ascending limb of the ethanol absorption phase captures pure stimulation, whereas longer test periods permit locomotor depression in response to ethanol to develop in some mouse strains (Phillips et al. 1995).
Based on the predicted amount of trait variation accounted for by this QTL (14–34%), a group size of 15–40 was predicted to be needed to confirm its presence in the introgressed region (Belknap & Atkins 2001). Twenty-six to 49 mice were tested per sex per strain in eight cohorts. In a representative number of animals from some cohorts, blood samples were obtained to determine if behavioral differences between the strains were associated with differences in blood ethanol concentrations (BECs). A blood sample (20 µl) was obtained from these mice from the retro-orbital sinus and processed following procedures standard in our laboratory (Boehm et al. 2000). BECs were determined by gas chromatography (Agilent 6890, Foster City, CA, USA) against a curve derived from a series of known standards in the expected range of concentrations.
All other behavioral tests used similar procedures. Differences in procedure from this experiment are noted below.
Experiment 2
Ethanol produces strain dependent biphasic effects on locomotor activity in mice (Crabbe et al. 1994; Dudek et al. 1991, 1994). While 2 g/kg ethanol in the D2 strain is stimulatory, in the B6 strain, this dose is more likely to produce locomotor depression (Crabbe et al. 1994; Dudek et al. 1991). A single QTL from the D2 genotype that confers increased sensitivity to ethanol stimulationmay not be able to overcome this alternate effect of ethanol that is characteristic of the B6 background strain. This possibility arose as an explanation for the results of experiment 1 and led us to reduce the ethanol dose used to test the chromosome 9 B6.D2 – short congenic and B6 controlmice in experiment 2. Thirty to 32 mice (half of each sex) were tested per genotype using the same behavioral procedure as in experiment 1, except that mice were given a 1.5 g/kg ethanol dose on day 3. All mice had blood taken after testing on day 3 for BEC analysis.
Experiment 3
Initial studies were conducted in a pre-existing B6 background congenic (B6.D2 – short) that possessed an introgressed D2 segment that was 13 cM shorter than the congenic segment in the D2.B6 strain (9–58 cM as compared to 9–71 cM, respectively). QTL capture was confirmed in the D2.B6, but not in B6.D2 – short congenic, mice for either dose of ethanol. We entertained the possibility that the QTL was not present in the shorter congenic region and thus created a new B6 background congenic with a longer 9–71 cM introgressed region (B6.D2 – long), comparable to that of the D2.B6 congenic. Forty to 41 chromosome 9 B6.D2 – long congenic and B6 controlmice were tested for ethanol-induced stimulation, using the dose of ethanol that was originally used to detect the QTL (2 g/kg). A blood sample was taken from all animals following testing on day 3 to examine BEC.
Experiment 4
All 260 congenic F2 mice had a small portion of the tail (approximately 2 mm) taken for DNA analysis at least 2 weeks prior to being tested for their acute locomotor response to ethanol (2 g/kg), using the procedures already described. F2 animals that had behavioral responses in the top or bottom 12.5% of the phenotypic distribution were genotyped and used for QTL analyses. There was an equal representation of each sex in both tails of the distribution. Genotyping this portion of the population (25% in total) provides the majority of linkage information available from the whole population (Darvasi & Soller 1992; Lander & Botstein 1989).
Experiment 5
The QTL for ethanol-induced locomotor stimulation on mouse chromosome 9 was captured on the D2 background, the background more prone to this effect of ethanol. We next explored the possibility that one or more of the nAChR subunit genes might play a role in the differential ethanol response by examining expression of the three nAChR subunit genes in the chromosome 9 D2.B6 congenic and D2 controlmice. In this initial analysis, whole brain tissue from naive male and female mice (n = 6 per D2.B6 and D2 strain) was used for qRT-PCR analysis of the Chrna5, Chrna3, and Chrnb4 genes.
Experiment 6
In experiment 5, only Chrna3 was differentially expressed between the D2 and D2.B6 congenic mice. To examine if Chrna3 was also differentially expressed in brain regions known to be important in the locomotor response to drugs of abuse including ethanol, the striatum and ventral midbrain were dissected for qRT-PCR. These regions were chosen because they are part of the motive circuit (Nestler 2005) known to modulate drug-induced locomotor stimulation (Wise & Bozarth 1987). These brain regions were dissected from naive male and female chromosome 9 D2.B6 congenic and D2 mice (n = 10–12 per strain), and Chrna3 expression was determined using qRT-PCR.
Experiment 7
In experiments 5 and 6, Chrna3 was differentially expressed between the D2 and D2.B6 congenic mice. To attempt to confirm a role for Chrna3 in ethanol-induced locomotor stimulation Chrna3 +/− and Chrna3 +/+ animals were tested for this trait. Fifteen to 16 mice per genotype and ethanol (1, 1.5, 2, or 2.75 g/kg) dose were tested. A range of ethanol doses was used because the mutation resides on a largely B6 background, and we wanted to maximize our chances of detecting a locomotor stimulant response difference. On the B6 background, doses of 2 g/kg and below were most likely to produce small stimulant effects during the first 5 min after injection, whereas locomotor depression was more likely at the 2.75 g/kg dose. On day 3, a blood sample was taken to examine BEC.
DNA isolation and genotyping
Genomic DNA was extracted from tail tissue using the Puregene Tissue and Mouse Tail Kit according to the manufacturer’s protocol (Qiagen, Valencia, CA, USA). Briefly, cells were placed overnight in cell lysis solution and proteinase K. RNA was removed with the addition of RNase, and then, proteins were precipitated with a protein precipitation solution provided in the kit. Finally, DNA was precipitated with isopropanol, washed and rehydrated in TE buffer (10 mM Tris, pH 7.5, and 1 mM ethylenediaminetetraacetic acid).
D2 × chromosome 9 D2.B6 (D9Mit90,18) congenic F2
Six microsatellite markers spaced approximately every 12 cM throughout the introgressed congenic region were genotyped in the F2 mice. To each DNA sample, 1.2 µl MgCl2 (25 mM), 1.5 µl buffer, 0.15 µl AmpliTaq Gold (5 U/ µl; Applied Biosystems, Foster City, CA, USA), 2.5 µl dNTPs (1.25 mM; Promega, Madison, WI, USA), 7.15 µl ddH20 and 0.3 µl forward and reverse primers (6.6 µM; Research Genetics, Inc., Huntsville, AL, USA) were added. The reaction was then amplified in a Perkin–Elmer PCR machine (Waltham, MA, USA) under the following conditions: 95°C for 10 min, 80°C for 5 min, 40 cycles of 94°C for 30 seconds, 53°C for 30 seconds, and 72°C for 30 seconds, followed by 72°C for 7 min and finally holding at 4°C. Amplified polymerase chain reaction (PCR) products were detected by ethidium bromide staining on 3% agarose gels.
B6.129S7-Chrna3tm1Bay/J heterozygous (Chrna3 +/−) and wild-type (Chrna3 +/+) mice
Chrna3 +/− and +/+ animals were genotyped using the standard procedures described above and forward and reverse primers (5′-CTAGGCCACAGAATTGAAAGATCT-3′ and 5′-GTAGGTGGAAATTCTAGCATCATCC-3′, respectively) to identify Chrna3 +/+ mice, and primers complimentary to the inserted mutant sequence to identify Chrna3 +/− animals (forward: 5′-TGTTCTCCTCTTCCTCATCTCC-3′ and reverse: 5′-ACCCTTTCCAAATCCTCAGC-3′; Integrated DNA Technologies; Coralville, IA, USA).
Tissue extraction
Whole brain
Mice were cervically dislocated and decapitated, and whole brains were removed. Brains were immediately frozen by submersion in cold isopentane, put into RNase-free tubes, and stored at −80°C until processed for gene expression analysis.
Microdissections
Whole brains were removed and placed on an ice-cold platform. The striatum was dissected from a slice between +1.75 and +0.25 bregma. The ventral midbrain (containing the ventral tegmental area, substantia nigra and interpeduncular nuclei) was dissected from a slice between −3.25 and −4.25 bregma. Each sample was placed into RNase-free tubes on dry ice after dissection, before being transferred to a −80°C freezer for storage until processed for gene expression analysis.
RNA extraction
RNA was extracted using the methods appropriate for either whole brain samples or microdissected tissue. For whole brain, RNA was extracted using the guanidinium isothiocyanate–phenol–chloroform extraction method (Chomczynski & Sacchi 1987, 2006), following procedures standard in our laboratory (Kamens & Phillips 2008). Briefly, RNA Stat-60 (Tel-Test, Inc., Friendswood, TX, USA) was used to extract RNA, which was then cleaned of DNA contamination. RNA quality was assessed with an A260 nm/A280 nm ratio by spectrophotometer (1.8–2 was considered good quality; Eppendorf, Hamburg, Germany; Chomczynski & Sacchi 2006) and confirmed by electrophoresis on a 1% agarose gel.
RNA from the microdissections was extracted using the Absolutely RNA Microprep Kit (Stratagene, La Jolla, CA, USA) and the manufacturer’s protocol. Briefly, β-mercaptoethanol/lysis buffer mixture (100 µl) was added to each sample before vortexing and manual homogenization. One hundred µl of 70% ethanol were then added to the mixture, which was vortexed (5 seconds) and then transferred to an RNA-binding spin cup. Following centrifugation (maximum speed, 60 seconds), 600 µl of a low-salt wash buffer was added to the spin cup before another round of centrifugation (60 seconds at maximum speed). The filtrate was then removed, and the column was dried by a 2-min spin at maximum speed. To remove DNA contamination, a 30 µl mixture of RNase-free DNAse I and DNase digestion buffer was added to the spin cup matrix, and the samples were incubated at 37°C for 15 min. Samples were then washed first with 500 µl of high-salt wash buffer, then 600 µl of low-salt wash buffer, and finally 300 µl of low-salt wash buffer, followed by centrifugation (60 seconds to 2 min) after each wash. To elute the RNA from the spin cup matrix, 30 µl of the elution buffer was added directly to the matrix, and the sample was centrifuged at maximum speed. The quality of the microdissection RNA samples was examined using the same criterion as the whole brain RNA samples.
Quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR)
Quantitative reverse transcriptase–polymerase chain reaction was performed in a two-step reaction using standard procedures in our laboratory (Kamens & Phillips 2008). Briefly, total RNA was reverse transcribed using a High-Capacity cDNA Archive Kit (Applied Biosystems) and the manufacturer’s specifications. Messenger RNA gene expression of the Chrna5, Chrna3 and Chrnb4 nAChR subunit genes and of the control Hprt1 (hypoxanthine guanine phosphoribosyltransferase 1) gene was analyzed using predesigned TaqMan gene expression assays (Applied Biosystems). The control gene Hprt1 was used because it has been validated as a reference gene in mouse models (Meldgaard et al. 2006). Quantitative PCR reactions were run using an iCycler (Bio-Rad, Hercules, CA, USA). Each sample was run in triplicate, and the average crossing threshold (Ct) for the Chrna5, Chrna3, Chrnb4 and Hprt1 genes was determined. For each sample, the Ct for Hprt1 was subtracted from the expression of the AChR subunits. Relative expression based on the ΔΔ Ct method (calculated as 2 to the negative power of the average expression of D2 mice minus each individual value) was used as the primary dependent variable.
Statistics
Behavioral and gene expression data from each congenic strain were compared independently to data from the relevant background strain, while Chrna3 +/− animals were compared to Chrna3 +/+ animals. Data were analyzed with two- or three-way factorial analysis of variance (anova). Strain, sex and ethanol dose were used as independent variables. Interactions were analyzed for simple main effects, while main effects were followed up by Newman–Keuls post hoc comparisons. statistica v. 6 (StatSoft, Tulsa, OK, USA) was used for all statistical analyses with an alpha level set at 0.05.
Effect size
The percentage of variation accounted for by the QTL was calculated for the congenic strain that was significantly different from its control strain (the one that captured the QTL). The QTL effect size (R2) was calculated from a one-way anova using the equation R2 = F/(F + dfw) (F = statistic from the one-way ANOVA; dfw = degrees of freedom within; Rosenthal 1994). The proportion of genetic variation account for was derived by taking the R2 divided by the heritability of the trait (0.49; Phillips et al. 1995).
QTL analysis
R/qtl was used to analyze the genotypic and phenotypic data from the congenic F2 study (Broman et al. 2003). Animals from the top and bottom 12.5% of the phenotypic distribution were analyzed with the ‘scanone’ command. This command identifies QTL using the expectation maximization algorithm.
Results
Experiment 1: confirmation of a QTL for ethanol-induced locomotor stimulation in a D2, but not B6, background congenic
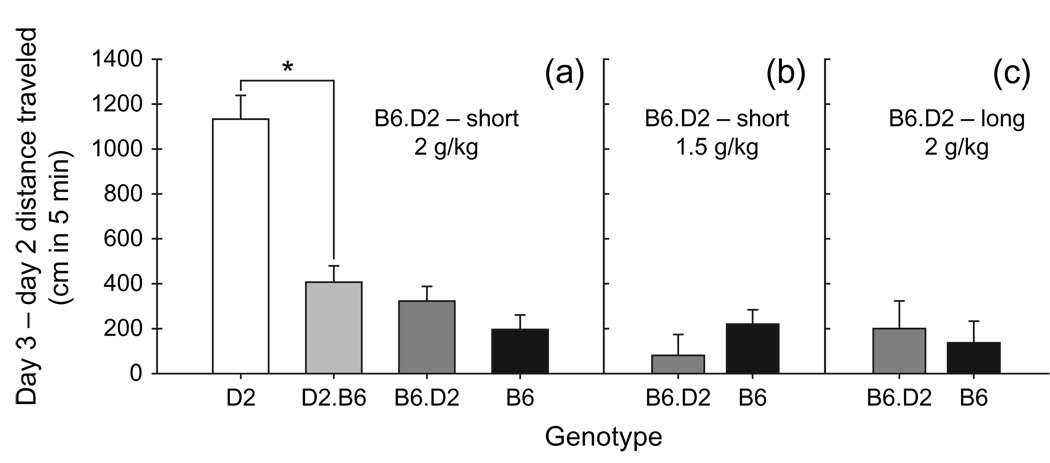
Chromosome 9 D2.B6 congenic mice exhibited an attenuated stimulant response to ethanol compared to D2 mice, confirming the presence of a QTL on chromosome 9 for ethanol-induced locomotor stimulation (Fig. 2a). Data from three animals were excluded fromthe analysis because of computer malfunction, illness or misplaced injection. A two-way anova of the acute ethanol response (day 3 – day 2) with strain and sex as independent variables revealed a significant main effect of strain (F1,128 = 34.4, P < 0.001) and sex (F1,128 = 7.7, P < 0.01; female more stimulated than male; 1009.8 ± 137.1 vs. 592.9 ± 71.9 cm, respectively) but no interaction of the two factors. This QTL accounted for 41%of the genetic variation in ethanol-induced stimulation. Examination of locomotor data from day 2 of saline baseline treatment identified no differences in locomotor behavior between the genotypes (data not shown). A subset of the animals tested for the acute response to ethanol (20–21 per strain) was used to assess BEC. There was no significant strain difference in BEC in the subset of animals sampled at the conclusion of the behavioral test (15 min after the 2 g/kg ethanol injection), but there was a significant main effect of sex (F1,37 = 6.0, P < 0.05). Female mice had higher BECs than male mice (2.15 ± 0.04 and 1.98 ± 0.02 mg/ml, respectively).
Figure 2. Capture of the chromosome 9 QTL for the locomotor response to ethanol is dependent on congenic background strain.
(a) The acute locomotor response to ethanol (2 g/kg) in male and female chromosome 9 D2.B6 (D9Mit90,18; 9–71 cM) congenic, chromosome 9 B6.D2 – short (D9Mit90,182; 9–58 cM) congenic, D2 and B6 strains. (b) The acute locomotor response to ethanol (1.5 g/kg) in male and female chromosome 9 B6.D2 – short (D9Mit90,182; 9–58 cM) congenic and B6 strains. (c) The acute locomotor response to ethanol (2 g/kg) in male and female chromosome 9 B6.D2 – long (D9Mit90,18; 9–71 cM) congenic and B6 strains. Data are for the first 5 min (mean ± SEM) of the 15-min test session. *P < 0.05.
The chromosome 9 B6.D2 – short congenic and B6 control mice were equally sensitive to the locomotor stimulant effect of ethanol (Fig. 2a). Data from one mouse were excluded because of a computer malfunction. A two-way anova of the acute ethanol response (day 3 – day 2) revealed a significant main effect of sex (F1,184 = 21.3, P < 0.01; female > male; 463.3 ± 66.4 and 61.8 ± 57.4 cm, respectively) but no other significant results. Examination of data from day 2 of saline treatment identified a significant main effect of strain (F1,184 = 11.0, P < 0.01), such that B6.D2 – short congenic mice had higher activity levels than B6 mice (1726.6 ± 48.6 and 1506.7 ± 45.6 cm, respectively). Female mice again had a significantly higher mean BEC compared to male mice (F1,43 = 4.3, P < 0.05; 2.03 ± 0.04 and 1.88 ± 0.06 mg/ml, respectively; n = 22–25 per strain), but there was no significant difference in BEC between the two strains.
Experiment 2: the B6 background congenic does not capture a QTL for the acute locomotor response to a 1.5 g/kg injection of ethanol
Chromosome 9 B6.D2 – short congenic and B6 mice were also equally sensitive to 1.5 g/kg ethanol, as indicated by no significant main effect of strain (Fig. 2b). Similar to results for the 2 g/kg ethanol dose, female mice were significantly more stimulated than male mice (F1,58 = 9.2, P < 0.01; 351.9 ± 96.6 and 26.3 ± 61.3 cm, respectively). When data from the second day of saline treatment were examined, the B6.D2 – short congenic and B6 mice had equivalent locomotor activity levels (data not shown). A two-way anova with strain and sex as independent variables detected no significant main effects or interactions for BEC.
Experiment 3: there is no evidence of a QTL for ethanol stimulation on distal chromosome 9 when tested on the B6 background
When the chromosome 9 B6.D2 – long congenic was tested for ethanol (2 g/kg)-induced locomotor stimulation, no difference in response compared to B6 control mice was observed (Fig. 2c). A two-way anova revealed a significant main effect of sex (F1 ,77 = 12.2, P < 0.001; female > male; 430.1 ± 102.3 and −86.0 ± 104.1 cm, respectively) that did not interact with strain. Chromosome 9 B6.D2 – long con-genic mice were more active on day 2 after a saline injection compared to B6 mice (F1,77 = 6.6, P < 0.05; 1724.8 ± 86.4 and 1429.6 ± 74.4 cm, respectively). When BEC values were examined, there was a significant strain × sex interaction (F1,77 = 4.6, P < 0.05) because of higher BEC only in male congenic compared to male B6 mice (P < 0.05; 2.34 ± 0.07 and 2.18 ± 0.08 mg/ml, respectively).
Experiment 4: the QTL for ethanol-induced locomotor stimulation resides between 23 and 69 cM
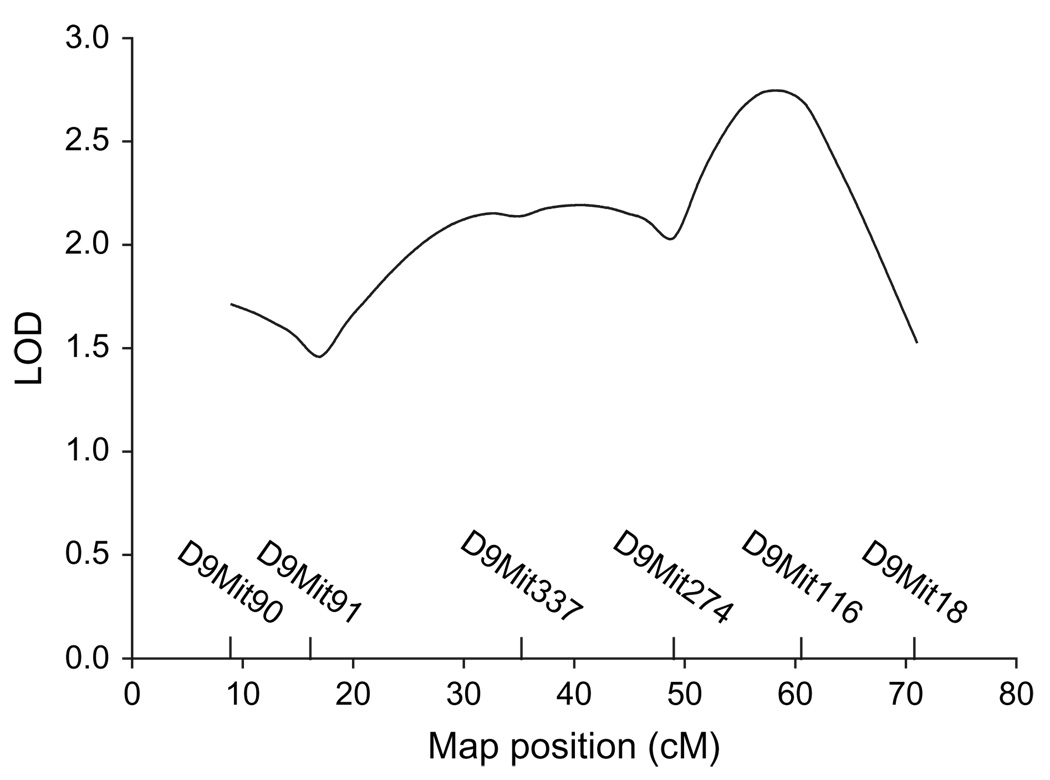
The congenic F2 mice provided some resolution of the QTL region. The data from one mouse were excluded because the animal was sick. When tested for the acute locomotor response to ethanol, the 259 congenic F2 mice showed a normal phenotypic distribution (data not shown). When the top and bottom 12.5% of the phenotypic population were analyzed using R/qtl, there was significant evidence of a QTL for ethanol stimulation on mouse chromosome 9 that exceeded Lander and Kruglyak (1995) guidelines for confirmation of a QTL previously identified in independent populations. The peak LOD score was at 59 cM with a 1 LOD support interval ranging from 23 to 69 cM (Fig. 3). This narrows the location of the QTL from a 62 cM region to a 46 cM region, which still contains the three genes encoding the nAChR subunits.
Figure 3. QTL mapping in a congenic F2 population reduces the QTL interval from a 62 cM region to a 46 cM region.
The peak LOD score for the chromosome 9 QTL is at 59 cM with a 1 LOD support interval of 23–69 cM. Shown are LOD scores representing the most likely location of the QTL for ethanol (2 g/kg)-induced stimulation on chromosome 9 based on interval mapping in the chromosome 9 D2.B6 (D9Mit90,18; 9–71 cM) congenic × D2 F2 mice. n used for mapping = 32 mice from the bottom and top tails of the phenotypic distribution curve.
Experiment 5: the α3 subunit of the nAChR is a candidate gene for ethanol stimulation
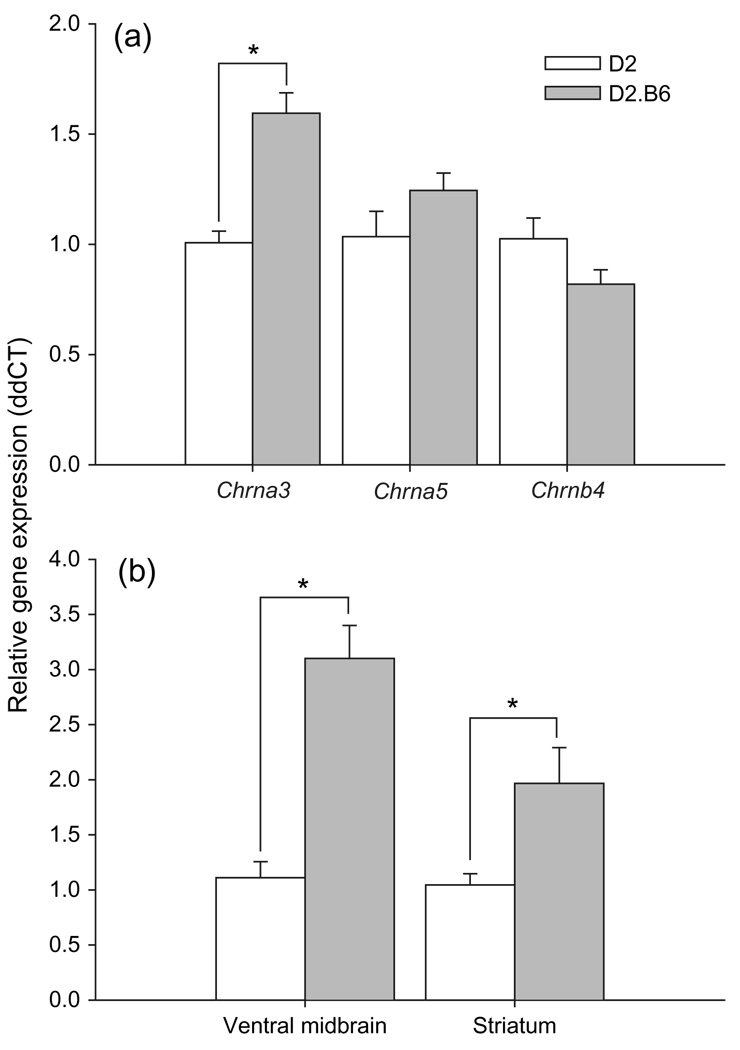
The α3, but not α5 or β4, subunit gene of the nAChR was differentially expressed between the chromosome 9 D2.B6 congenic and D2 mice (Fig. 4a). Congenic mice had significantly more Chrna3 RNA expression than D2 mice (F1,8 = 29.1, P < 0.001), independent of sex.
Figure 4. Greater whole brain and region-specific Chrna3 expression corresponds with reduced locomotor response to ethanol.
(a) Whole brain mean relative Chrna3, Chrna5 and Chrnb4 expression in the chromosome 9 D2.B6 (D9Mit90,18; 9–71 cM) congenic and D2 strains of mice. (b) Ventral midbrain and striatum mean relative Chrna3 expression in the chromosome 9 D2.B6 (D9Mit90,18; 9–71 cM) congenic and D2 strains of mice. *P < 0.05.
Experiment 6: strains of mice that differ in response to an acute injection of ethanol also differ in Chrna3 expression in brain regions involved in this response
Chromosome 9 D2.B6 congenic mice had significantly more Chrna3 RNA expression than D2 mice in both the ventral midbrain (F1,18 = 35.0, P < 0.001) and the striatum (F1,18 = 5.9, P < 0.05), independent of sex (Fig. 4b).
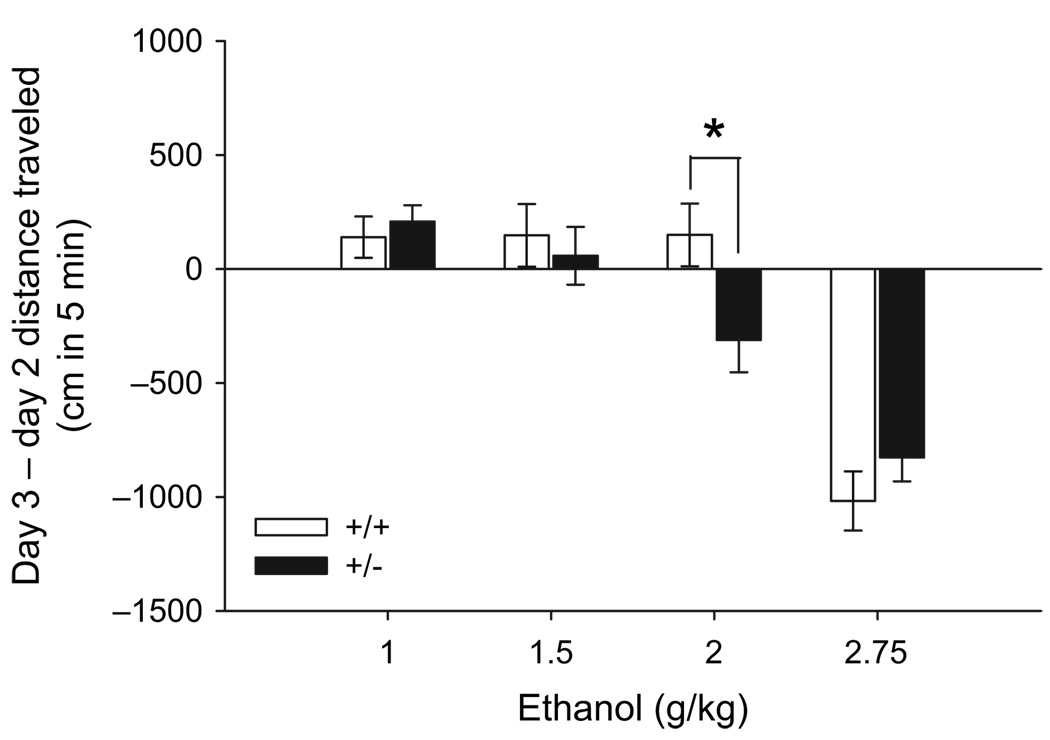
Experiment 7: Chrna3 +/− mice exhibit greater locomotor depression compared to Chrna3 +/+ mice after 2 g/kg ethanol
Data for three animals were excluded because their behavioral data points were statistical outliers (+/− 2.5 SD from the mean). Because sex was not a significant factor, nor did it interact with strain or dose, analyses were performed with data collapsed on sex. Chrna3 +/− mice were more sensitive to the locomotor depressant effects of 2 g/kg ethanol compared to +/+ animals, but these strains were equally sensitive to ethanol at all other doses tested (line × dose interaction; F3,113 = 2.7, P < 0.05; Fig. 5). BEC increased in a dose-dependent manner (F3,108 = 140.4, P < 0.001; 1 g/kg = 1.09 ± 0.04, 1.5 g/kg = 1.58 ± 0.05, 2 g/kg = 2.18 ± 0.07, and 2.75 g/kg = 3.18 ± 0.11mg/ml), independent of strain.
Figure 5. Strain differences in ethanol-induced locomotor activity between Chrna3 +/+ and +/− mice are dependent upon ethanol dose.
Data are for the first 5 min (mean ± SEM) of the 15-min test session. *P < 0.05.
Discussion
The current studies confirmed the presence of a gene on mouse chromosome 9 that accounts for 41% of the genetic variation in the acute locomotor response to ethanol. These studies also provided evidence that the gene expressing the α3 subunit of the nAChR is a stronger candidate for this QTL than are the α5 and β4 subunits genes, based on expression profiles in mice that differ greatly in the stimulant response to ethanol.
Prior mapping data provided evidence of a QTL for ethanol-induced stimulation on chromosome 9 (Cunningham 1995; Palmer et al. 2006). The presence of a QTL in this region was confirmed at a P value that exceeds Lander and Kruglyak’s (1995) threshold for a significant QTL in the D2 background congenic. We were unable to detect the QTL for ethanol stimulation using two B6 background congenics. We considered and tested two possible reasons for this lack of confirmation.
First, because B6 mice are particularly sensitive to the inhibitory effects of ethanol on locomotor behavior, use of the 2 g/kg dose of ethanol may have masked detection. However, when the B6.D2 – short congenic was tested at a lower ethanol dose (1.5 g/kg), the QTL for ethanol stimulation was still not detected. It should be noted that the QTL was originally mapped using a 2 g/kg ethanol dose and drug QTL are often dose dependent (Phillips et al. 1998). Second, we considered the length of the congenic region by creating a longer segment B6.D2 congenic. However, the QTL was not captured in this congenic either.
These data do not rule out that a gene influencing ethanol stimulation resides in the congenic region on the B6 background. A third possible explanation for our results is that this gene epistatically interacts with another locus (or multiple loci) elsewhere in the genome and that expression of the trait is dependent on the presence of a particular allele at this other locus that is not present in the B6 mouse. In other words, the presence of a D2 allele at another locus may be required in combination with the chromosome 9 D2 allele(s) to obtain a phenotypic effect. Such an epistatic interaction (i.e. background effect) involving the 5-HT1B receptor gene (Htr1b), for example, has been observed for ethanol consumption (Phillips & Belknap 2002; Phillips et al. 1999). A QTL for ethanol stimulation has been confirmed on mouse chromosome 2 in both a D2 and a B6 background congenic (Palmer et al. 2006). Creating double B6.D2 congenics (i.e. mice carrying introgressed D2 segments on both chromosomes 2 and 9) may address the possibility of epistatic interactions or more simple additive effects of trait-relevant genes.
In multiple cases, we found a significant main effect of sex that did not interact with genotype. In all cases, females were more stimulated by ethanol compared to males, a finding that is consistent with prior literature (Kamens et al. 2006; Palmer et al. 2006). Given that a significant interaction with genotype was never observed, these data suggest that this QTL is not sex specific.
To our knowledge, this is the first attempt to fine map a QTL using a congenic F2 population. We were able to narrow the location of the gene from a 62 cM region to a 46 cM region. The effect size of this QTL is large for a behavioral QTL and in the range of effect sizes of QTL that have been fine mapped to smaller regions (Flint et al. 2005). Additional strategies, such as the use of interval-specific congenic strains (Darvasi 1997; Fehr et al. 2002), might be useful for finer mapping of the location of this QTL. While Chrna3 was differentially expressed, there may be other genes in this region that are also differentially expressed and influence this trait. If finer mapping could be accomplished, the number of genes under consideration would be reduced, and this would be beneficial for more rigorous candidate gene identification. However, if more than one gene underlies this large-effect QTL, when the genes are separated via a finer mapping process in smaller segment congenics, the phenotype alteration may become undetectable.
Nicotinic acetylcholine receptors have been implicated in ethanol stimulation (Blomqvist et al. 1992; Kamens & Phillips 2008; Larsson et al. 2002). The non-specific nAChR antagonist mecamylamine attenuated ethanol-induced stimulation in D2 mice (Kamens & Phillips 2008). While mecamylamine is regarded as non-specific, some suggest that this drug is most potent at inhibiting α3β4 nAChR (Papke et al. 2001). Furthermore, in NMRI outbred mice, mecamylamine attenuated ethanol stimulation and the involvement of α4β2 and α7 nAChR was excluded because the nAChR antagonists, dihydro-β-erythroidine (α4β2 specific) and methyllycaconitine (α7 specific) had no effect on this trait (Larsson et al. 2002). Additionally, α-conotoxin MII (α3β2, β3 andα6 specific), but not α-conotoxin PIA analogue (α6 specific), attenuated ethanol-induced locomotor stimulation, providing evidence that α3β2- or β3-containing nAChRs are involved in this response (Jerlhag et al. 2006; Larsson et al. 2004). Recent evidence in humans showed that single nucleotide polymorphisms within the α3/α5/β4 gene cluster are associated with the age of initiation of alcohol use (Schlaepfer et al. 2008) and alcohol dependence (Wang et al. 2008).
Our data nominate α3 as the best candidate of the α3/α5/β4 genetic cluster, but it is possible that α5 or β4 may be involved in this ethanol response. Given the fact that these subunits are highly coregulated (Xu et al. 2006) and that recent studies have found associations of alcohol traits with genetic variants in CHRNA5 and CHRNB4, in addition to CHRNA3 (Schlaepfer et al. 2008; Wang et al. 2008), we cannot completely rule out the possibility that the other subunits are involved. It is possible that we would find differential α5 or β4 expression if we looked in distinct brain regions, a future direction we will explore.
Homozygous α3 knockout (−/−) mice have growth impairments and die within weeks after birth (Xu et al. 1999), but heterozygous knockout (+/−) mice are less sensitive to nicotine-induced seizures than +/+ mice (Salas et al. 2004). We found that Chrna3 +/− exhibited locomotor depression to a 2 g/kg ethanol dose, whereas +/+ mice were insensitive to this effect at this dose of ethanol. We found no significant difference in ethanol stimulation between these genotypes, but we also observed little or no ethanol-induced stimulation. These findings are consistent with our results showing lack of confirmation of the ethanol stimulation QTL on the B6 background. We are currently backcrossing the null mutation onto the D2 background to better address the hypothesis that Chrna3 is involved in ethanol stimulation. However, it is possible that even on this background, an effect will not be seen in +/− mice with loss of only a portion of the α3 subunits.
18-Methoxycoronaridine appears to be specific for α3β4 nAChR but is not commercially available. To our knowledge, the effect of 18-methoxycoronaridine on ethanol-induced stimulation has not been tested, but this drug was shown to decrease ethanol consumption and preference (Rezvani et al. 1997). Intracerebral administration of conotoxins that are specific for α3-containing nAChR may help to elucidate the role of these receptors in ethanol stimulation with regard to neuroanatomical location (Clark et al. 2006; Talley et al. 2006), another future direction.
Differences in Chrna3 expression between chromosome 9 D2.B6 congenic and D2 control mice were observed, but further work is needed to determine if these strains differ in α3 protein levels. The assays that have been used to measure α3 protein levels have some inadequacies. Cytisine-resistant epibatidine binding has been the assay typically used (Perry et al. 2002), but there is evidence that this assay is not detecting exclusively α3-containing receptors (Kuryatov et al. 2000;Marks et al. 2006; Parker et al. 1998; Xiao & Kellar 2004). Furthermore, antibodies for the α3 subunit are non-specific (Moser et al. 2007).
In lines of mice selectively bred for extreme sensitivity or insensitivity to the locomotor stimulant effects of ethanol, Chrna3 was not differentially expressed (Kamens & Phillips 2008). It is not uncommon to obtain different results when examining genetic relationships in panels of inbred strains vs. selected lines (Crabbe et al. 1990). There may be more than one mechanism by which an extreme ethanol stimulation phenotype can arise, and selective breeding may have favored a mechanism that is not dependent on Chrna3, whereas this phenotype in the D2 strain may be more strongly associated with level of Chrna3 expression. However, only whole brain expression has been examined in the selected lines.
The α3 subunit gene is expressed in midbrain dopamine and GABA (gamma-aminobutyric acid) neurons (Azam et al. 2002; Klink et al. 2001). Mouse synaptosome preparations have provided evidence that receptors containing the α3 subunit are not directly involved in striatal dopamine release (Salminen et al. 2004), but acetylcholine receptors containing this subunit that are located on dopamine cell bodies could modulate dopamine release. Mecamylamine administered directly into the ventral tegmental area decreases both ethanol-induced locomotor stimulation and elevations in dopamine levels in the nucleus accumbens in response to ethanol (Blomqvist et al. 1997). Because the ventral tegmental area is a major dopamine cell body-containing region, these data suggest the hypothesis that α3-containing nAChR on dopamine cell bodies may modulate these ethanol responses. The expression differences observed in the chromosome 9 D2.B6 congenic and control mice are consistent with this hypothesis. D2 control mice had less Chrna3 expression than D2.B6 congenic mice. The lower level of Chrna3 expression in D2 mice may be the result of greater endogenous acetylcholine levels. This would be consistent with the work of Schwartz and Kellar (1983) who showed that treatment with a cholinesterase inhibitor, which had the effect of increasing acetylcholine levels, decreased nAChR levels.
In humans, administration of ethanol in the laboratory can produce self-reported ratings of stimulation and euphoria, effects that are decreased by pretreatment with mecamylamine (Chi & de Wit 2003; Young et al. 2005). These data suggest that nAChR may be involved in the acute response to ethanol in both humans and mouse models. Improvements in the detection of nAChR constructed from specific subunits will lead to a better understanding of the role of these receptors in addiction-related processes, including sensitivity traits that may serve as risk factors.
Supplementary Material
Acknowledgments
These studies were performed with support from the Department of Veterans Affairs (T.J.P. and J.K.B.), P60 AA10760 (T.J.P.) and F31 AA015822 (H.M.K.). The authors thank Dr Kristine Wiren for use of her laboratory equipment for the qRT-PCR study. Additionally, we thank Joel Hashimoto and Sue Burkhart-Kasch for their technical assistance.
References
- Azam L, Winzer-Serhan UH, Chen Y, Leslie FM. Expression of neuronal nicotinic acetylcholine receptor subunit mRNAs within midbrain dopamine neurons. J Comp Neurol. 2002;444:260–274. doi: 10.1002/cne.10138. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Bergstrom HC, Palmer AA, Wood RD, Burkhart-Kasch S, McKinnon CS, Phillips TJ. Reverse selection for differential response to the locomotor stimulant effects of ethanol provides evidence for pleiotropic genetic influence on locomotor response to other drugs of abuse. Alcohol Clin Exp Res. 2003;27:1535–1547. doi: 10.1097/01.ALC.0000091226.18969.B9. [DOI] [PubMed] [Google Scholar]
- Blomqvist O, Soderpalm B, Engel JA. Ethanol-induced locomotor activity: involvement of central nicotinic acetylcholine receptors? Brain Res Bull. 1992;29:173–178. doi: 10.1016/0361-9230(92)90023-q. [DOI] [PubMed] [Google Scholar]
- Blomqvist O, Ericson M, Engel JA, Soderpalm B. Accumbal dopamine overflow after ethanol: localization of the antagonizing effect of mecamylamine. Eur J Pharmacol. 1997;334:149–156. doi: 10.1016/s0014-2999(97)01220-x. [DOI] [PubMed] [Google Scholar]
- Boehm SL, II, Schafer GL, Phillips TJ, Browman KE, Crabbe JC. Sensitivity to ethanol-induced motor incoordination in 5-HT(1B) receptor null mutant mice is task-dependent: implications for behavioral assessment of genetically altered mice. Behav Neurosci. 2000;114:401–409. [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Chi H, de Wit H. Mecamylamine attenuates the subjective stimulant-like effects of alcohol in social drinkers. Alcohol Clin Exp Res. 2003;27:780–786. doi: 10.1097/01.ALC.0000065435.12068.24. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nature Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- Clark RJ, Fischer H, Nevin ST, Adams DJ, Craik DJ. The synthesis, structural characterization, and receptor specificity of the alpha-conotoxin Vc1.1. J Biol Chem. 2006;281:23254–23263. doi: 10.1074/jbc.M604550200. [DOI] [PubMed] [Google Scholar]
- Collins AC, Romm E, Selvaag S, Turner S, Marks MJ. A comparison of the effects of chronic nicotine infusion on tolerance to nicotine and cross-tolerance to ethanol in long- and short-sleep mice. J Pharmacol Exp Ther. 1993;266:1390–1397. [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Kosobud A, Belknap JK. Estimation of genetic correlation: interpretation of experiments using selectively bred and inbred animals. Alcohol Clin Exp Res. 1990;14:141–151. doi: 10.1111/j.1530-0277.1990.tb00461.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Gallaher ES, Phillips TJ, Belknap JK. Genetic determinants of sensitivity to ethanol in inbred mice. Behav Neurosci. 1994;108:186–195. doi: 10.1037//0735-7044.108.1.186. [DOI] [PubMed] [Google Scholar]
- Cunningham CL. Localization of genes influencing ethanol-induced conditioned place preference and locomotor activity in BXD recombinant inbred mice. Psychopharmacology (Berl) 1995;120:28–41. doi: 10.1007/BF02246142. [DOI] [PubMed] [Google Scholar]
- Darvasi A. Interval-specific congenic strains (ISCS): an experimental design for mapping a QTL into a 1-centimorgan interval. Mamm Genome. 1997;8:163–167. doi: 10.1007/s003359900382. [DOI] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Selective genotyping for determination of linkage between a marker locus and a quantitative trait locus. Theor Appl Genet. 1992;85:353–359. doi: 10.1007/BF00222881. [DOI] [PubMed] [Google Scholar]
- Dudek BC, Phillips TJ, Hahn ME. Genetic analyses of the biphasic nature of the alcohol dose-response curve. Alcohol Clin Exp Res. 1991;15:262–269. doi: 10.1111/j.1530-0277.1991.tb01867.x. [DOI] [PubMed] [Google Scholar]
- Dudek BC, Tritto T, Underwood KA. Genetic influences on locomotor activating effects of ethanol and sodium pentobarbital. Pharmacol Biochem Behav. 1994;48:593–600. doi: 10.1016/0091-3057(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Erblich J, Earleywine M. Behavioral undercontrol and subjective stimulant and sedative effects of alcohol intoxication: independent predictors of drinking habits? Alcohol Clin Exp Res. 2003;27:44–50. doi: 10.1097/01.ALC.0000047300.46347.CE. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fiebre CM, Medhurst LJ, Collins AC. Nicotine response and nicotinic receptors in long-sleep and short-sleep mice. Alcohol. 1987;4:493–501. doi: 10.1016/0741-8329(87)90092-9. [DOI] [PubMed] [Google Scholar]
- de Fiebre CM, Marks MJ, Collins AC. Ethanol-nicotine interactions in long-sleep and short-sleep-mice. Alcohol. 1990;7:249–257. doi: 10.1016/0741-8329(90)90014-4. [DOI] [PubMed] [Google Scholar]
- de Fiebre CM, Romm E, Collins JT, Draski LJ, Deitrich RA, Collins AC. Responses to cholinergic agonists of rats selectively bred for differential sensitivity to ethanol. Alcohol Clin Exp Res. 1991;15:270–276. doi: 10.1111/j.1530-0277.1991.tb01868.x. [DOI] [PubMed] [Google Scholar]
- de Fiebre NC, Dawson R, Jr, de Fiebre CM. The selectively bred high alcohol sensitivity (HAS) and low alcohol sensitivity (LAS) rats differ in sensitivity to nicotine. Alcohol Clin Exp Res. 2002;26:765–772. [PubMed] [Google Scholar]
- Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- Gabbay FH. Family history of alcoholism and response to amphetamine: sex differences in the effect of risk. Alcohol Clin Exp Res. 2005;29:773–780. doi: 10.1097/01.alc.0000164380.16043.4f. [DOI] [PubMed] [Google Scholar]
- Grant BF, Hasin DS, Chou SP, Stinson FS, Dawson DA. Nicotine dependence and psychiatric disorders in the United States: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry. 2004;61:1107–1115. doi: 10.1001/archpsyc.61.11.1107. [DOI] [PubMed] [Google Scholar]
- National Institute of Health. Bethesda, MD: National Institute of Health; Principles of Laboratory Animal Care. 1985 Publication 85-23.
- Holdstock L, King AC, de Wit H. Subjective and objective responses to ethanol in moderate/heavy and light social drinkers. Alcohol Clin Exp Res. 2000;24:789–794. [PubMed] [Google Scholar]
- Istvan J, Matarazzo JD. Tobacco, alcohol, and caffeine use: a review of their interrelationships. Psychol Bull. 1984;95:301–326. [PubMed] [Google Scholar]
- Jerlhag E, Grotli M, Luthman K, Svensson L, Engel JA. Role of the subunit composition of central nicotinic acetylcholine receptors for the stimulatory and dopamine-enhancing effects of ethanol. Alcohol Alcohol. 2006;41:486–493. doi: 10.1093/alcalc/agl049. [DOI] [PubMed] [Google Scholar]
- Kamens HM, Phillips TJ. A role for neuronal nicotinic acetylcholine receptors in ethanol-induced stimulation, but not cocaine- or methamphetamine-induced stimulation. Psychopharmacology (Berl) 2008;196:377–387. doi: 10.1007/s00213-007-0969-7. [DOI] [PubMed] [Google Scholar]
- Kamens HM, Burkhart-Kasch S, McKinnon CS, Li N, Reed C, Phillips TJ. Ethanol-related traits in mice selectively bred for differential sensitivity to methamphetamine-induced activation. Behav Neurosci. 2006;120:1356–1366. doi: 10.1037/0735-7044.120.6.1356. [DOI] [PubMed] [Google Scholar]
- King AC, Houle T, de Wit H, Holdstock L, Schuster A. Biphasic alcohol response differs in heavy versus light drinkers. Alcohol Clin Exp Res. 2002;26:827–835. [PubMed] [Google Scholar]
- Klink R, de Kerchove d’Exaerde A, Zoli M, Changeux JP. Molecular and physiological diversity of nicotinic acetylcholine receptors in the midbrain dopaminergic nuclei. J Neurosci. 2001;21:1452–1463. doi: 10.1523/JNEUROSCI.21-05-01452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov A, Olale F, Cooper J, Choi C, Lindstrom J. Human alpha6 AChR subtypes: subunit composition, assembly, and pharmacological responses. Neuropharmacology. 2000;39:2570–2590. doi: 10.1016/s0028-3908(00)00144-1. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Larsson A, Svensson L, Soderpalm B, Engel JA. Role of different nicotinic acetylcholine receptors in mediating behavioral and neurochemical effects of ethanol in mice. Alcohol. 2002;28:157–167. doi: 10.1016/s0741-8329(02)00244-6. [DOI] [PubMed] [Google Scholar]
- Larsson A, Jerlhag E, Svensson L, Soderpalm B, Engel JA. Is an alpha-conotoxin MII-sensitive mechanism involved in the neurochemical, stimulatory, and rewarding effects of ethanol? Alcohol. 2004;34:239–250. doi: 10.1016/j.alcohol.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Madden PA, Heath AC. Shared genetic vulnerability in alcohol and cigarette use and dependence. Alcohol Clin Exp Res. 2002;26:1919–1921. doi: 10.1097/01.ALC.0000040960.15151.30. [DOI] [PubMed] [Google Scholar]
- Madden PA, Bucholz KK, Martin NG, Heath AC. Smoking and the genetic contribution to alcohol-dependence risk. Alcohol Res Health. 2000;24:209–214. [PMC free article] [PubMed] [Google Scholar]
- Marks MJ, Whiteaker P, Collins AC. Deletion of the alpha7, beta2, or beta4 nicotinic receptor subunit genes identifies highly expressed subtypes with relatively low affinity for [3H]epibatidine. Mol Pharmacol. 2006;70:947–959. doi: 10.1124/mol.106.025338. [DOI] [PubMed] [Google Scholar]
- Meldgaard M, Fenger C, Lambertsen KL, Pedersen MD, Ladeby R, Finsen B. Validation of two reference genes for mRNA level studies of murine disease models in neurobiology. J Neurosci Methods. 2006;156:101–110. doi: 10.1016/j.jneumeth.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Moser N, Mechawar N, Jones I, Gochberg-Sarver A, Orr-Urtreger A, Plomann M, Salas R, Molles B, Marubio L, Roth U, Maskos U, Winzer-Serhan U, Bourgeois JP, Le Sourd AM, De Biasi M, Schroder H, Lindstrom J, Maelicke A, Changeux JP, Wevers A. Evaluating the suitability of nicotinic acetylcholine receptor antibodies for standard immunodetection procedures. J Neurochem. 2007;102:479–492. doi: 10.1111/j.1471-4159.2007.04498.x. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Is there a common molecular pathway for addiction? Nat Neurosci. 2005;8:1445–1449. doi: 10.1038/nn1578. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Lessov-Schlaggar CN, Ponder CA, McKinnon CS, Phillips TJ. Sensitivity to the locomotor-stimulant effects of ethanol and allopregnanolone: a quantitative trait locus study of common genetic influence. Genes Brain Behav. 2006;5:506–517. doi: 10.1111/j.1601-183X.2005.00198.x. [DOI] [PubMed] [Google Scholar]
- Papke RL, Sanberg PR, Shytle RD. Analysis of mecamylamine stereoisomers on human nicotinic receptor subtypes. J Pharmacol Exp Ther. 2001;297:646–656. [PubMed] [Google Scholar]
- Parker MJ, Beck A, Luetje CW. Neuronal nicotinic receptor beta2 and beta4 subunits confer large differences in agonist binding affinity. Mol Pharmacol. 1998;54:1132–1139. [PubMed] [Google Scholar]
- Perry DC, Xiao Y, Nguyen HN, Musachio JL, Davila-Garcia MI, Kellar KJ. Measuring nicotinic receptors with characteristics of alpha4beta2, alpha3beta2 and alpha3beta4 subtypes in rat tissues by autoradiography. J Neurochem. 2002;82:468–481. doi: 10.1046/j.1471-4159.2002.00951.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK. Complex-trait genetics: emergence of multivariate strategies. Nat Rev Neurosci. 2002;3:478–485. doi: 10.1038/nrn847. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson M, Gwiazdon C, Burkhart-Kasch S, Shen EH. Effects of acute and repeated ethanol exposures on the locomotor activity of BXD recombinant inbred mice. Alcohol Clin Exp Res. 1995;19:269–278. doi: 10.1111/j.1530-0277.1995.tb01502.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson MG, McKinnon CS. Localization of genes mediating acute and sensitized locomotor responses to cocaine in BXD/Ty recombinant inbred mice. J Neurosci. 1998;18:3023–3034. doi: 10.1523/JNEUROSCI.18-08-03023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Hen R, Crabbe JC. Complications associated with genetic background effects in research using knockout mice. Psychopharmacology (Berl) 1999;147:5–7. doi: 10.1007/s002130051128. [DOI] [PubMed] [Google Scholar]
- Rezvani AH, Overstreet DH, Yang Y, Maisonneuve IM, Bandarage UK, Kuehne ME, Glick SD. Attenuation of alcohol consumption by a novel nontoxic ibogaine analogue (18-methoxycoronaridine) in alcohol-preferring rats. Pharmacol Biochem Behav. 1997;58:615–619. doi: 10.1016/s0091-3057(97)10003-x. [DOI] [PubMed] [Google Scholar]
- Rosenthal R. Parametric measures of effect size. In: Cooper H, Hedges LV, editors. The Handbook of Research Synthesis. New York: Russell Sage Foundation; 1994. pp. 231–244. [Google Scholar]
- Salas R, Cook KD, Bassetto L, De Biasi M. The alpha3 and beta4 nicotinic acetylcholine receptor subunits are necessary for nicotine-induced seizures and hypolocomotion in mice. Neuropharmacology. 2004;47:401–407. doi: 10.1016/j.neuropharm.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, Grady SR. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol Pharmacol. 2004;65:1526–1535. doi: 10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–1046. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RD, Kellar KJ. Nicotinic cholinergic receptor binding sites in the brain: regulation in vivo. Science. 1983;220:214–216. doi: 10.1126/science.6828889. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58:182–190. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- Talcott GW, Poston WS, II, Haddock CK. Co-occurrent use of cigarettes, alcohol, and caffeine in a retired military population. Mil Med. 1998;163:133–138. [PubMed] [Google Scholar]
- Talley TT, Olivera BM, Han KH, Christensen SB, Dowell C, Tsigelny I, Ho KY, Taylor P, McIntosh JM. Alpha-conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as alpha3beta2 and alpha7 nicotinic acetylcholine receptors. J Biol Chem. 2006;281:24678–24686. doi: 10.1074/jbc.M602969200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk HE, Scherrer JF, Bucholz KK, Todorov A, Heath AC, Jacob T, True WR. Evidence for specificity of transmission of alcohol and nicotine dependence in an offspring of twins design. Drug Alcohol Depend. 2007;87:225–232. doi: 10.1016/j.drugalcdep.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.42. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RA, Bozarth MA. A psychomotor stimulant theory of addiction. Psychol Rev. 1987;94:469–492. [PubMed] [Google Scholar]
- Xiao Y, Kellar KJ. The comparative pharmacology and up-regulation of rat neuronal nicotinic receptor subtype binding sites stably expressed in transfected mammalian cells. J Pharmacol Exp Ther. 2004;310:98–107. doi: 10.1124/jpet.104.066787. [DOI] [PubMed] [Google Scholar]
- Xu W, Gelber S, Orr-Urtreger A, Armstrong D, Lewis RA, Ou CN, Patrick J, Role L, De Biasi M, Beaudet AL. Megacystis, mydriasis, and ion channel defect in mice lacking the alpha3 neuronal nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A. 1999;96:5746–5751. doi: 10.1073/pnas.96.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Scott MM, Deneris ES. Shared long-range regulatory elements coordinate expression of a gene cluster encoding nicotinic receptor heteromeric subtypes. Mol Cell Biol. 2006;26:5636–5649. doi: 10.1128/MCB.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young EM, Mahler S, Chi H, de Wit H. Mecamylamine and ethanol preference in healthy volunteers. Alcohol Clin Exp Res. 2005;29:58–65. doi: 10.1097/01.alc.0000150007.34702.16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.