Abstract
While much has been known about the mutualistic associations between the sepiolid squid Euprymna tasmanica and the luminescent bacterium, Vibrio fischeri, less is known about the connectivity between the microscopic and molecular basis of initial attachment and persistence in the light organ. Here, we examine the possible effects of two symbiotic genes on specificity and biofilm formation of V. fischeri in squid light organs. Uridine diphosphate-dehydrogenase (UDPDH) and mannose-sensitive hemagglutinin (mshA) mutants were generated in V. fischeri to determine whether each gene has an effect on host colonization, specificity, and biofilm formation. Both squid light organ colonization assays and transmission electron microscopy (TEM) confirmed differences in host colonization between wild-type and mutant strains, and also demonstrated the importance of both UDPDH and mshA gene expression for successful light organ colonization, and furthers our understanding of the genetic factors playing important roles in this environmentally transmitted symbiosis.
Introduction
Mutualistic associations that occur in the marine environment have provided numerous examples of beneficial capabilities that aid in the success of both host and symbiont (Boucher, et al., 1982). One such relationship that has been studied over the past 20 years is the mutualistic association between sepiolid squids (Cephalopoda: Sepiolidae) and their Vibrio bacteria (γ Proteobacteria: Vibrionaceae). During the onset of the symbiosis, free-living Vibrio bacteria are environmentally acquired by aposymbiotic juvenile squids through initial invasion and colonization steps, and are eventually housed in a specialized bilobed light organ (Montgomery & McFall-Ngai, 1993). Once Vibrio bacteria successfully colonize the naïve squid light organ, those bacteria are able to bioluminescence via quorum sensing, and eventually produce diffuse, downward focused light, which is used by the squid in a behavior known as counterillumination (Jones & Nishiguchi, 2004). In return, the host provides a haven for the bacteria to reproduce at a faster rate, and eventually aid in the daily release of bacteria into the environment through a venting behavior that occurs with the onset of dawn (Lee & Ruby, 1994).
During the initial infection and colonization process Vibrio bacteria have developed sophisticated mechanisms to invade the host light organ and eventually persist in this complex (Nyholm & McFall-Ngai, 2004; Nyholm & Nishiguchi, 2008). During such infections they are involved in inducing a variety of extra-cellular factors such as adhesions, synthesis of polysaccharides, and toxin regulators (Montgomery & Kirchman, 1993). Specifically, adhesion and biofilm formation are two mechanisms that are considered to be crucial for initial colonization (Darnell et al., 2008; Geszvain & Visick 2008; Hussa et al., 2008; O'Toole et al., 2000). Genes that encode for proteins used in adhesion and biofilm production are oftentimes unique, and can be easily distinguished by their differential gene expression during symbiosis (prior or during colonization; Jones & Nishiguchi, 2006). Two such examples of genes that are differentially expressed by V. fischeri are uridine diphosphate glucose-6-dehydrogenase (UDPDH), and mannose-sensitive hemagglutinin (mshA). UDPDH is considered to be a main factor for biofilm formation (Nesper, et al., 2001), while mshA is believed to be essential for initial adhesion and colonization (Bomchil, et al., 2003). MshA has also been demonstrated as a necessary component in V. cholerae for initial attachment and biofilm formation to abiotic substrates using the type IV pili (Watnick & Kolter, 1999). Earlier studies using a technique termed Selective Capture Of Transcribed Sequences (SCOTS) have shown the expression of UDPDH solely in the light organ, whereas mshA is exclusively expressed under natural environmental conditions (Jones & Nishiguchi, 2006). Therefore, the goals of this study were to determine whether both UDPDH and mshA were necessary for successful colonization by V. fischeri in the sepiolid squid Euprymna tasmanica. Mutations were initiated to disrupt gene function for both genes and to determine whether colonization efficiency was decreased when squids were infected with either UDPDH or mshA mutants. We also examined whether these mutations had a decreased ability to attach to the brush border epithelia of E. tasmanica light organs. Finally, complementation of both mutant strains was used to regain loss of function, both in vitro and in colonization of naïve juvenile squids.
Materials and Methods
Strains, plasmids, and growth conditions
All strains used in this study are listed in Table 1. Wild type V. fischeri strain ETJB1H was isolated from the light organs of E. tasmanica from Jervis Bay, Australia as previously described (Jones, et al., 2006). All V. fischeri and E. coli strains used in this study were grown in seawater tryptone (SWT) and Luria Broth (LB) at 28 and 37°C, and supplemented with appropriated antibiotics for selection of mutant strains.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description |
|---|---|
| V. fischeri strains | |
| ETJB1H | Wildtype |
| UDPDH− | UDP-glucose-6-dehydrogenase mutant |
| mshA − | Mannose sensitive hemagglutinin mutant |
| UDPDH+ | UDP-glucose-6-dehydrogenase complement |
| mshA + | Mannose sensitive hemagglutinin complement |
| Plasmids | |
| pEVS122 | R6K, ErmR |
| pVSV105 | pES213 replicon, CmR |
ErmR (Erythromycin resistance), CmR (Chloromphenical resiatance)
PCR
All loci in this study were amplified using 50 μL volume PCR reactions as previously described (Jones, et al., 2006; Table 2).
Table 2.
UDPDH−, mshA−, UDPDH+, and mshA+ primer sequences used for constructing mutant strains (F = forward primer sequence, R = reverse primer sequence, bp = base pairs).
| Primer | Primer sequence 5'-3' | PCR product Size |
|---|---|---|
| mshA-forward | AGC AGA TCT TTT ATG GTA AAG CCG CGA TT | ~180 |
| mshA-Reverse | AGC AGA TCT GCT GCA GTT GGG TTA TCT GA | ~180 |
| UDP-Forward | AGC AGA TCT AA AAT CGC CTA TTT CAG ATG T | ~160 |
| UDP-Reverse | AGC AGA TCT GCT TCA ACA GAG CCC GTA TT | ~160 |
| mshA-Comp-F | CCC GGG GAT CAG TGA GAA TGG CCG TA | ~450 |
| mshA-Comp-R | CCC GGG CGA TTG TTG ATA CGC CAG AA | ~450 |
| UDP-Comp-F | CCC GGG ATT CAG GTC GCA GGT TTC AG | ~700 |
| UDP-Comp-R | CCC GGG TTC AGA TTG CTC ACC CAC AA | ~700 |
Mutant construction
Mutants were constructed using plasmid insertion where, an internal (partial) fragment of each targeted gene was PCR amplified and cloned directly into pEVS 122. This plasmid was introduced into V. fischeri by tri-parental mating (Stabb & Ruby, 2002). The V. fischeri strains that had undergone homologous recombination between the genome and the internal gene fragment were selected on SWT-Erythromycin plates and were verified via Southern blots.
Biofilm assays
All bacterial strains (wild-type, mutant and complement) were grown and biofilm assays were performed as previously described (Nair, 2006). Biofilm assays for every strain were completed in triplicate and statistically analyzed by students t-test.
Motility assays
All bacterial strains were grown overnight at 28°C and 250 rpm. The following day 20 μL of bacterial culture was inoculated and strains were re-grown in fresh 32 ppt SWT media to a 0.1 OD600. 10 μL of each culture was spotted onto an SWT plate containing 5% agar. Motility was determined by measuring the diameter of the spot after 24 hours. Each motility assay for every strain was completed in triplicate and statistically analyzed by students t-test.
Colonization assay
To determine the colonization efficiency between mutant and wild-type bacterial strains, colonization assays of different V. fischeri strains were completed as previously described (Nishiguchi, 2002). Briefly, both wild-type and mutant strains were grown in 5 mL SWT overnight in shaking incubator (250 rpm) at 28°C. Strains were transferred the next morning and re-grown in 5 mL of fresh SWT media to 0.3–0.5 OD600. Cultures were diluted to approximately 1 × 103 CFU/mL and used to inoculate 5 mL of sterile seawater with axenic juvenile squids. After inoculation, seawater in all vials was changed every 12 hours and bioluminescence was measured using a luminometer (Turner Designs., Sunnyvale, CA) over a period of 48 hours. During incubation, infected animals were kept in 12/12 h light/dark cycle. Infected juvenile squids were sacrificed at 48 hours and plated on SWT agar. The number of CFUs on each plate represented the colonization efficiency of each strain in a squid light organ. Results were analyzed by students t-test.
Electron Microscopy
Juvenile squids infected for 48 hours with either strain were fixed and processed for transmission electron microscopy as previously described (Nair, 2006).
All bacterial strains examined for SEM were grown overnight at 250 rpm and 28°C in 5 mL SWT. The following day, 20 μL of each bacterial culture was used to inoculate a new test tube with 3 mL sterile SWT media and re-grown at 250 rpm and 28°C, until each culture reached 0.1 OD600. One mL of each culture was transferred into 50 mL falcon tubes with a sterile cover slip half immersed in the media and incubated 24 hours without shaking at 28°C. Cover slips were then washed with 32 ppt seawater to remove excess particles other than the biofilm formed by each bacterial culture. Excess seawater was blotted dry, and all cover slips were incubated in 1 mL of a 0.2% aqueous solution of crystal violet at room temperature for one hour. Cover slips from each strain were viewed with the Hitachi S3400 scanning electron microscope (Hitachi, Schaumburg, IL, USA).
Results and Discussion
Growth curves and bioluminescence
V. fischeri ETJB1H and the mutant strains generated in this study are listed in Table 1. Growth analyses were completed for both UDPDH− and mshA− mutants and compared to V. fischeri ETJB1H. ETJB1H exhibited a doubling time of 25.7 minutes compared to UDPDH− and mshA−, which had doubling times of 76.2 and 56.4 minutes. Simultaneously, luminescence of both mutant strains and the wild-type ETJB1H was measured. Comparisons between all 3 strains demonstrated that mutants had lower levels of bioluminescence/OD values at 13.7 relative light units (RLU) for mshA− and 181 RLU for UDPDH− compared to values of more than 7205 RLU for ETJB1H.
In addition, growth analyses were also completed for complemented UDPDH+ and mshA+ strains and compared to their respective mutant strains. UDPDH+ exhibited a doubling time of 29.6 minutes, which was similar to the rate of the wild-type parental strain. MshA+ had a doubling time of approximately 33.3 minutes. Luminescence of UDPDH+ and mshA+ strains was also measured simultaneously. Interestingly, luminescence/OD of complemented strains exhibited a lower amount of bioluminescence/OD compared to the parental strain, with values of 18 RLU for UDPDH+ and 3.4 RLU for mshA+.
In vitro biofilm assays
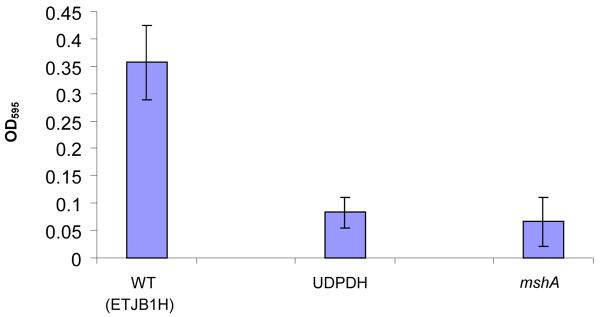
Both V. fischeri UDPDH− and mshA− strains exhibited a deficiency in biofilm formation with values of 0.1 at OD595 when eluted with the alcohol: acetone mixture (Figure 1A). These values were compared to the parental wild-type, which had values of 0.35 OD595 (Figure 1A). Test analysis between V. fischeri ETJB1H UDPDH− and mshA− compared to parental wild-type showed that average mean values were significantly different.
Figure 1.
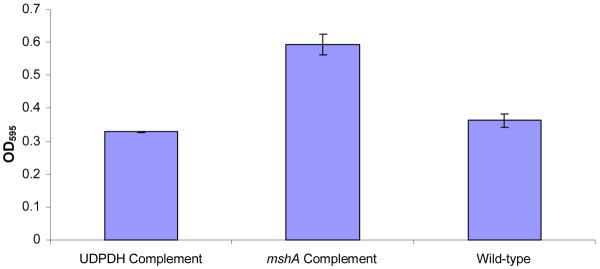
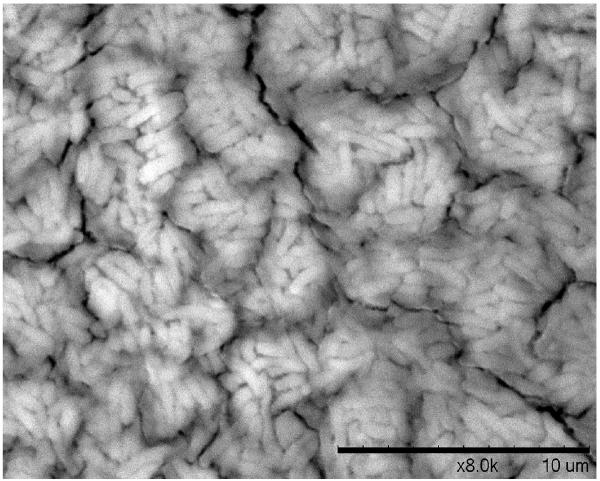
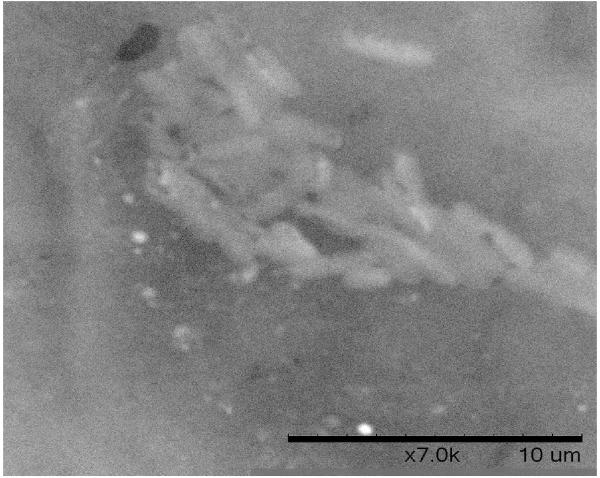
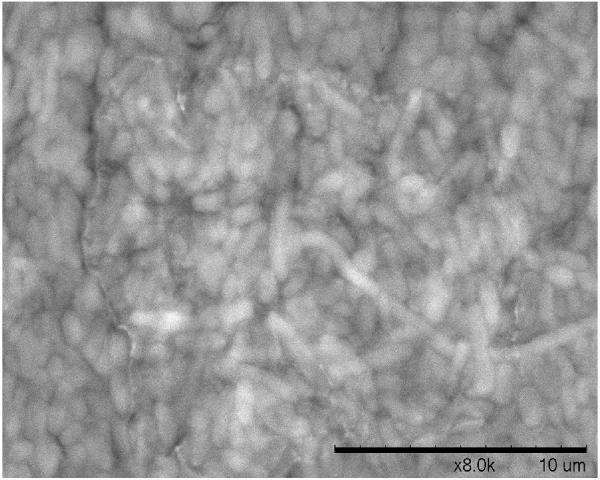
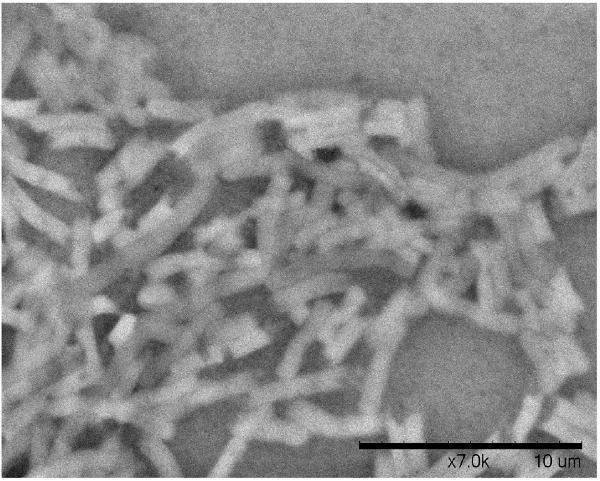
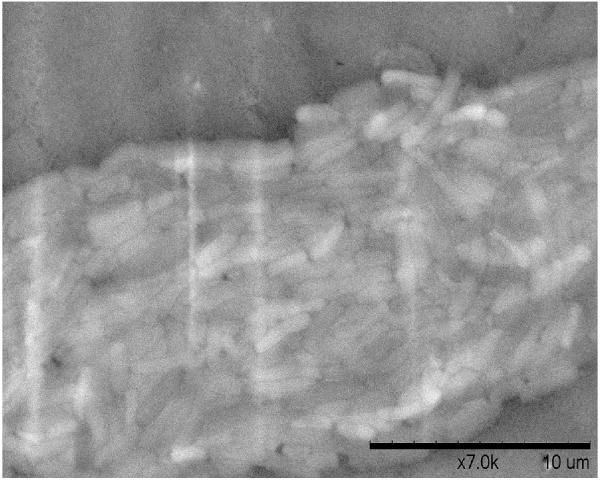
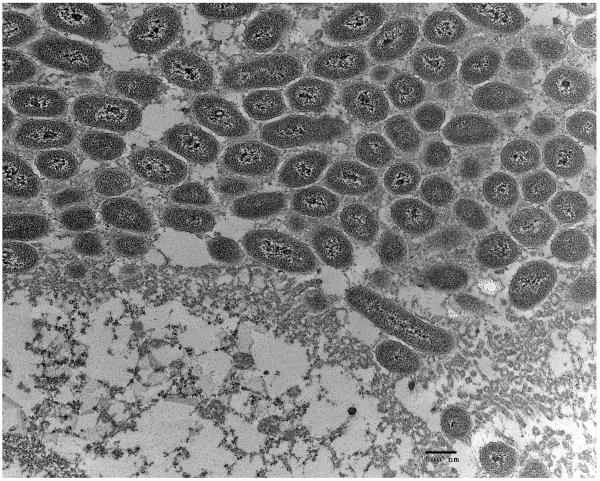
A. Biofilm produced by V. fischeri ETJB1H wild-type and V. fischeri ETJB1H UDPDH− had a p value of 0.0034 at α = 0.05 and mshA− mutant strains had a p value of 0.005 (α = 0.05) (bars represent ±1 standard error). B. Biofilm produced by V. fischeri ETJB1H parental wild-type and both V. fischeri ETJB1H UDPDH+ and mshA+ complemented strains (bars represent ±1 standard error). C. Scanning electron micrograph of biofilm formed by the wild-type strain V. fischeri ETJB1H. Scale bar = 10 μm. D. Scanning electron micrograph of biofilm formed by V. fischeri ETJB1H UDPDH− strain. Scale bar = 10 μm. E. Scanning electron micrograph of biofilm formed by V. fischeri ETJB1H mshA− strain. Scale bar = 10 μm. F. Scanning electron micrograph of biofilm formed by V. fischeri ETJB1H UDPDH+ complement strain. Scale bar = 10 μm. G. Scanning electron micrograph of biofilm formation by V. fischeri ETJB1H mshA+ complement strain. Scale bar = 10 μm
UDPDH+ and mshA+ complemented strains also had an increase in biofilm formation with mean values of 0.3 at OD595. MshA+ (Figure 1B) had a greater value than parental wild-type V. fischeri ETJB1H. Statistical analysis of biofilm production gave significant differences between UDPDH− and UDPDH+ strains, with p values of 0.0001 at α=0.05. Additionally, the mshA− and the mshA+ strains exhibited significant differences, with p values of 0.002 (α=0.05).
Biofilm morphology
Biofilm produced from V. fischeri ETJB1H parental and mutant strains were visualized using SEM. Significant morphological differences were observed between parental V. fischeri ETJB1H (Figure 1C) and mutants UDPDH− (Figure 1D) and mshA− (Figure 1E). V. fischeri ETJB1H exhibited compact bacterial cells that were attached to one another in a 3-dimensional structure. This was in contrast to both mutant strains, which were thinner and less dense than the wild-type. Additionally, bacterial biofilms from the complemented UDPDH+ (Figure 1F) and mshA+ strains (Figure 1G) regained the ability to form well organized biofilms, and appeared to form complexes similar to their wild-type parent (Figure 1C).
Motility assay
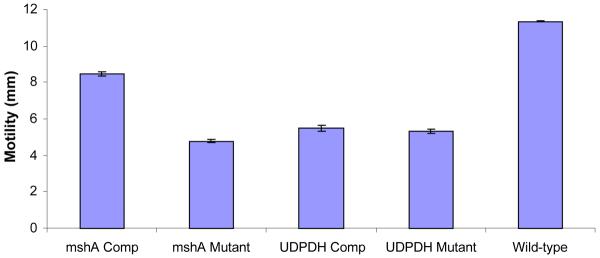
Wild-type ETJB1H exhibited the highest degree of motility compared to both mutant strains (Figure 2). Both UDPDH− and the wild-type strains exhibited significant differences (students t-test). Motility of complemented strains was also measured and exhibited greater motility compared to their respective mutant strains.
Figure 2.
Mean average motility between V. fischeri ETJB1H mshA− and mshA+ strains, V. fischeri ETJB1H UDPDH− and UDPDH+ strains and the wild-type V. fischeri ETJB1H strains. Each sample was run in triplicate. Both UDPDH− and mshA− strains exhibited p values of 0.001 and 0.0007 accordingly at α = 0.05 compared to the wild-type strains. The UDPDH− and UDPDH+ did not show significant difference, where as the mshA− and mshA+ had significant p values of 0.001 at α = 0.05. Standard error ±1.
Colonization assay
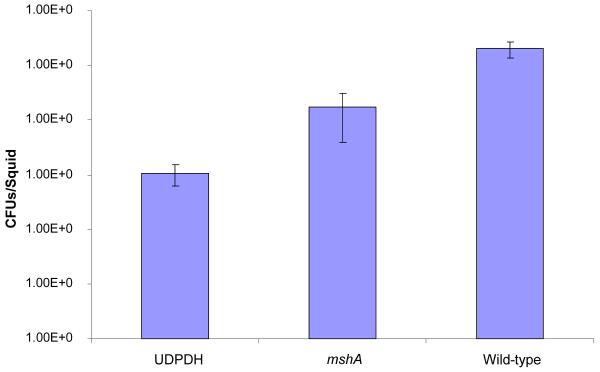
Axenic juvenile E. tasmanica squids were infected with both mutants as well as wild-type V. fischeri ETJB1H. Seven animals were used in this study for each strain. Single strain infections were measured by homogenizing juvenile squid after 48 h of infection and calculating the number of colony forming units (CFUs/mL; Nishiguchi, 2002). V. fischeri ETJB1H parental wild-type had a mean colonization efficiency of 1 × 105 CFUs/mL compared to UDPDH− and mshA−, which had values of 1 × 103CFU/mL and 1 × 104 CFU/mL respectively (Figure 3).
Figure 3.
Colonization assay 48 hours post infection of juvenile E. tasmanica by the parental wild-type V. fischeri ETJB1H and both V. fischeri ETJB1H mutant strains (UDPDH− and mshA−). Both UDPDH− and mshA− compared to the wild-type strains exhibited significant differences with p values of 1.4E-10 and 7.8E-5 at α = 0.05. Bars within each graph represent the standard error within ±1 of the mean. (CFU= colony forming units)
Transmission Electron Microscopy
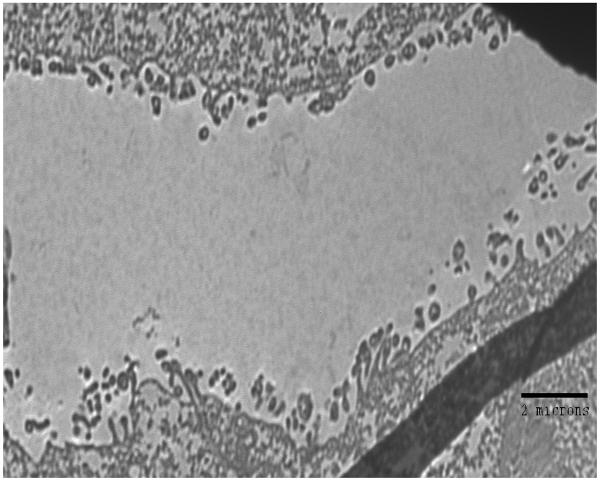
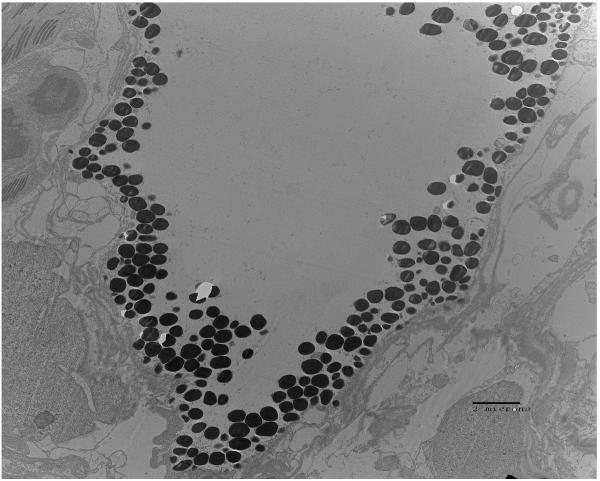
Examination of the crypt regions of juvenile E. tasmanica that were infected with either parental wild-type V. fischeri ETJB1H, V. fischeri ETJB1H UDPDH− or V. fischeri ETJB1H mshA− was completed using transmission electron microscopy. Parental wild-type ETJB1H (Figure 4A) had complete colonization of the crypt region, whereas V. fischeri ETJB1H UDPDH− (Figure 4B) exhibited little or no change in microvillar density to the brush border epithelia. V. fischeri ETJB1H mshA− mutant (Figure 4C) showed deficiency in colonization of the crypt region, where few bacteria were found along the brush border epithelial lining.
Figure 4.
Transmission electron micrographs of the squid light organ infected by V. fischeri ETJB1H wild-type strain (A) bar = 500 nm, V. fischeri ETJB1H UDPDH− mutant (B) bar = 2 μm and V. fischeri ETJB1H mshA− (C) mutant strains bar = 2 μm.
In this study we examined how mutations in either UDPDH and mshA genes in Vibrio fischeri affect infection and colonization in juvenile E. tasmanica light organs. As an environmentally transmitted bacterium, symbiotic V. fischeri must be capable of adapting to the selective environment of the squid light organ upon initial infection. Following successful colonization, squids will vent 90%–95% of the bacteria from their crypt spaces every morning at dawn (McFall-Ngai, 2000), which may allow the streamlining of better adapted V. fischeri strains to be maintained in the colonized light organ. Earlier studies have provided evidence that bacterial biofilms are present in squid light organs (Nair, 2006). Therefore, the question of whether biofilm formation is important for colonization and persistence within the squid light organ may be of interest for determining specificity.
UDPDH
Biofilm formation has been shown to be one factor responsible for persistence of the bacterium P. aeruginosa within infected lungs of human patients (Lam et al., 1980). In addition, studies using E. coli and P. aeruginosa indicate the importance of colanic acid biosynthesis for capsular formation, which increases the ability of those bacteria to adhere to substrates. Colonic acid production is also believed to enhance the ability of bacteria to form biofilm production upon initial infection of epithelial cells in the lungs of cystic fibrosis patients (Davies, et al., 1993). Biosynthesis of this acid first begins with the pathways of four different nucleotide sugars (UDP-galactose, UDP-glucose, UDP-D-glucuronate and Guanine-Di-Phosphate-L-fucose, where UDPDH catalyzes UDP-glucose to UDP-D-glucuronic acid (Stevenson, et al., 1996). Interestingly, this gene was only expressed in the symbiotic state in both E. tasmanica and E. scolopes light organs (Jones & Nishiguchi, 2006). Since biofilm production is also linked to increased infectivity in a number of pathogenic bacteria (Austin & Zhang, 2006), it may also be involved in colonization by mutualistic bacteria such as V. fischeri. Additional studies have demonstrated that biofilms in V. fischeri are highly regulated and important for symbiosis to occur in the E. scolopes light organs (Darnell et al., 2008; Geszvain & Visick, 2008; Hussa et al., 2008; Yip et al., 2006; Yip et al., 2005). Thus, biofilms are an integral part of the infection, colonization, and persistence processes of mutualistic vibrios in this dynamic symbiosis.
Previous studies in V. cholerae also indicate the importance of UDP glucose dehydrogenase for the synthesis of lipopolysaccharide (LPS) in addition to colanic acid biosynthesis. UDP glucose dehydrogenase mutant strains also exhibit a deficiency in capsular formation in pathogenic strains of V. cholerae (Nesper, et al., 2001). Similar results were observed during colonization assays with V. fischeri ETJB1H UDPDH− mutant strains, which exhibited a 10−2 decrease in colonization compared to wild-type parental strains (Figure 3). This may provide initial evidence that UDPDH has an additional role in the biosynthetic pathway producing LPS via glucuronic acid synthesis in V. fischeri. Previous studies have shown the role between LPS and peptidoglyan that together trigger morphogenesis in juvenile squid light organs (Koropatnick et al., 2004; Foster et al., 2000). Peptidoglycan is also known to be essential for mucin synthesis (Nyholm, et al., 2002; Nyholm et al., 2000). Induction of mucus secretion in E. scolopes demonstrates the presence of gram-negative bacteria such as V. fischeri. Together the morphogenesis and mucus production enhances the capability of specific V. fischeri to recognize and infect their squid hosts. This is one of the mechanisms that is responsible for selecting symbiotically competent bacteria from all others in the surrounding seawater.
TEM observations in this study also support the hypothesis that UDPDH is responsible for colanic acid biosynthesis thereby affecting capsular and biofilm formation in symbiotic Vibrio fischeri. UDPDH− mutant V. fischeri were not found in any part of the crypt region of the light organs upon examination (Figures 4B). In addition, the ability to produce biofilm in vitro was measured to verify whether biofilm production was reduced in the UDPDH mutant strains. In vitro biofilm assays exhibited a 3-fold reduction in biofilm production by V. fischeri ETJB1H UDPDH− mutants compared to parental wildtype V. fischeri ETJB1H (Figure 1). Similar deformations in biofilm production were observed in UDP glucose mutants in V. cholerae where mutant strains are not capable of inducing biofilm production (Nesper, et al., 2001).
Additionally, motility assays were completed to verify whether the UDPDH gene is linked to motility, since motility is associated with a decrease in colonization by V. fischeri (Millikan & Ruby, 2002). V. fischeri UDPDH− mutants did exhibit significant differences in motility compared to the wild-type parental strain. This result indicated that mutating the UDPDH locus did effect the motility of the bacterium, providing evidence that colonization deficiencies at this locus were linked not only to the decreases in biofilm production but also motility.
mshA
Studies examining V. cholerae have provided information regarding the role of mshA during initial infection prior to biofilm formation (Watnick, et al., 1999). This gene is responsible for the formation of type IV bundle forming pili, which are crucial for the initial attachment to abiotic substrates. In addition, type IV bundle pili are vital for twitching motility in P. aeruginosa, which is considered to be an essential trait for the spread of infection following initial attachment and biofilm production (O'Toole & Kolter, 1998; Skerker & Berg, 2001). Earlier studies using selective capture of transcribed sequences (SCOTS) in symbiotic V. fischeri detected two genes (pilM and mshA) that were responsible for pili formation and attachment, and are solely expressed in seawater cultures (Jones & Nishiguchi, 2006). Expression of both pilM and mshA prior to colonization (in the free-living stage) suggests that both have important roles in initiating symbiosis with E. tasmanica. For example, another pil locus (pilA) has decreased colonization when mutant (pilA−) V. fischeri were infected in E. scolopes juveniles (Stabb & Ruby, 2003), as well as establishing specificity during the early stages of colonization in enteropathogenic E. coli (Hicks, et al., 1998). It has also been demonstrated that a high degree of variability exists at this operon when comparing symbiotic and free-living strains of V. fischeri (Browne-Silva & Nishiguchi, 2008). This is yet another example of how subtle differences at a particular symbiotic locus can determine specificity in this association, and may be responsible for the differences observed in infection (commencement of the interaction) and colonization (growth and persistence of the interaction) between a number of symbiotically competent V. fischeri isolated (Nishiguchi et al., 1998; Nishiguchi, 2002).
Studies have also shown that juvenile E. scolopes contain mannose along the crypt region of the light organs, enhancing colonization by V. fischeri (Visick & McFall-Ngai, 2000). Symbiotic V. fischeri were also able to agglutinate to guinea pig red blood cells and that exogenous mannose blocked colonization, indicating the presence of mannose recognizing adhesions (McFall-Ngai, et al., 1998). In addition, further studies have provided evidence for mannose residues on the contact surface of host epithelial lining, implicating a receptor-ligand interaction between symbionts and the brush border epithelia of squid light organs (Visick & McFall-Ngai, 2000). Interactions such as those between symbiotic bacteria and mannose or other sugar containing residues are commonly found in environmentally transmitted associations (Nishiguchi et al., 2008; Nyholm and Nishiguchi, 2008), providing yet another mechanism for recognizing specific partners. This is particularly important in associations where a few bacteria are responsible for initiating the colonization of a new host amidst a number of abiotic and biotic forces that may delay or deter colonization from ever occurring (Soto et al., 2009a; 2009b
In this study, mshA mutants were generated to examine whether this gene was involved in colonization processes. Results indicate that V. fischeri ETJB1H mshA− mutants had deficiencies in their colonization ability compared to the parental wild-type, with a reduction of mutants observed inside the crypt region after infection (Figure 4C). V. fischeri ETJB1H mshA− mutants also exhibited severe deficiencies in biofilm production when compared to the wild-type parent. These results were similar to V. cholerae mshA− mutants where the mshA− mutants have a lack of biofilm production and are severely deficient in motility (Watnick, et al., 1999). Motility is believed to be essential for the colonization of the squid light organ, since non-motile V. fischeri are incapable of successful colonization (Millikan & Ruby, 2002). The lack of motility in V. fischeri mshA− mutants is additional support that movement to the light organ pores as well as through the duct region is required to colonize the crypts of the light organ after initial infection (Millikan & Ruby, 2002, Millikan & Ruby, 2003).
Complementation of mutant strains
Both V. fischeri UDPDH+ and mshA+ were able to regain the loss of function significantly for biofilm production, growth rate, and motility. The only phenotype that was not regained by complementation was bioluminescence with either UDPDH+ or mshA+. This suggests that both UDPDH and mshA genes may be linked to luminescence related behavior (quorum sensing), and disruption of these genes interferes not only with bioluminescence production, but the ability for bacteria to quorum sense. Quorum sensing is activated through a cascade of genes in the lux operon, so that disruption of UDPDH may have possibly interfered with other potential downstream genes, regulated through bioluminescence production (Callahan & Dunlap, 2000). Although lux genes were not found to be downstream from either UDPDH or mshA in V. fischeri ES114 (Ruby et al., 2005), it may differ in the V. fischeri ETJB1H strain that was used in this study. Further studies examining whether UDPDH is transcriptionally or translationally regulated via the lux operon would be required to verify the presence of either cis or trans acting regulatory elements linked to quorum sensing.
Conclusions
This study has examined whether UDPDH and mshA genes were involved in the early events of colonization of squid light organs. Results from our experiments indicate that both genes are involved in the establishment of the mutualism between V. fischeri and E. tamanica, where early attachment, colonization, and biofilm production are important for a successful environmentally transmitted symbiosis to occur. Experimental comparisons of environmentally expressed genes to those that are expressed solely in symbiosis can help us better understand whether trade-offs evolve to benefit either situation, such as the expression of mshA in the free-living state and UDPDH in the squid light organ, and how both genes are beneficial during different steps in establishing mutualistic relationships. Additionally, our understanding of how different pathways are co-opted from either a benign or pathogenic association can provide clues as to the evolution of virulence, and whether those genes involved in the symbiosis have been selected for amongst a wide variety of genotypes (Nishiguchi et al., 2008). The selective pressures that bacteria are exposed to (abiotic and biotic) have an incredible amount of influence that determines the evolutionary trajectory of a specific genotype; balancing those forces is one feat that symbiotic bacteria have overcome to be successful in nearly all ecological niches, including those that invade eukaryotic host tissues (Soto et al., 2009a, 2009b). Continued research in the genetic basis behind V. fischeri's adaptations to biotic and abiotic factors will continue to illuminate the underpinnings of symbiosis in the years to come.
Acknowledgements
The authors would like to thank A. Dunn and E. Stabb for their gift of the plasmids used in this study. This work was supported by NIH SO6 GM008136-32S2-1, NIH-NIAID 1SC1AI081659-01, and NSF IOS-0744498 to M.K.N.
References
- Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, Post JC, Sauer K. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J Bacteriol. 2006;188:2325–2335. doi: 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin B, Zhang XH. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol. 2006;43:119–124. doi: 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- Bomchil N, Watnick P, Kolter R. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J Bacteriol. 2003;185:1384–1390. doi: 10.1128/JB.185.4.1384-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher DH, James S, Keeler KH. The ecology of mutualism. Annu Rev Ecol Syst. 1982;13:315–347. [Google Scholar]
- Browne-Silva J, Nishiguchi MK. Gene sequences of the pil operon reveal relationships between symbiotic strains of Vibrio fischeri. Int J Syst Evol Microbiol. 2008;58:1292–1299. doi: 10.1099/ijs.0.65370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan SM, Paul V. Dunlap. LuxR- and Acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J Bacteriol. 2000;182:2811–2822. doi: 10.1128/jb.182.10.2811-2822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell CL, Hussa EA, Visick KL. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, sypG and vpsR. J Bacteriol. 2008;190:4941–4950. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms; substratum activation of alginate gene expression by Psuedomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Martin MO, Stabb EV. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid. 2005;54:114–134. doi: 10.1016/j.plasmid.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Stabb EV. The twin arginine translocation system contributes to symbiotic colonization of Euprymna scolopes by Vibrio fischeri. FEMS Microbiol Lett. 2008;279:251–258. doi: 10.1111/j.1574-6968.2007.01043.x. [DOI] [PubMed] [Google Scholar]
- Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Bio. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- Geszvain K, Visick KL. The hybrid sensor kinase RscS integrates positive and negative signals to modulate biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4437–4446. doi: 10.1128/JB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks S, Frankel G, Kaper JB, Dougan G, Phillips AD. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, Darnell CL, Visick KL. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Nishiguchi MK. Counterillumination in the bobtail squid, Euprymna scolopes (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–1155. [Google Scholar]
- Jones BW, Nishiguchi MK. Differentially expressed genes reveal adaptations between free-living and symbiotic niches of Vibrio fischeri in a fully established mutualism. Can J Microbiol. 2006;52:1218–1227. doi: 10.1139/w06-088. [DOI] [PubMed] [Google Scholar]
- Jones BW, Lopez JE, Huttenburg J, Nishiguchi MK. Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Mol Ecol. 2006;15:4317–4329. doi: 10.1111/j.1365-294X.2006.03073.x. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial Factor-Mediated Development in a Host-Bacterial Mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Lam J, Chan R, Lam K, Costerton JW. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-H, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. Negotiations between animals and bacteria: the `diplomacy' of the squid-vibrio symbiosis. Comp Biochem Physiol A. 2000;126:471–480. doi: 10.1016/s1095-6433(00)00233-6. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Brennan C, Weis V, Lamarcq LH. Mannose adhesin-glycan interactions in the Euprymna scolopes-Vibrio fischeri symbiosis. In: Le Gal Y, Halvorson HO, editors. New Developments in Marine Biotechnology. Plenum Press; N.Y.: 1998. pp. 273–276. [Google Scholar]
- Millikan DS, Ruby EG. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl Environ Microbiol. 2002;68:2519–2528. doi: 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan DS, Ruby EG. FlrA, a o54-dependent transcriptional activator in Vibrio fischeri, is required for mobility and symbiotic light organ colonization. J Bacteriol. 2003;185:3547–3557. doi: 10.1128/JB.185.12.3547-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- Montgomery MT, Kirchman DL. Role of chitin-binding proteins in the specific attachment of the marine bacterium Vibrio harveyi to chitin. Appl Environ Microbiol. 1993;59:373–379. doi: 10.1128/aem.59.2.373-379.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S. Vinod. PhD dissertation. New Mexico State University; 2006. The evolution of symbiosis in Vibrionaceae and factors influencing host colonization in squid-vibrio mutualism. [Google Scholar]
- Nesper J, Lauriano CM, Klose KE, Kapfhammer D, Krai A, Reidl J. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: Influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun. 2001;69:435–445. doi: 10.1128/IAI.69.1.435-445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi MK, DeVinney R, Hirsch AM, Riley M, Mansky L, Vendatum G. Perspective: Evolution of virulence: Deciphering the mechanisms between pathogenic and benign symbioses. Vie et Milieu. 2008;58:87–106. [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi MK. Host recognition is responsible for symbiont composition in environmentally transmitted symbiosis. Microbiol. Ecol. 2002;44:10–18. doi: 10.1007/BF03036870. [DOI] [PubMed] [Google Scholar]
- Nishiguchi MK, Jones BW. Microbial biodiversity within the Vibrionaceae. In: Seckbach J, editor. Origins. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. pp. 531–548. [Google Scholar]
- Nishiguchi MK, Ruby EG, McFall-Ngai MJ. Competitive dominance during colonization is an indicator of coevolution in an animal-bacterial symbiosis. Appl Environ Microbiol. 1998;64(9):3209–13. doi: 10.1128/aem.64.9.3209-3213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Nishiguchi MK. The evolutionary ecology of a sepiolid squid-Vibrio association: From cell to environment. Vie et Milieu. 2008;58:175–184. [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The Winnowing: Establishing the Squid–Vibrio Symbiosis. Nat. Rev. Microbiol. 2004;2(8):632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol. 2003;69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. Proc Nat Acad Sci USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;68:5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- O'Toole G, Kolter R. Flagellar and twitching motility are necessary for Psuedomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Whistler K, Visick C, Greenberg EP. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Nat Acad Sci. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto W, Gutierrez J, Remmenga MR, Nishiguchi MK. Synergistic affects of temperature and salinity in competing strains of symbiotic. Vibrio fischeri. Microbiol Ecol. 2009a;57:140–150. doi: 10.1007/s00248-008-9412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto W, Lostroh CP, Nishiguchi MK. Physiological responses to stress in the Vibrionaceae: Aquatic microorganisms frequently affiliated with hosts. In: Seckbach J, Grube M, editors. Cooperation and stress in Biology. Springer; New York, NY: 2009b. [Google Scholar]
- Stabb EV, Ruby EG. RP-4 Based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl Environ Microbiol. 2003;69:820–826. doi: 10.1128/AEM.69.2.820-826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, McFall-Ngai MJ. An exclusive contract: Specificity in the Vibrio fischeri-Euprymna scolopes partnership. J Bacteriol. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]