Abstract
Background
Dexamethasone contributes to high cure rates in pediatric acute lymphoblastic leukaemia (ALL) but significantly and adversely alters sleep and fatigue. Herein we explored three mechanisms (pharmacokinetics, serum albumin, and pharmacogenetics) through which dexamethasone may cause debilitating fatigue and disrupted sleep.
Methods
We enrolled 100 patients on a 10-day study: 5 days of no dexamethasone (OFF DEX) followed by 5 days of dexamethasone (ON DEX) during continuation chemotherapy. Sleep variables were collected with continuous actigraphy on Days 1 through 5, both OFF DEX and ON DEX. On Days 2 and 5 of each 5-day period, parents and patients 7 years of age and older completed a sleep diary and Fatigue Scale questionnaire. Blood was collected at 0 (pre-dexamethasone), 1, 2, 4, and 8 hours after the first oral dexamethasone dose for pharmacokinetic analysis. Serum albumin concentration was retrospectively analyzed in stored samples. Patient DNA was genotyped for 99 polymorphic loci in candidate genes associated with glucocorticoid metabolism.
Results
Dexamethasone clearance was significantly greater in younger patients than in older ones and in lower risk patients. In multiple regression models, risk group was significantly related to pharmacokinetic parameters. We found that polymorphisms in three genes (AHSG, IL6, POLDIP3) were significantly associated with sleep measures but not fatigue.
Conclusion
Risk group had the most significant relationship with disrupted sleep in patients while on dexamethasone. Serum albumin levels had neither a direct relationship with sleep or fatigue variables nor an indirect relationship through systemic exposure to dexamethasone. We identified candidate genes that may help explain the adverse events of disrupted sleep in pediatric patients receiving dexamethasone.
Keywords: sleep, fatigue, paediatric acute lymphoblastic leukaemia, dexamethasone, pharmacokinetics, pharmacogenomics, single nucleotide polymorphism (SNP)
Introduction
Survival rates for childhood acute lymphoblastic leukaemia (ALL) now exceed 80%, and with current therapy, these rates are expected to rise to nearly 90%.1 These high cure rates can be attributed to multiagent chemotherapy regimens that include a backbone of glucocorticoid treatment throughout remission induction chemotherapy and intermittent dosing of glucocorticoids during maintenance/continuation therapy (∼2 years).1-3 Dexamethasone is the most effective glucocorticoid used to treat ALL, because it has higher systemic potency and activity, direct apoptotic effects, and higher cerebrospinal fluid–to-plasma ratios than does prednisone.4, 5 Despite decades of use, we still do not fully understand the pharmacodynamics of dexamethasone, which shows a wide range of side effect severity due to large intra- and inter-individual variability in systemic exposure.6-8 Notable adverse events of glucocorticoid treatment include severe avascular necrosis, altered behavior (e.g., emotional lability, inattention/hyperactivity, mania, psychosis), significantly disturbed sleep, and increased fatigue.9-11
Sleep is essential for cellular restoration, tissue renewal, wound healing, and immune functioning.12-14 Sleep disturbances in healthy children and those with cancer have been associated with decreased cognitive functioning, increased fatigue affecting daily activities, and lower perceived quality of life.14-16 Children and adolescents receiving glucocorticoid therapy can experience serious and persistent sleep disturbances.9, 10 Sleep disturbances have been reported among ALL survivors with as many as 50% reporting sleep problems more than 10 years after completion of therapy.17 Paediatric protocols, including Children's Oncology Group (COG) 9904 and 9905 and St. Jude Children's Research Hospital (St. Jude) Total XV, prescribe dexamethasone with doses varying from 30 to 60 mg/m2 over 5- to 7-day pulses during maintenance chemotherapy. The immediate and cumulative effects of this prolonged use of dexamethasone in patients with ALL and its resultant effects on sleep and fatigue are questions of research interest.
A recently published study by Hinds and colleagues10 established the correlation between dexamethasone and adverse health behaviours associated with sleep quality and fatigue in 100 children and adolescents treated on COG and St. Jude protocols in continuation therapy for ALL between weeks 50 and 76 at one of three specified points in treatment. This study collected actigraphic data and child fatigue and parent sleep and fatigue data during the 5 days that the child received dexamethasone (ON DEX) and the 5 days that they did not (OFF DEX). Even at baseline OFF DEX, patients had poor sleep quality with significantly poorer average sleep efficiency average of 84% (sleep efficiency = time asleep/total time in bed) than is considered the “acceptable normal” in children and adolescents. These patients also had more nocturnal awakenings, averaging 12 to 16 per night, compared to healthy, age-matched cohort which awakens 1 to 5 times per night.18 Recommended normal sleep time in a child age 3-5 years is 11 to 13 hours daily, school aged child 10 to 11 hours, and an adolescent should have about 8.5 to 9.5 hours.19 Study findings also indicated that nocturnal sleep minutes during ON DEX were lower than the recommended daily hours by 2 to 3 hours for each age cohort and similarly were less during the OFF DEX period for each age cohort by 100 to 140 minutes on average.10 In comparison, during ON DEX, patients had progressively worse sleep, stayed in bed longer with longer periods of wake time after sleep onset (WASO) and had significantly increased daytime fatigue.10, 20 Sex-related differences in sleep were also found in this cohort, i.e., girls took more daytime naps and had less fragmented sleep than did boys, after controlling for age and risk group.21 During ON DEX, the higher total dose of dexamethasone and the ALL risk group adversely affected sleep; however, these factors did not explain the striking changes and extreme inter-patient variability in sleep and fatigue during that period.
The objective of this study was to investigate potential mechanisms through which dexamethasone disrupted sleep and increased fatigue in the previously described patient cohort; those mechanisms included dexamethasone pharmacokinetics (PK), serum albumin levels, and pharmacogenomics. We hypothesized that dexamethasone PK, including the area under the concentration-vs-time curve (AUC), achieving a targeted threshold, clearance (CL) of dexamethasone, or dosing regimen could explain the variability in sleep and fatigue patterns. Yang et al.8 found that albumin is the greatest measurable factor in dexamethasone PK variability in paediatric oncology patients during re-induction. Low serum albumin correlates with and/or predicts severe fatigue in adult patients with cancer at baseline and during treatment. 22-24 Patients with a low serum albumin are four times more likely to be fatigued.23 We looked directly for associations between serum albumin levels and sleep and fatigue measurements and indirectly through PK studies. Lastly, there is emerging evidence that patients at risk for toxicities of dexamethasone during treatment of ALL may be identified through pharmacogenomics.25 Knowledge of the glucocorticoid pathway as an inducer and substrate of cytochrome P4503A (CYP3A)7, 26, provided the rationale for exploration of genes involved in CYP3A, P-glycoprotein,27 and glucocorticoid receptor28 function, that may be predictive of sleep quality. Therefore, we explored the association of candidate gene polymorphisms with sleep and fatigue variables OFF DEX and ON DEX.
Methods
Participants
One hundred outpatient paediatric patients with low- or standard-risk ALL were enrolled at three different institutions (St. Jude in Memphis, TN, Texas Children's Cancer Center in Houston, TX, and The Hospital for Sick Children in Toronto, Ontario) under three treatment protocols (St. Jude Total XV, COG 9904, or COG 9905). The cohort was divided into four subgroups: St. Jude low-risk, St. Jude standard-risk, COG low-risk, and COG standard-risk. Low risk patients for St. Jude and COG included those patients with initial white blood count of less than 50×109, ages 1-9.9 years, no CNS3 disease, no overt testicular disease, positive for TEL-AML1 fusion, and no adverse genetic features. St. Jude low risk criteria also included patients with DNA index ≥1.16 and excluded poor early responders. COG low risk also included criteria for the presence of trisomies of chromosomes 4 and 10. St. Jude Standard Risk category included all T-cell or B-cell ALL that did not meet criteria for low-risk, high-risk, or very-high-risk ALL while the COG standard was the same but only for B-lineage ALLs. Institutional review board approval was obtained at each of the three study sites.
Study Design
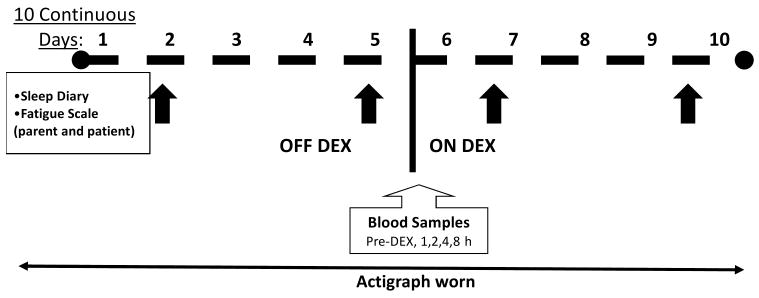
Hinds and colleagues10 enrolled 100 patients for the continuous 10-day study that included 5 days OFF DEX followed by 5 days ON DEX. Each patient wore a Micro-mini (Ambulatory Monitoring Inc., Ardsley, NY) wrist actigraph 24 hours per day, which recorded sleep characteristics including nocturnal awakenings, sleep duration (time spent in bed at night), sleep efficiency, WASO, actual sleep minutes (time sleeping at night), total daily sleep minutes, and total daily nap minutes. On Days 2 and 5 of each period, all parents and patients at least 7 years of age completed fatigue instruments: Fatigue Scale-Child, Fatigue Scale-Adolescent, or Fatigue Scale-Parent.29, 30 The sleep diary completed by parents is a brief questionnaire about the parent's perceptions of their child's sleep and nap patterns during the previous 24 hours. Figure 1 illustrates the time points of data and blood collection.
Figure 1.
Illustration of study regimen. Sleep was monitored by a wrist actigraph continuously for 10 days. Sleep diaries and fatigue scales (black arrows) were completed on Days 2 and 5 of each period. Blood samples were collected on Day 1 of the ON DEX period.
Dexamethasone Pharmacokinetics
Blood (2 mL) was collected at 0 (pre-dexamethasone), 1, 2, 4, and 8 hours after the first oral dose of dexamethasone on Day 1 of ON DEX and analyzed as previously described by Yang and colleagues8 Plasma was stored at −80 °C until processing. Dexamethasone and triamcinolone acetonide (10 μL) were extracted from plasma by solid-phase reversed-phase C18 columns. The supernatant was placed into a high-performance liquid chromatography system with a diode array detector (Shimadzu, Columbia, MD) and a 150?2.0 mm Phenomenex Luna C18 (2) column (Phenomenex, Torrance, CA). The assay was linear from 10 to 200 nmol/L with recovery greater than 95%. The inter- and intra-day coefficients of variation were less than 10%.
The PK parameters were estimated via ADAPT II (Biomedical Simulations Resource, Los Angeles, CA)31 by fitting a one-compartment PK model to the plasma concentration data, as previously described.8 The PK parameters estimated included: Apparent volume (V/F), the first-order absorption rate constant (ka), the elimination rate constant (ke). In addition, apparent clearance (CL/F; CL/F=ke * V/F), and half-life (t1/2; t1/2=0.693/ke) were determined. The AUC was estimated with the model-fitted curve for each patient from 0 to 8 hours.
Serum Albumin Concentration
To accurately measure serum albumin levels per the St. Jude Department of Pathology's standard testing methods, the sample volume must be more than 0.1 mL. Of the 100 patients originally enrolled, 74 had an adequate serum sample obtained before dexamethasone was given (pre-dex) on Day 1 of the ON DEX week. The normal range of serum albumin is 3.7 to 5.4 g/dL. Because albumin has a t1/2 of 21 days, minor daily changes in albumin concentration during the study period were not a concern.
Genotyping
Germline DNA was extracted from peripheral white blood cells. Genotyping was completed with greater than 90% call rates for 99 polymorphic loci in candidate genes associated with glucocorticoid metabolism, including genes involved in CYP3A, P-glycoprotein, and glucocorticoid receptor function. Genotyping for the following 16 polymorphic loci was performed as described by Owawa et al 30: CYP3A4*1B (A>G at position 392) and CYP3A5*3 (G>A at position 22893), GSTP1 313A>G, GSTM1 deletion, GSTT1 deletion, MDR1 exon 21 (2677G>T/A), MDR1 exon 26 (3435C>T), MTHFR 677C>T, MTHFR 1298A>C, NR3C1 1088A>G, RFC (SLC19A1) 80A>G, TPMT (238G>C, 460G>A, 719A>G), TYMS enhancer repeat, UGT1A1 promoter repeat, VDR intron 8 G>A, and VDR FokI (start site) T>C. The remaining loci were genotyped by DNAPrint Genomics (Sarasota, FL).
Statistical Analyses
Sleep quality and fatigue data analyses have been previously described.10 In 12 patients, sleep data were not evaluable due to actigraph failure or insufficient recordings; thus, those data were excluded from the sleep variable analysis (n=88). Fatigue data were analyzed by calculating the summed score for each fatigue questionnaire at each time point. Patient characteristic variables that were considered included risk group (St. Jude low- and standard-risk and COG low- and standard-risk), sex, race (white vs other), and age as a continuous variable.
Linear regression models were used to describe the relation between PK and patient characteristic variables. To assess the relationship between PK and sleep or fatigue, we used the average of each sleep variable and fatigue score during the ON DEX week. A univariate linear regression model was used first and then multiple regression models were applied to adjust for age and risk group.
Pearson correlation coefficient statistics were done to evaluate the relations between serum albumin concentration and sleep, fatigue, or PK variables in the 74 patients with pre-dex albumin levels and sleep data. The correlations were further adjusted for patient characteristics by using a multiple linear regression model.
Because race was a significant confounding factor for the association between genotypes of most single nucleotide polymorphisms (SNPs) and sleep measures, only data from the white patients (n=72) were included in the genotype analysis. A one-factor linear regression model was applied to determine whether patient characteristics were significantly associated with each sleep measurement. Multiple linear regression models were then applied to determine whether genotypes of a SNP had independent significant association with a sleep measurement by including the significant patient characteristics and one SNP as explanatory variables in the models. If no patient characteristic was associated with the sleep measurement, then only the SNP was taken as the explanatory variable in the model. For those SNPs that were significantly associated with a sleep measurement, genotypes were pooled into binary groups for statistical analysis to test for dominant or recessive genetic effect. The analyses were implemented for sleep measurements of OFF DEX and ON DEX, and those sleep measures that showed a significant difference between the two 5-day periods. Similar analyses were carried out for fatigue scores.
Longitudinal methods incorporating the correlation among each day's sleep/fatigue variables were also applied, and the same results were obtained as OFF DEX and ON DEX periods (data not shown). The criterion for significance for all analyses was a p-value at the level of α=0.05. All analyses were performed with SAS, Release 9.1.3 (SAS Institute, Cary, NC).
Results
The mean age of this cohort was 9.24 ± 3.23 years (range, 5.03-18.14 years). All patients were past Week 50 and prior to Week 72 of the continuation chemotherapy phase of their treatment for ALL. The majority of the patients were white (79%) and male (62%) with standard-risk ALL (63%).
Pharmacokinetics of dexamethasone and Sleep and Fatigue Measures
The estimates of dexamethasone PK parameters in the 100 enrolled patients were as follows: the mean CL/F was 20.9 L/h per m2; mean V/F, 48.5 L/m2; mean ka, 2.03 h-1; mean ke, 0.54 h-1; and the t1/2 was 1.27 h. We noted substantial (54%) inter-patient variability of clearance across the entire cohort (Table 1); that of the COG low- and standard-risk groups was 56% (3 mg/m2 per dose); St. Jude low-risk group, 51% (2.67 mg/m2 per dose); and St. Jude standard-risk group, 36% (4 mg/m2 per dose). The AUC increased with age (p=0.02) and was significantly higher in patients in the standard-risk group than in those in the low-risk group (p <0.001). Clearance was greater in patients with low-risk ALL (p=0.02) and in patients younger than 10 years (p=0.0014).
Table 1.
Dexamethasone pharmacokinetics in 100 paediatric patients with ALL
| Variable | Minimum | Median | Maximum | Mean | SD | CV |
|---|---|---|---|---|---|---|
| AUC1 | 96.27 | 369.36 | 1119.84 | 404.68 | 186.54 | 0.461 |
| Cumulative AUC | 1347 | 5779 | 25,887 | 6951 | 4227 | 0.608 |
| Elimination rate constant (Ke) | 0.065 | 0.388 | 2.069 | 0.544 | 0.42 | 0.767 |
| Apparent volume | 11.93 | 43.97 | 140.97 | 48.47 | 24.50 | 0.505 |
| Absorption rate constant (Ka) | 0.16 | 2.15 | 4.75 | 2.03 | 1.25 | 0.618 |
| Clearance | 3.96 | 18.82 | 75.72 | 20.86 | 11.26 | 0.540 |
Area under the concentration vs time curve (AUC) was calculated during the first 8 hours of exposure to dexamethasone.
As the dexamethasone AUC increased, sleep efficiency and actual sleep time decreased in the univariate models (p=0.016 and p=0.033, respectively) (Table 2). WASO increased as AUC increased (p=0.048). However, after controlling for risk group and age, there was no statistical evidence that AUC could significantly explain the variations in sleep measurements. Attaining a threshold of 100 nM dexamethasone was not related to the fatigue variables; however, for the sleep variables there was a trend of increasing sleep efficiency (p=0.08) and a significance in actual sleep time (p=0.05) as the cumulative time above the threshold decreased. Clearance was not directly associated with sleep variables in univariate analysis and in the multivariable model, risk group was significant (p=0.026).
Table 2.
Relations between dexamethasone pharmacokinetics and sleep measures in 88 paediatric patients with ALL
| Dexamethasone pharmacokinetic parameters | ||||||
|---|---|---|---|---|---|---|
| AUC | Threshold | Clearance | ||||
| Sleep Variable | r | p-value | r | p-value | r | p-value |
| Sleep efficiency | −0.26 | 0.014 | −0.19 | 0.08 | 0.16 | 0.14 |
| Actual sleep time | −0.24 | 0.027 | −0.22 | 0.05 | 0.075 | 0.51 |
| Sleep duration | −0.18 | 0.09 | −0.18 | 0.12 | 0.029 | 0.82 |
| Nocturnal awakenings | 0.11 | 0.32 | 0.14 | 0.20 | −0.051 | 0.66 |
| WASO | 0.21 | 0.048 | 0.14 | 0.20 | 0.15 | 0.14 |
Abbreviation: r, Pearson correlation coefficient; WASO, wake after sleep onset
When comparing sleep variables based on dexamethasone dosing, we found that the mean sleep efficiency of patients in the St. Jude low-risk group (8 mg/m2 per day) was 84%; that of the St. Jude standard-risk group (12 mg/m2 per day) was 78%; and that of patients in the combined COG low- and standard-risk (6 mg/m2 per day) was 87% (p=0.0015). We also compared outcome variables of 22 patients on St. Jude low-risk study to those of the 39 patients on the COG low- and standard-risk studies to control for single-dose effects. The St. Jude low-risk group (2.67 mg/m2 per dose, 3 times per day [TID]) had a higher average number of nocturnal awakenings (16.2 vs 12.6; p=0.021) compared to the COG low- and standard-risk group (3 mg/m2 per dose, twice per day [BID]) The St. Jude standard-risk group (4.0 mg/m2 per dose, 3 times per day [TID]) had a higher WASO (110.8 vs 68.8), lower sleep efficiency (77.61% vs 87.47%) and lower actual sleep time than (409.5 vs 510.7) when compared to the COG low- and standard risk group (all p<0.01, Table 3). There were no PK associations with fatigue when controlling for age and sex.
Table 3.
The effects of dexamethasone dosage on the 88 patients with actigraph sleep measures
| Study Group (n) | Daily dose (mg/m2) | Dosage schedule | WASO (min) | Mean Sleep Measures (SE) | Nocturnal awakenings (No.) | ||
|---|---|---|---|---|---|---|---|
| Sleep Efficiency (%) | Actual sleep time (min) | Sleep duration (min) | |||||
| COG low- and standard-risk (39) | 6 | BID | 68.84 (49.30) | 87.47 (9.63) | 510.67 (126.71) | 596.30 (102.58) | 12.63 (6.20) |
| St. Jude low-risk (22) | 8 | TID | 88.97 (35.31) | 84.13 (6.83) | 477.72 (88.72) | 589.87 (64.06) | 16.23 (6.20)* |
| St. Jude standard-risk (27) | 12 | TID | 110.84 (59.46)* | 77.61 (13.82)* | 409.55 (141.60)* | 551.80 (100.68) | 15.43 (7.05) |
Abbreviations: BID, twice per day; COG, Children's Oncology Group; No, number; TID, three times per day; WASO, wake after sleep onset;
P-value <0.05 for comparing with COG low- and standard- risk.-
Effects of Albumin on Pharmacokinetic and Sleep and Fatigue Measures
The median serum albumin concentration in the 74 samples was 4.3 g/dL (range, 3-5.5 g/dL). Only 5 of 74 (6.8%) samples were below the St. Jude laboratory's low normal of 3.7 g/dL. Albumin concentration was negatively associated with the dexamethasone AUC (ρ=−0.027); however, this relation was not significant (p=0.82). Also, we found no statistically significant relations between the serum albumin values and sleep or fatigue data in these 74 patients.
Genotype Analysis
Our comparison of genotypes with sleep and fatigue measures revealed four genotype markers corresponding with three genes, α2-Heremans-Schmid glycoprotein (AHSG), interleukin-6 (IL6), and polymerase delta-interacting protein 3 (POLDIP3), that were significantly associated with sleep measures (Table 4). The AHSG C>G (Thr238Ser) exon 7 genotype (rs4918) was found to effect sleep both OFF and ON DEX, with significance found between the OFF DEX and ON DEX weeks and was a reliable predictor of WASO and sleep duration. During OFF DEX, patients with the CC genotype were found to have a shorter WASO (p=0.024) and a suggestive trend of increased sleep efficiency (p=0.054). Those with the GG and CG genotype had lower sleep efficiency and higher WASO than the C C genotype. During ON DEX, patients with the GG genotype experienced a longer sleep duration or time in bed (p=0.04) and had longer actual sleep minutes (p=0.032) than those with the CC and CG genotype. Between OFF DEX and ON DEX weeks, we also observed significant differences in sleep duration (p=0.023) and actual sleep time (p=0.005) for patients with the GG genotype.
Table 4.
Genotypes associated with sleep measures in 72 paediatric patients with ALL
| Genotype | Study week | Sleep measure | Genotype grouping | Mean (SD) | P-value |
|---|---|---|---|---|---|
| AHSG C>G (T238S) exon 7 (rs4918) | OFF DEX | ↑ Sleep efficiency | CC vs CG + GG | 87.0 (9.3) vs 81.8 (12.1) | 0.054 |
| ↓ WASO | CC vs CG + GG | 65.4 (44.0) vs 93.4 (54.7) | 0.024 | ||
| ON DEX | ↑ Sleep duration | GG vs CC + CG | 651.4 (151) vs 590.3 (68.1) | 0.040 | |
| ↑ Actual sleep time | GG vs CC + CG | 566.8 (171.5) vs 476.4 (106.9) | 0.032 | ||
| OFF/ON DIF | ↑ Sleep duration | GG vs CC+CG | 110.5 (123.8) vs 44.5 (72.2) | 0.023 | |
| ↑ Actual sleep time | GG vs CC+CG | 116.3 (129.9) vs 33.5 (70.5) | 0.005 | ||
| IL-6 (G174C) promoter (rs 1800795) | OFF DEX | ↑ Actual sleep time | GG vs CG + CC | 465.5 (93.1) vs 430.1 (89.1) | 0.026 |
| ↑ Total daily sleep | GG vs CG + CC | 583.8 (79.8) vs 569.7 (58.7) | 0.034 | ||
| IL-6 (C634G) (rs13447445) | OFF DEX | ↑ Actual sleep time | CG vs CC | 485.4 (90.1) vs 435.6 (90.4) | 0.040 |
| ↑ Total daily sleep | CG vs CC | 604.9 (56.0) vs 569.2 (68.7) | 0.021 | ||
| POLDIP3 (rs1771889) | OFF DEX | ↑ Actual sleep time | AA + AG vs GG | 453.2 (82.5) vs 392.4 (124.0) | 0.026 |
| ↑ Sleep efficiency | AA + AG vs GG | 85.2 (9.5) vs 76.7 (17.0) | 0.019 | ||
| ↓ WASO | AA vs AG + GG | 68.9 (56.1) vs 89.4 (48.5) | 0.035 | ||
| ↓ Nocturnal awakenings | AA vs AG + GG | 13.2 (7.9) vs 17.1 (6.7) | 0.004 |
The IL-6 G174C (promoter region) genotype was found to be associated with sleep during OFF DEX, with patients carrying the GG genotype experiencing longer actual sleep time (p=0.026) and increased daily sleep minutes (p=0.034) when compared to those with the CG and CC genotypes. Another IL-6 genotype, IL-6 C634G, was significant during OFF DEX with a longer actual sleep time (p=0.04) and increased daily sleep minutes (p=0.02) found in those with the CG genotype in comparison to the CC genotype. However, there was no significant association between genotype and the difference in sleep measures between the two treatment weeks.
During OFF DEX, the POLDIP3 genotypes AA and AG were significantly associated with higher actual sleep time (p=0.026) and higher sleep efficiency (p=0.019). Additionally, those with the AA genotype had fewer nocturnal awakenings (0.004) and shorter WASO (p=0.035) than those carrying the AG and GG genotype.
Discussion
The mechanisms underlying the negative effect and high variability of systemic behavioural side effects of glucocorticoid therapy are not completely understood. This study is the first to explore possible PK, serum albumin, and pharmacogenomic mechanisms that contribute to poor sleep and increased fatigue in children with ALL receiving dexamethasone. As one would expect, dexamethasone clearance was higher in younger children and decreased with age. This lower clearance and higher dosing of steroids in the higher risk groups resulted in a higher exposure (AUC) in older children and adolescents and in those in the higher-risk groups.
The mean apparent oral clearance of dexamethasone was slightly higher in our study (20.9 L/h/m2) than in the only previously published dexamethasone PK study of children with ALL by Yang and colleagues8 In that study, the 214 patients were in reinduction therapy and received more intensive therapy, including recent administration of asparaginase. Clearance in that patient group was 14 L/h per m2 and most likely reflective of poorer organ function due to more intensive treatment during remission induction. In that earlier study, the variability of dexamethasone PK parameters (46% inter-patient, 53% intra-patient) was comparable to that in our results.
In our study, older participants were usually in the standard-risk group, and they received higher doses of dexamethasone; thus, it was difficult to distinguish between the effects of dose, disease risk, and age. In an exploratory analysis, we attempted to control for dosing schedules (BID vs TID) and observed that children on TID dosing had more awakenings and a longer duration of WASO. It is difficult to determine whether the difference in dose or the timing of the third dose closer to bedtime explains the difference in sleep outcomes. The actual timing of the last dose was not recorded in this study. We theorize that the later the dose is given in the evening, the more likely the peak of dexamethasone will interfere with sleep. The current data cannot test if dose frequency (BID vs TID) has an independent effect on sleep, while adjusting for the total dose, because of multicolinearity. Future studies should explore the effect of timing of dexamethasone dosing on disturbed sleep and fatigue.
We retrospectively measured albumin levels in stored serum from time 0 samples. In the Yang study8, albumin concentration was the largest covariate contributing to dexamethasone clearance. In our patient population, most albumin levels were in the normal range, and PK analysis did not find a significant correlation with albumin. Children with lower albumin levels appeared to experience greater fatigue, but our small patient group and normal albumin levels prevented us from determining whether a true relationship exists. In patients with a normal albumin level, other factors (i.e., age, risk group, etc.) most likely account for the large inter-patient variability of systemic dexamethasone side effects.
Using candidate genes, we explored 99 gene loci and their association with sleep and fatigue. There were no genotypes associated with fatigue; however, four genotypes on three genes to be associated with sleep measures. The AHSG C>G (Thr238Ser) exon 7 genotype (rs4918) showed a significant association with sleep measures both OFF and ON DEX with a significant difference in sleep duration and actual sleep time for those with the GG genotype. While the effect of the described SNP on gene expression within this patient population is not known, a previous genomic analysis identified two common SNPs associated with circulating AHSG levels, one being the C/G in exon 7 (rs4918), as described in this study.32
ASHG is a major hepatic protein that regulates inflammation and recovery and calcium levels. ASHG also inhibits insulin receptor tyrosine kinase activity, thereby regulating insulin signaling and energy homeostasis.33 ASHG has been identified as a susceptibility locus for type 2 diabetes and the metabolic syndrome.34-36 A study by Siddiq and colleaugues37 found the AHSG SNP rs1071592 to be associated with type 2 diabetes and the SNPs rs2248690 and rs4918 to have borderline association. Along with insulin resistance, high AHSG plasma levels have been implicated in the accumulation of fat in the human liver.38 Wöltje and colleagues39 found that in the presence of dexamethasone, the expression of AHSG was upregulated indirectly via glucocorticoid hormone–induced transcriptional upregulation in the CCAAT enhancer binding protein β (C/EBP-β) in mouse hepatocytes and mouse hepatoma cells. This murine model may explain a possible upregulation of expression of AHSG in the presence of dexamethasone, which is further potentiated in the presence of the GG genotype.
We found the wild-type (normal) AHSG CC genotype to be significantly associated with better sleep during the OFF DEX week, providing sleep protection with higher sleep efficiency and shorter WASO. During the ON DEX period, the homozygous variant AHSG GG genotype was associated with longer actual sleep time and longer sleep duration. Patients with the AHSG GG genotype experienced significantly longer sleep duration during ON DEX than during OFF DEX. These sleep parameters, longer actual sleep time and longer sleep duration have been associated with fatigue during dexamethasone use in treatment of paediatric ALL.10 The rs4918 polymorphism interaction with dexamethasone may further increase the expression of AHSG, which may result in longer sleep duration in compensation for the negative effect of fatigue.
Cytokines, most specifically IL-6, have been implicated in the inflammatory process and contribute to cancer-related symptomatology.40, 41 IL-6 is a known mediator of sleepiness and increased levels may increase one's drive for sleep.42 IL-6 (174) wild type G allele has been associated with higher production of the cytokine.43, 44 Our analysis found that OFF DEX, patients with the IL-6 (174) GG genotype experienced longer actual sleep time and longer total daily sleep minutes. These results suggest that IL-6 (174) GG contributes to an increased need for sleep. However, ON DEX there was no significant difference in sleep measures and IL-6 genotype. The IL-6 (C634G) CG genotype was also associated with longer actual sleep time and longer total daily sleep minutes during the OFF DEX week than those with the CC genotype. This specific IL-6 genotype has not yet been associated with sleep as the IL-6(174). The association of IL-6 and sleep disappeared during the ON-DEX week, suggesting that the anti-inflammatory actions of steroids may influence IL-6 interactions with sleep.
Lastly, the POLDIP3 AA and AG genotypes were protective with significantly longer actual sleep time and better sleep efficiency during the OFF DEX week. The POLDIP3 AA genotype also was associated with fewer nocturnal awakenings and decreased WASO during the OFF DEX week. Although the POLDIP3 gene has not been previously associated with sleep, it plays a role in autoimmune disorders.45 This is an exciting new finding as the POLDIP3 A allele was significantly associated four sleep variables, but further exploration of this gene and its role in sleep is necessary.
This study has several limitations. First, the original study was powered to elucidate the effect of dexamethasone on sleep and fatigue variables; the additional analysis presented here is lacking in power, most specifically for discerning the effects of the associated genotypes. Second, the analysis of serum albumin concentration was conducted retrospectively; thus, adequate samples were not available from all 100 participants.
In conclusion, risk group had the most significant influence on sleep variables; however, it is impossible to distinguish the true impact of PK factors with risk group being highly correlated to dexamethasone dose and age, influencing both AUC and clearance. We did not identify a direct relationship of fatigue to dexamethasone pharmacokinetics or serum albumin. However, we identified candidate genes that may help explain the adverse events of disrupted sleep and increased fatigue in patients on dexamethasone. Most specifically, the regulatory effects of AHSG expression on insulin metabolism and energy homeostasis may play a role in the increased fatigue and poorer sleep quality that was noted by Hinds and colleagues10 in patients prior to dexamethasone. Dexamethasone may then potentiate AHSG expression and contribute to poor sleep quality, prolonged sleep as a compensatory mechanism to increased fatigue during dexamethasone treatment and account for the significant differences in sleep quality in these patients between the OFF DEX and ON DEX weeks. Further genetic exploration is needed to validate the association between AHSG expression and sleep in paediatric patients with ALL.
Acknowledgments
We sincerely appreciate the review by Angela McArthur, PhD, ELS, the mentorships of Dr. Ching-Hon Pui, Dr. Mary Relling, Dr. Deo Kumar Srivastava, and preliminary analysis by James Okuma. We would like to acknowledge Kathy McCarthy, BSN and Heather Jones, MSN who contributed to data collection and other aspects of the original study. This study was supported in part by Cancer Center Core Grant CA 21765 from the National Institutes of Health, RO1NR007610 from the National Institute of Nursing Research, and the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Financial disclosure: No author has potential conflict of interest or financial disclosures for this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006 Jan 12;354(2):166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 2.Gaynon PS, Trigg ME, Heerema NA, et al. Children's Cancer Group trials in childhood acute lymphoblastic leukemia: 1983-1995. Leukemia. 2000 Dec;14(12):2223–2233. doi: 10.1038/sj.leu.2401939. [DOI] [PubMed] [Google Scholar]
- 3.Veerman AJ, Hahlen K, Kamps WA, et al. High cure rate with a moderately intensive treatment regimen in non-high-risk childhood acute lymphoblastic leukemia. Results of protocol ALL VI from the Dutch Childhood Leukemia Study Group. J Clin Oncol. 1996 Mar;14(3):911–918. doi: 10.1200/JCO.1996.14.3.911. [DOI] [PubMed] [Google Scholar]
- 4.Balis FM, Lester CM, Chrousos GP, Heideman RL, Poplack DG. Differences in cerebrospinal fluid penetration of corticosteroids: possible relationship to the prevention of meningeal leukemia. J Clin Oncol. 1987 Feb;5(2):202–207. doi: 10.1200/JCO.1987.5.2.202. [DOI] [PubMed] [Google Scholar]
- 5.Jones B, Freeman AI, Shuster JJ, et al. Lower incidence of meningeal leukemia when prednisone is replaced by dexamethasone in the treatment of acute lymphocytic leukemia. Med Pediatr Oncol. 1991;19(4):269–275. doi: 10.1002/mpo.2950190411. [DOI] [PubMed] [Google Scholar]
- 6.Czock D, Keller F, Rasche FM, Haussler U. Pharmacokinetics and pharmacodynamics of systemically administered glucocorticoids. Clin Pharmacokinet. 2005;44(1):61–98. doi: 10.2165/00003088-200544010-00003. [DOI] [PubMed] [Google Scholar]
- 7.Richter O, Ern B, Reinhardt D, Becker B. Pharmacokinetics of dexamethasone in children. Pediatr Pharmacol (New York) 1983;3(3-4):329–337. [PubMed] [Google Scholar]
- 8.Yang L, Panetta JC, Cai X, et al. Asparaginase may influence dexamethasone pharmacokinetics in acute lymphoblastic leukemia. J Clin Oncol. 2008 Apr 20;26(12):1932–1939. doi: 10.1200/JCO.2007.13.8404. [DOI] [PubMed] [Google Scholar]
- 9.Drigan R, Spirito A, Gelber RD. Behavioral effects of corticosteroids in children with acute lymphoblastic leukemia. Med Pediatr Oncol. 1992;20(1):13–21. doi: 10.1002/mpo.2950200104. [DOI] [PubMed] [Google Scholar]
- 10.Hinds PS, Hockenberry MJ, Gattuso JS, et al. Dexamethasone alters sleep and fatigue in pediatric patients with acute lymphoblastic leukemia. Cancer. 2007 Nov 15;110(10):2321–2330. doi: 10.1002/cncr.23039. [DOI] [PubMed] [Google Scholar]
- 11.Stuart FA, Segal TY, Keady S. Adverse psychological effects of corticosteroids in children and adolescents. Arch Dis Child. 2005 May;90(5):500–506. doi: 10.1136/adc.2003.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adam K, Oswald I. Protein synthesis, bodily renewal and the sleep-wake cycle. Clin Sci (Lond) 1983 Dec;65(6):561–567. doi: 10.1042/cs0650561. [DOI] [PubMed] [Google Scholar]
- 13.Akerstedt T, Palmblad J, de la TB, Marana R, Gillberg M. Adrenocortical and gonadal steroids during sleep deprivation. Sleep. 1980;3(1):23–30. doi: 10.1093/sleep/3.1.23. [DOI] [PubMed] [Google Scholar]
- 14.Mindell JA, Owens JA. A Clinical Guide to Pediatric Sleep: Diagnosis and Management of Sleep Problems. Philadelphia, PA: Lippincott, Williams & Wilkins Publishers; 2003. [Google Scholar]
- 15.Hart CN, Palermo TM, Rosen CL. Health-related quality of life among children presenting to a pediatric sleep disorders clinic. Behav Sleep Med. 2005;3(1):4–17. doi: 10.1207/s15402010bsm0301_3. [DOI] [PubMed] [Google Scholar]
- 16.Sadeh A, Gruber R, Raviv A. Sleep, neurobehavioral functioning, and behavior problems in school-age children. Child Dev. 2002 Mar;73(2):405–417. doi: 10.1111/1467-8624.00414. [DOI] [PubMed] [Google Scholar]
- 17.Meeske KA, Siegel SE, Globe DR, Mack WJ, Bernstein L. Prevalence and correlates of fatigue in long-term survivors of childhood leukemia. J Clin Oncol. 2005 Aug 20;23(24):5501–5510. doi: 10.1200/JCO.2005.03.210. [DOI] [PubMed] [Google Scholar]
- 18.Mindell JA, Owens JA, Carskadon MA. Developmental features of sleep. Child Adolesc Psychiatr Clin N Am. 1999 Oct;8(4):695–725. [PubMed] [Google Scholar]
- 19.Meltzer LJ, Mindell JA. Sleep and sleep disorders in children and adolescents. Psychiatr Clin North Am. 2006 Dec;29(4):1059–1076. doi: 10.1016/j.psc.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Sadeh A, Hauri PJ, Kripke DF, Lavie P. The role of actigraphy in the evaluation of sleep disorders. Sleep. 1995 May;18(4):288–302. doi: 10.1093/sleep/18.4.288. [DOI] [PubMed] [Google Scholar]
- 21.Sanford SD, Okuma JO, Pan J, et al. Gender differences in sleep, fatigue, and daytime activity in a pediatric oncology sample receiving dexamethasone. J Pediatr Psychol. 2008 Apr;33(3):298–306. doi: 10.1093/jpepsy/jsm110. [DOI] [PubMed] [Google Scholar]
- 22.Shafqat A, Einhorn LH, Hanna N, et al. Screening studies for fatigue and laboratory correlates in cancer patients undergoing treatment. Ann Oncol. 2005 Sep;16(9):1545–1550. doi: 10.1093/annonc/mdi267. [DOI] [PubMed] [Google Scholar]
- 23.Wang XS, Giralt SA, Mendoza TR, et al. Clinical factors associated with cancer-related fatigue in patients being treated for leukemia and non-Hodgkin's lymphoma. J Clin Oncol. 2002 Mar 1;20(5):1319–1328. doi: 10.1200/JCO.2002.20.5.1319. [DOI] [PubMed] [Google Scholar]
- 24.Wratten C, Kilmurray J, Nash S, et al. Fatigue during breast radiotherapy and its relationship to biological factors. Int J Radiat Oncol Biol Phys. 2004 May 1;59(1):160–167. doi: 10.1016/j.ijrobp.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Relling MV, Yang W, Das S, et al. Pharmacogenetic risk factors for osteonecrosis of the hip among children with leukemia. J Clin Oncol. 2004 Oct 1;22(19):3930–3936. doi: 10.1200/JCO.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999 Oct 15;286(5439):487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 27.Ueda K, Yoshida A, Amachi T. Recent progress in P-glycoprotein research. Anticancer Drug Des. 1999 Apr;14(2):115–121. [PubMed] [Google Scholar]
- 28.Huizenga NA, Koper JW, De Lange P, et al. A polymorphism in the glucocorticoid receptor gene may be associated with and increased sensitivity to glucocorticoids in vivo. J Clin Endocrinol Metab. 1998 Jan;83(1):144–151. doi: 10.1210/jcem.83.1.4490. [DOI] [PubMed] [Google Scholar]
- 29.Hinds PS, Hockenberry-Eaton M, Gilger E, et al. Comparing patient, parent, and staff descriptions of fatigue in pediatric oncology patients. Cancer Nurs. 1999 Aug;22(4):277–288. doi: 10.1097/00002820-199908000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Hinds PS, Quargnenti A, Bush AJ, et al. An evaluation of the impact of a self-care coping intervention on psychological and clinical outcomes in adolescents with newly diagnosed cancer. Eur J Oncol Nurs. 2000 Mar;4(1):6–17. doi: 10.1054/ejon.1999.0051. [DOI] [PubMed] [Google Scholar]
- 31.D'Argenio D, Schumitzky A. ADAPT II user's guide. Los Angeles: Biomedical Simulations Resource, USC; 2008. [Google Scholar]
- 32.Osawa M, Umetsu K, Ohki T, Nagasawa T, Suzuki T, Takeichi S. Molecular evidence for human alpha 2-HS glycoprotein (AHSG) polymorphism. Hum Genet. 1997 Jan;99(1):18–21. doi: 10.1007/s004390050302. [DOI] [PubMed] [Google Scholar]
- 33.Inoue M, Takata H, Ikeda Y, et al. A promoter polymorphism of the alpha2-HS glycoprotein gene is associated with its transcriptional activity. Diabetes Res Clin Pract. 2008 Jan;79(1):164–170. doi: 10.1016/j.diabres.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Francke S, Manraj M, Lacquemant C, et al. A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet. 2001 Nov 15;10(24):2751–2765. doi: 10.1093/hmg/10.24.2751. [DOI] [PubMed] [Google Scholar]
- 35.Kissebah AH, Sonnenberg GE, Myklebust J, et al. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vionnet N, Hani EH, Dupont S, et al. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am J Hum Genet. 2000 Dec;67(6):1470–1480. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiq A, Lepretre F, Hercberg S, Froguel P, Gibson F. A synonymous coding polymorphism in the alpha2-Heremans-schmid glycoprotein gene is associated with type 2 diabetes in French Caucasians. Diabetes. 2005 Aug;54(8):2477–2481. doi: 10.2337/diabetes.54.8.2477. [DOI] [PubMed] [Google Scholar]
- 38.Stefan N, Hennige AM, Staiger H, et al. Alpha2-Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care. 2006 Apr;29(4):853–857. doi: 10.2337/diacare.29.04.06.dc05-1938. [DOI] [PubMed] [Google Scholar]
- 39.Woltje M, Tschoke B, von B V, et al. CCAAT enhancer binding protein beta and hepatocyte nuclear factor 3beta are necessary and sufficient to mediate dexamethasone-induced up-regulation of alpha2HS-glycoprotein/fetuin-A gene expression. J Mol Endocrinol. 2006 Apr;36(2):261–277. doi: 10.1677/jme.1.02001. [DOI] [PubMed] [Google Scholar]
- 40.Lee BN, Dantzer R, Langley KE, et al. A cytokine-based neuroimmunologic mechanism of cancer-related symptoms. Neuroimmunomodulation. 2004;11(5):279–292. doi: 10.1159/000079408. [DOI] [PubMed] [Google Scholar]
- 41.Wood LJ, Nail LM, Gilster A, Winters KA, Elsea CR. Cancer chemotherapy-related symptoms: evidence to suggest a role for proinflammatory cytokines. Oncol Nurs Forum. 2006 May;33(3):535–542. doi: 10.1188/06.ONF.535-542. [DOI] [PubMed] [Google Scholar]
- 42.Vgontzas AN, Bixler EO, Lin HM, Prolo P, Trakada G, Chrousos GP. IL-6 and its circadian secretion in humans. Neuroimmunomodulation. 2005;12(3):131–140. doi: 10.1159/000084844. [DOI] [PubMed] [Google Scholar]
- 43.Belluco C, Olivieri F, Bonafe M, et al. -174 G>C polymorphism of interleukin 6 gene promoter affects interleukin 6 serum level in patients with colorectal cancer. Clin Cancer Res. 2003 Jun;9(6):2173–2176. [PubMed] [Google Scholar]
- 44.Hoffmann SC, Stanley EM, Darrin CE, et al. Association of cytokine polymorphic inheritance and in vitro cytokine production in anti-CD3/CD28-stimulated peripheral blood lymphocytes. Transplantation. 2001 Oct 27;72(8):1444–1450. doi: 10.1097/00007890-200110270-00019. [DOI] [PubMed] [Google Scholar]
- 45.Avila J, Acosta E, Machargo MD, et al. Autoantigenic nuclear proteins of a clinically atypical renal vasculitis. J Autoimmune Dis. 2008;5:3. doi: 10.1186/1740-2557-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]