Abstract
A simple two-step protocol for modification of atomic force microscopy tip and substrate by using a “click reaction” has been developed. The modified tip and substrate were applied to detect trace amounts of ricin by using single molecule recognition force microscopy (SMRFM1). A key feature of the approach is the use of a PEG derivative functionalized with a thiol and azide group end group. This compound could be attached to a gold coated AFM tip by a strong Au-thiol bond. The azide of the immobilized PEG was used for the attachment of an anti-ricin antibody modified with an alkyne group using a ‘click reaction’. The latter reaction is highly efficient, compatible with the presence of many functional groups and proceeds under mild reaction conditions. In a separate step, ricin was immobilized on the gold substrate surface that was modified by active esters. For this process, a novel bifunctional reagent was employed containing an active ester and a thioctic acid moiety. The SMRFM results showed a sub fg/mL level of detection sensitivity. The unbinding force between the anti-ricin antibody and ricin was studied by the force-distance curves (F-D). The unbinding force between the ricin and the antibody was determined to be 64.89±1.67 pN by constructing a force histogram.
1. INTRODUCTION
Atomic force microscopy, which was first described by Binning et al. in 1986 1, has found wide application in the field of molecular recognition 2. In particular, single molecular recognition force microscopy, which is a technique based on AFM, is gaining increasing attention 3–5. SMRFM combines chemical force microscope and magnetic alternative current (MAC) imaging mode AFM, thereby producing both images and force spectroscopic curves. It offers a rapid and sensitive tool for molecular identification and interaction studies 6. Due to its high sensitivity and fast imaging process, SMRFM could be a high-resolution alternative for detection of deadly biological warfare agents (BWA), such as ricin toxin.
Ricin toxin, which is a byproduct of castor oil production, was discovered in the seeds of the castor bean plant, Ricinis communis. It is a heterogeneous proteinaceous toxin, which consists of many different proteins 7,8. The average lethal dose of ricin in humans is approximately 0.2 mg, which makes it 6000 times more toxic than cyanide and 12000 times more lethal than rattlesnake venom by weight. There is no antidote for ricin toxin once introduced above the lethal dosage 9. In addition, ricin toxic can be formulated in a variety of physical forms including powder, mist, pellet, or dissolved in water or weak acids. Due to its ready availability, high toxicity, and stability, ricin has the potential to be used in bioterrorism attacks. For this reason, a rapid, sensitive, and quantitative detection method for trace amounts of the toxin is needed. 9,10.
The current detection approach for ricin is based on traditional immunological methods, such as radioimmunoassay (RIA) 11, enzyme-linked immunosorbent assay (ELISA) 12, and enhanced colorimetric and chemi- luminescence ELISA13. However, lengthy assay times and limited throughput make these methods impractical for rapid responses. Numerous alternative techniques have been developed, and example include capillary electrophoresis (CE) 14, hybrid combination of quadrupole with time-of-flight (Qq TOF) 15, and hydrogel-based protein microchips 16. Although the detection time is greatly decreased, these methods still do not meet all the requirements for inexpensive, rapid, and sensitive detection of toxins.
Tip modification is a critical step for qualitative detection by SMRFM 17,18. Several protocols have been applied to chemically functionalize AFM tips, and generally entail three steps, namely modification of a tip surface, grafting of a cross-linker, and conjugation of a biomolecule 19–21. There are three main classes of methods for carrying out the first step: i) silanization of the tip surface 20; ii) hydrosilylation and polymerization on the tip surface 22; and iii) formation of a self assembled monolayer (SAM) on a metal coated tip surface 23. The latter approach is the most popular and convenient one 18.
We report here a simplified method to modify AFM tip for SMRFM based on the powerful “click chemistry” 24,25, which facilitated practical and sensitive detection of toxins. A key feature of the approach involves the use of a polyethylene glycol derivative (compound 6, Scheme 1) modified with a thiol and azide end group. The thiol moiety of the PEG derivative enables attachment to a gold-coated tip without the need for initial tip modification. The azide of the immobilized PEG can then be employed in a CuI catalyzed 1,3-dipolar cyclization with an alkyne moiety of an appropriate biomolecule. The cycloaddition reaction, which has been coined ‘click chemistry”, is attractive because it can be performed at room temperature to provide stereospecifically stable triazoles in high yield and is compatible with a variety of solvents and functional groups 26. We have applied the technology for immobilization of an anti-ricin antibody (anti-ricin Ab), which made it possible to qualitatively and quantitatively detect ricin with a greater sensitivity than traditional approaches 9. In addition, the interaction force between these two biomolecules could be measured.
Scheme 1.
Synthesis of bifunctionalized PEG linker 6 and succinimidyl active esters 8 and 10. Reagents and conditions: (a) TsCl, Py, 0 °C; (b) KSAc, CH3OH, reflux; (c) TsCl, Py, 0 °C; (d) NaN3, DMF, 60 °C; (e) NaOCH3, CH3OH, rt. a) EDC, NHS, DCM, rt; (b) DCC, NHS, dioxane, rt;
2. EXPERIMENTAL SECTION
2.1. Materials
Fluorescein labeled Ricinus Communis Agglutinin II (RCA II, RCA60, ricin) (Vector Laboratories Inc.), affinity purified goat anti-ricinus communis agglutinin I&II (1mg/ml, Vector Laboratories AS-2084), regenerated cellulose dialysis membrane (MWCO=3500) (Membrane Filtration Products, Inc.), 1-(5-[1,2]dithiolan-3-yl-pentanoyl)-pyrrolidine-2,5-dione (LA-NHS, as described), pentynoic acid 2,5-dioxo-pyrrolidin-1-yl ester (Alkyne-NHS, as described), HS–PEG(2000)–N3 (as described), PNS buffer (10mM phosphate, pH=7.8, 0.15M NaCl, 0.08% Sodium Azide), CS-10 silicon AFM probe (Nanoscience Instruments).
All chemicals used for synthesis were purchased from Aldrich or Fluka. Dichloromethane was distilled from CaH2 and stored over 4 Å molecular sieves. Pyridine was distilled from P2O5 and stored over 4 Å molecular sieves. Prior use THF was distilled from sodium. Other chemicals were used without further purification.
All reactions were performed in anhydrous conditions under an atmosphere of Ar. Reactions were monitored using thin layer chromatography (TLC) on Kieselgel 60 F254 (Merck). Detection was performed by examination under UV light (254 nm) or by charring with 5% sulfuric acid in methanol. Flash chromatography was performed on silica gel (Merck, 70–230 mesh). Iatrobeads (60 µm) were purchased from Bioscan. 1H NMR (1D, 2D) and 13C NMR were recorded on a Varian Merc 300 spectrometer as well as Varian 500 and 600 MHz spectrometers equipped with Sun workstations. Negative ion matrix assisted laser desorption ionization time of flight (MALDI-TOF) measurements were recorded on a VOYAGER-DE Applied Biosystems using dihydrobenzoic acid as a matrix. High-resolution mass spectra were obtained using a VOYAGER-DE Applied Bio-systems in the positive mode by using 2,5-dihydroxyl-benzoic acid in THF as matrix.
2.2. Synthesis of N3(CH2CH2O)44CH2CH2SAc (5)
TsCl (190 mg, 1.0 mmol) was added to a solution of an uncapped PEG diol (2.0 g, average molecular weight 2000 Da, 1.0 mmol) in pyridine (15 mL). The resulting mixture was stirred at 0 °C for 12 h. The precipitate was removed by filtration and the solvent of the filtrate evaporated. The residue and potassium thioacetate (0.26 g, 2.0 mmol) in anhydrous DMF (5 mL) was stirred at 80 °C for 4 h. DCM (25 mL) was added and the resulting solution was washed with water (5 mL). The organic layer was dried (MgSO4), and the solvents were removed in vacuo. The residue was dissolved in pyridine (15 mL) and TsCl (190 mg, 1.0 mmol) was added. The resulting mixture was stirred at 0 °C for 12 h. Next, the mixture was filtered and the residue was concentrated under reduced pressure. The residue and sodium azide (130 mg, 2.0 mmol) in anhydrous DMF (5 mL) were stirred at 80 °C for 8 h. CH2Cl2 (50 mL) was added and the resulting solution was washed with water (5 mL). The organic layer was dried (MgSO4) and the solvents removed under reduced pressure. The crude product was purified by silica gel chromatography (DCM: CH3OH 10:1) to afford 5 (43% over four-steps). 1H NMR (CDCl3, 300 MHz) δ : 3.87 (m, 2H), 3.83–3.60 (m, 80H), 3.41 (m, 2H), 2.07 (s, 3H).
2.3. Synthesis of N3(CH2CH2O)44CH2CH2SH (6)
NaOCH3 in CH3OH (1M) was added to a solution of 5 (0.4 mmol) in CH3OH to adjust the pH value to 10, under an atmosphere of Ar. The resulting reaction mixture was stirred at room temperature for 5h. The solvent was evaporated and the residue was purified by silica gel chromatography (CH2Cl2:CH3OH, 8:1) to afford 6 (78%). 1H NMR (CDCl3, 300 MHz) δ : 3.87 (m, 2H), 3.83–3.60 (m, 80H), 3.31 (m, 2H).
2.4. Synthesis of 1-(5-[1,2]dithiolan-3-yl-pentanoyl)-pyrrolidine-2,5-dione (LA-NHS) (8)
N-hydroxysuccinimide (0.310 g, 2.7 mmol) and DCC (0.56 g, 2.7 mmol) was added to a stirred solution of thioctic acid (0.5 g, 2.6 mmol) in dioxane (10 mL) cooled on an ice bath. The resulting mixture was stirred at room temperature for 48 hours and then filtered through a pad of Celite. The filtrate was concentrated in vacuo and the obtained residue was re-dissolved 2-propanol (5 mL) and left overnight at 4 °C, to give a white precipitate which filtered off, dried in vacuo to give 8 (57%). 1H NMR (DMSO) δ : 2.78 (t, 4H), 2.42–2.51 (m, 3H), 2.29 (t, 2H), 1.84 (m, 2H), 1.37–1.62 (m, 4H), 1.25 (m, 2H), 13C NMR (DMSO) δ : 176.0, 166.4, 53.7, 42.9, 37.3, 36.8, 32.6, 25.9, 24.1, 21.3.
2.5. Synthesis of pentynoic acid 2,5-dioxo-pyrrolidin-1-yl ester (Alkyne-NHS) (10)
N,N-dicyclohexyl-carbdiimide (DCC) (0.42 g, 2.2 mmol) was added to a stirred solution of 4-Pentynoic acid (0.2 g, 2 mmol) and NHS (0.25 g, 2.2 mmol) in CH2Cl2 (10 mL). The resulting mixture was stirred at room temperature overnight and was then filtered through a pad of Celite. The filtrate was concentrated in vacuo and the resulting residue was purified by silica gel chromatography (hexane: ethyl acetate = 2: 1) to afford 10 (85%). 1H NMR (500 Hz, CDCl3) δ : 2.88–2.83 (m, 6 H), 2.60 (td, J1 = 2.4 Hz, J2 = 7.8 Hz, 2 H), 2.04 (t, J = 2.4 Hz, 1 H).
2.6. Alkyne modification of Anti-Ricin Ab
Commercial anti-ricin Ab in a PNS buffer (200 µL, 1 mg/mL) was dialyzed using a regenerated dialysis membrane (MWCO:3500 Da, two Quick Seps) and purified water (18.2 MΩ) at 0 °C for 3~5 hours. Salt removal was deemed important to avoid possible inference with the conjugation reaction. The resulting anti-ricin Ab (200 µL) was mixed with active ester 8 (200 µL, approx 1mg/mL) in dimethyl sulphoxide/CH2Cl2 (1:1). The mixture was stirred at 0 °C for 1 hour. Next, the solution was washed with ethyl acetate (3 × 400 µL) and the aqueous solution was placed in Quick Seps and dialyzed against purified water at 0 °C for 3~5 hours.
2.7. Functionalization of AFM tip
Tips were coated with 20~30 nm of gold using an E-beam evaporator and then immersed in HS–PEG(2000)–N3 (6) in chloroform (5 mg/mL) at 4 °C for 10~12 h to form a compact SAM 27. The PEG modified tips were washed with dimethyl sulphoxide followed by purified water three times. The tips were placed into alkyne modified antibody solution (200 µL, 0.1 mg/mL) and then sodium ascorbate (3 µL, 1 M) and copper (II) sulfate (1µL, 0.3 M) in water were added to the solution 28 and the click reaction was allowed to perform for 10~12 hours in the dark at 0 °C. Finally, the tips were washed by PNS buffer three times and stored in PNS buffer at 4 °C.
2.8. Ricin immobilized on the functionalized gold substrate
A fresh thermal gold-coated mica substrate was annealed by a hydrogen flame for 2 min, and then active ester 10 in dichloromethane (200 µL, 5mg/ml) was immediately added to the surface. The substrate was kept at 4 °C for 3 hours 29. Following a cleaning by dimethyl sulphoxide and purified water, 200 µL (24 fg/mL) of ricin solution were put on the surface. The mixture was maintained at 4 °C for 30 min.
2.9. AFM experimental setup
An Agilent 5500 AFM system equipped with an inverted light microscope (ILM) system (Agilent, Chandler, AZ) was used. An Agilent multi-purpose AFM scanner was used for scanning an area of 10 µm2. Silicon cantilever tips with a nominal spring constant of about 0.1 N/m were used throughout the experiments. All the images were taken in water using single-molecule recognition imaging microscopy 20 based on Agilent magnetic AC (MAC) mode AFM with a magnetically-coated lever. The detailed parameters for the image are as follows: drive is approximately 46%, resonance gain is 2, resonance frequency is 12.95 KHz, resonance amplitude is 1.68 v, and scan rate is 1.22 line/s.
3. RESULTS AND DISCUSSION
3.1. Tip and substrate modification
A novel approach for AFM tip modification has been developed employing bifunctional PEG derivative 6. The thiol moiety of 6 allows convenient attachment to a Au modified tip because of the high binding energy of the Au-S bond which ranges from 20–35 kcal/mol (85–145 kJ/mol) and its ability to form compact monolayers 29. The azido moiety of the resulting SAM can then be employed in click reactions with biomolecules modified with alkynes.
The bifunctional PEG derivative 6 was prepared by a convenient four-step procedure in an overall yield of 43%. Thus, commercially available PEG diol 1 (2,000 Da) was desymmetrized by reaction with one equivalent of p-toluenesulfonyl chloride in pyridene to give a monotosylate 2, which could easily be separated from starting material and disubstituted product using silica gel column chromatography (Scheme 1). The resulting monotosylate 2 was converted into thioacetate 3 by nucleophilic displacement with the potassium salt of thioacetic acid in DMF. The alcohol of 3 was tosylated using standard conditions to give 4, which was subjected to a displacement reaction with sodium azide in DMF to yield compound 5. Prior to SAM formation, the thioacetyl ester of 5 was saponified using sodium methoxide to give target compound 6.
An anti-ricin Ab was modified by alkyne groups by aminolysis with NHS ester of 4-pentynoic acid (8). The NHS ester could easily be prepared by the esterification of the carboxylic acid with hydroxysuccinimide using EDC (Scheme 1). Alkyne modified anti-ricin Ab was then attached to the SAM using a Cu (I) catalyzed reaction between azide of tip and alkyne of the toxin.
For ricin immobilization to gold-coated mica substrate surface, novel compound 10 was employed which contains a thioctic acid moiety for immobilization to the gold surface and an activated ester for capture of proteins. The thioctic was deemed attractive because it forms more stable complexes with gold with two Au-S bond compared to monothiol ligands 29. Compound 10 was easily prepared in good yield by the esterification of the carboxylic acid of 9 with NHS using DCC (Scheme 1).
3.2. Ricin detection
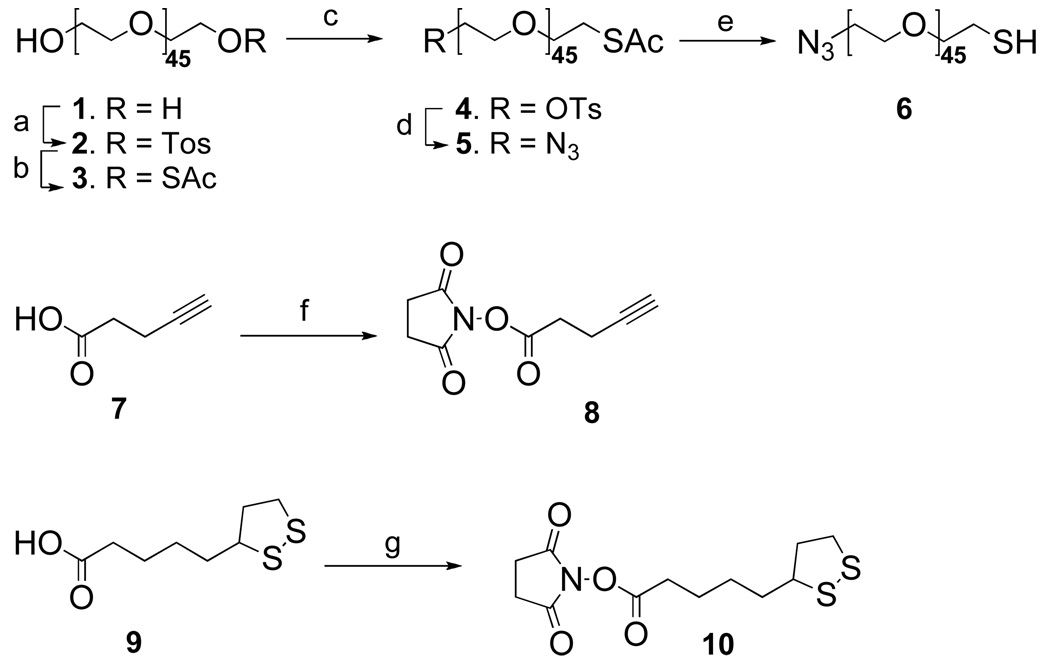
During scanning, the anti-ricin antibody functionalized tip interacts stronger with ricin immobilized to gold modified mica than with other parts of the surface. Therefore, in the recognition image, the ricin molecules are visualized as black dots. The scanned topography image was simultaneously recorded to provide further proof of detection. By using this method, one could not only detect the presence of the ricin, but also estimate the quantity of the ricin. Thus, 200 µL of a ricin solution (24 fg/mL) was kept on the antibody modified gold surface for 30 min; then the chip was placed into the SMRFM system for scanning. Clear topography, amplitude, and recognition images were obtained (Figure 3A-C, respectively). As can be seen in Figure 3, each ricin molecule has a spherical shape (dimensions 20~30nm). By comparing the topographical and recognition images, we could easily identify which feature belonged to a ricin molecule: each bright spot in the topographic image, if represents a ricin molecule, will result in a dark spot in the recognition image at the same location. Based on the obtained results, it was concluded that the detection method based on SMRFM technique could easily be achieved in the femto- or sub femto-level. Compared with other detection methods, the new method is easy and faster, and offers a significant advantage in both sensitivity and resolution.
Figure 3.
(A) Topographical image for 24 fg/mL; (B) Amplitude image for 24 fg/mL; (C) Recognition image for 24 fg/mL;
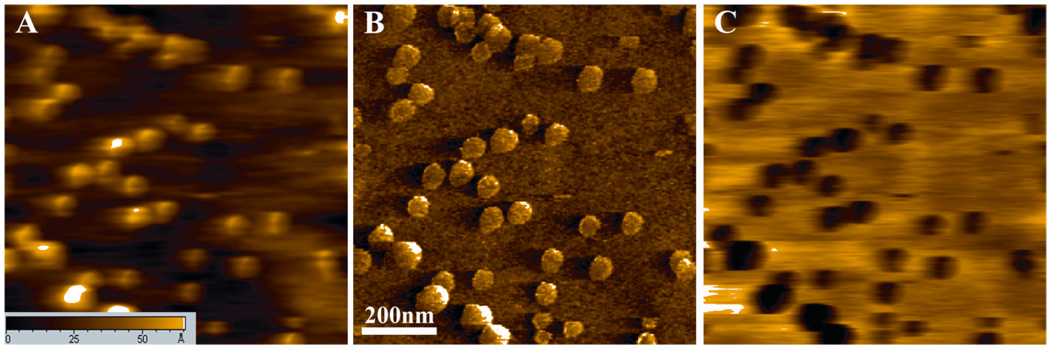
To confirm the detection results, a control experiment was carried out adopting a flow-cell technique 20. When the SMRFM system was stable, 200 µL of ricin protein (24 pg/mL) was injected into the liquid cell using an inlet syringe. Meanwhile, 200 µL of water in the liquid cell was drawn out by the outlet syringe to prevent the liquid in the cell from overflowing. The real time changes in the recognition image were monitored at the same location. It was observed that features were still clear in the topography image (Figure 4, A, C). However, the corresponding black dots fully disappeared in the recognition image (Figure 4 B, D). This observation can be explained by the binding of the injected ricin molecules to the anti-ricin Ab on the tip and blocking the interaction between the anti-ricine Ab and the ricin on the surface. These results strongly support that the detection results (Figure 3) were reliable and indicate that SMRFM is a powerful technique to provide insight into molecular binding events.
Figure 4.
Topographic image before (A) and after (C) the blocking of antibodies on AFM tip and the corresponding recognition image before (B) and after (D) the blocking.
3.3. Binding Force study
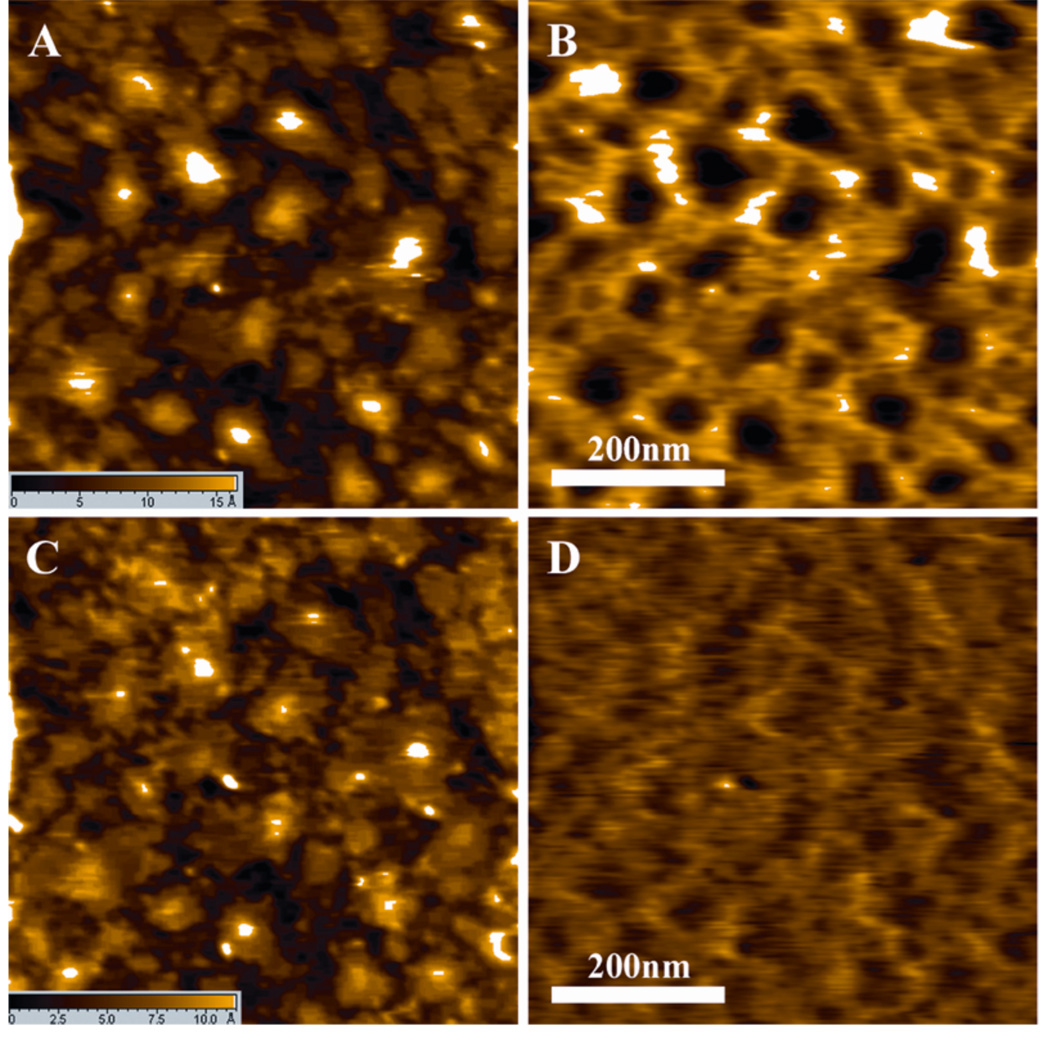
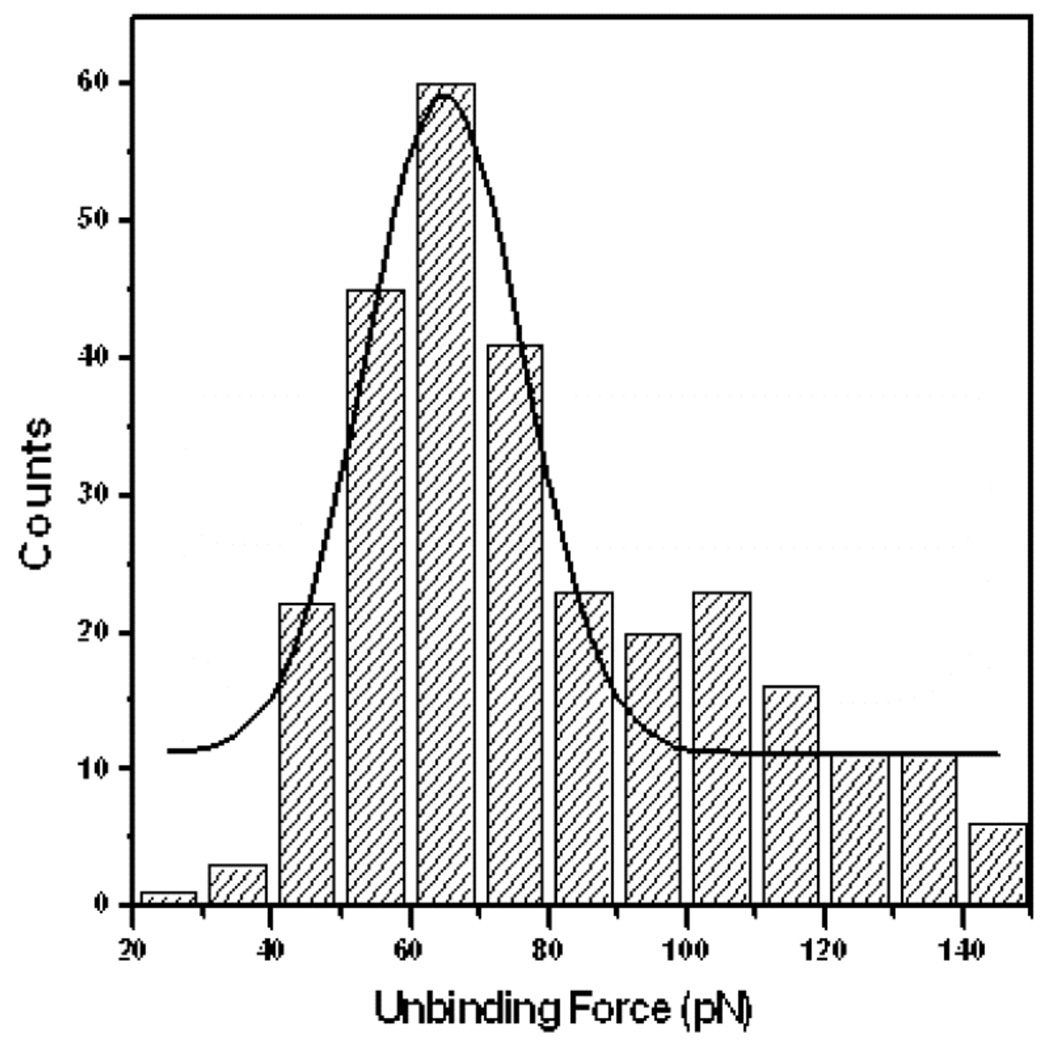
In addition to images, the SMRFM technique can also provide force measurements of pico-Newton accuracy. Therefore, during the detection of ricin, the unbinding force was also measured through recording the force-distance curve (F-D curve). To measure the unbinding force as accurately as possible, the spring constant of the AFM cantilever was calibrated through the resonance frequency changes which were induced by small mass 30. The force measurement was accomplished under the MAC mode to ensure that the tip was directly positioned above the ricin molecules. The typical unbinding force in relation to the distance curves is shown in Figure 5. The measured force-distance curves agreed with a one-barrier bond dissociation model 31,32. Figure 5 shows that the typical F-D curves have small variations little bit, which are probably due to microscopic complexity. To determine accurately the unbinding force, the force histogram was built from 3000 F-D curves and Gaussian-fitting was applied to identify the peak values (Figure 6), which gave an expected unbinding force of 64.89±1.67 pN.33.
Figure 5.
The typical unbinding force versus distance curves
Figure 6.
The unbinding force distribution and its Gaussian fitted curve (black solid line)
4. CONCLUSIONS
A new and efficient protocol for tip functionalization for SMRFM has been developed using a PEG derivative containing a thiol and azide end group. This bifunctional linker could be employed for SAM formation on a gold modified tip and the azides of the resulting SAM could be exploited for the immobilization of an anti-ricin Ab modified by alkynes using a click reaction. The high efficiency, mild reaction condition, and simple operation of the click reaction greatly simplified the tip modification process. Furthermore, ricin could be immobilized on a gold-coated mica substrate by employing another novel bifuntional composed of a N-hydroxysuccinimidyl ester and a thioctic acid moiety. The thiotic acid moiety of this reagent binds strongly with a gold-coated mica substrate and the active esters of the resulting surface can then be reacted with amines of the protein. Using the immobilization approach, qualitative and quantitative information could be easily obtained from the images attained by the SMRFM system. Compared with other detection methods, this approach has significantly higher sensitivity (sub-femtogram level), a faster responses, and easier operation protocol. Blocking experiment confirmed the detection results. Therefore, we expected this SMRFM technique to be an attractive method for single molecule toxin detection as well as for studies of molecular recognition events.
The approach also allowed the measurement of the unbinding force of the Ab - ricin interaction. The obtained F-D curves were consistent with the well-established one-barrier bond dissociation model 31,32. The average unbinding force obtained from the histogram was 64.89±1.67 pN, which is in good agreement with values for antibody and antigen unbinding events. Therefore this SMRFM technique has provided a detailed understanding of antigen-antibody interactions.
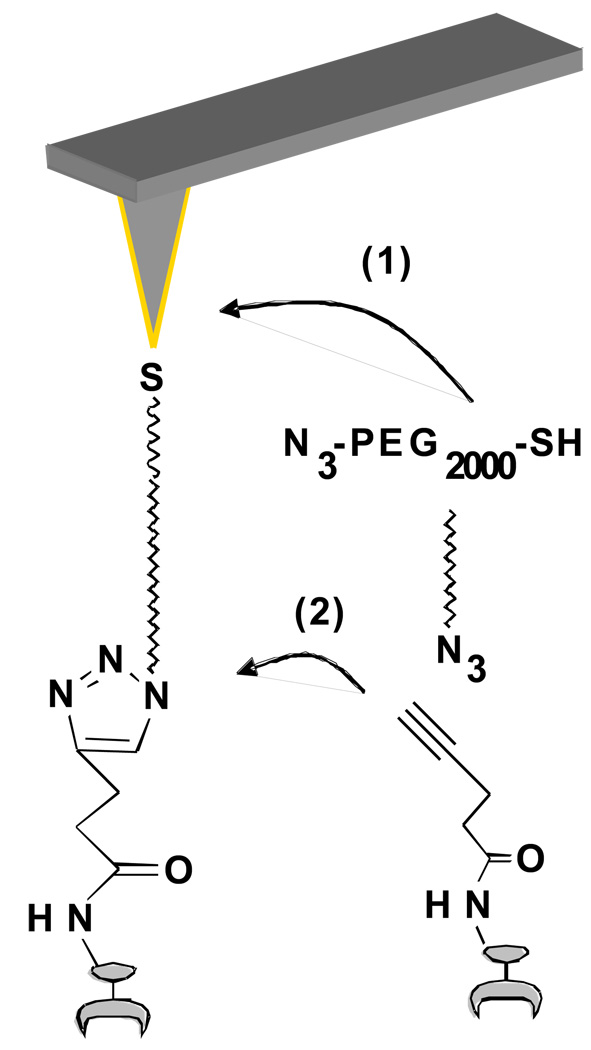
Figure 1.
Schematic representation of AFM tip modification. (1) The thiol moiety of PEG2000 formed a SAM on the gold-coated tip; (2) the anti-ricin Ab attached to the tip through click chemistry with an alkyne modified protein.
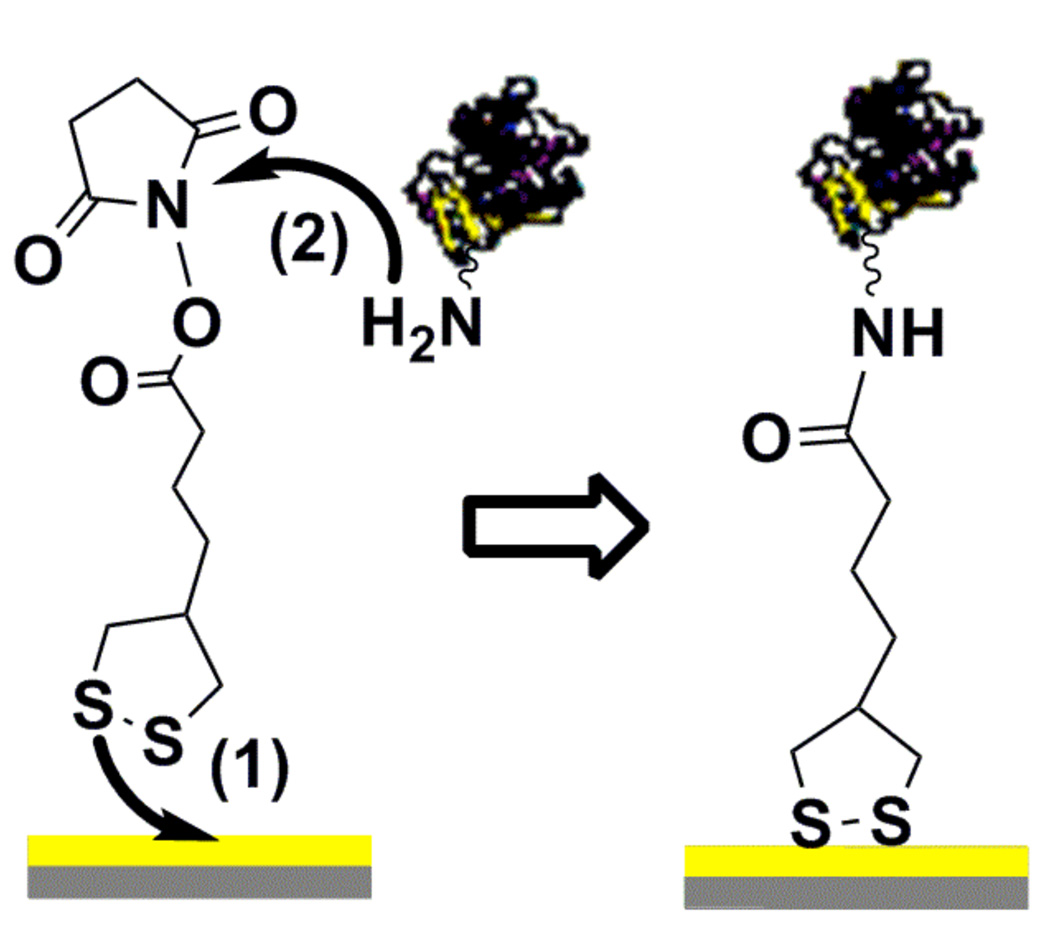
Figure 2.
Schematic representation of ricin immobilization on the gold surface. (1) active ester 10 attached to the gold coated mica surface by the thioctic acid moiety; (2) ricin binds to active ester 10 by forming amide bond with amine groups of the protein.
ACKNOWLEDGEMENT
We thank Dr. Anna Jagielska for helpful discussions. Bingqian Xu acknowledges University of Georgia UGARF faculty research fund and the National Science Foundation (ECCS-0823849) for financial support. Geert-Jan Boons acknowledges support by the Research Resource Center for Biomedical Complex Carbohydrates (P41-RR-5351).
Footnotes
Key Words and Abbreviations: SMRFM, single molecular recognition force microscopy; AFM, atomic force microscopy; PEG, poly ethylene glycol; SAM, self-assembled monolayer;
REFERENCES
- 1.Binnig G, Quate CF, Gerber C. Phys. Rev. Lett. 1986;56:930. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 2.Morris VJ, Kirby AR, Gunning AP. Atomic Force Microscopy for Biologists. London: Imperial College Press; 2004. [Google Scholar]
- 3.Zlatanova J, Lindsay SM, Leuba SH. Prog. Biophys. Mol. Biol. 2000;74:37–61. doi: 10.1016/s0079-6107(00)00014-6. [DOI] [PubMed] [Google Scholar]
- 4.Janshoff A, Neitzert M, Oberdorfer Y, Fuchs H. Angew. Chem. Int. Ed. 2000;39:3213–3237. doi: 10.1002/1521-3773(20000915)39:18<3212::aid-anie3212>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 5.Allen S, Rigby-Singleton SM, Harris H, Davies MC, O'Shea P. Biochem. Soc. Trans. 2003;31:1052–1057. doi: 10.1042/bst0311052. [DOI] [PubMed] [Google Scholar]
- 6.Ebner A, Kienberger F, Kada G, Stroh CM, Geretschlager M, Kamruzzahan ASM, Wildling L, Johnson WT, Ashcroft B, Nelson J, Lindsay SM, Gruber HJ, Hinterdorfer P. Chemphyschem. 2005;6:897–900. doi: 10.1002/cphc.200400545. [DOI] [PubMed] [Google Scholar]
- 7.Olsnes S, Kozlov JV. Toxicon. 2001;39:1723–1728. doi: 10.1016/s0041-0101(01)00158-1. [DOI] [PubMed] [Google Scholar]
- 8.Lord JM, Roberts LM, Robertus JD. FASEB J. 1994;8:201–208. [PubMed] [Google Scholar]
- 9.Lubelli C, Chatgilialoglu A, Bolognesi A, Strocchi P, Colombatti M, Stirpe F. Anal. Biochem. 2006;355:102–109. doi: 10.1016/j.ab.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Ler SG, Lee FK, Gopalakrishnakone P. J. Chromatogr. A. 2006;1133:1–12. doi: 10.1016/j.chroma.2006.08.078. [DOI] [PubMed] [Google Scholar]
- 11.Ramakrishnan S, Eagle MR, Houston LL. Biochim. Biophys. Acta, Gen. Subj. 1982;719:341–348. doi: 10.1016/0304-4165(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 12.Koja N, Shibata T, Mochida K. Toxicon. 1980;18:611–618. doi: 10.1016/0041-0101(80)90088-4. [DOI] [PubMed] [Google Scholar]
- 13.Poli MA, Rivera VR, Hewetson JF, Merrill GA. Toxicon. 1994;32:1371–1377. doi: 10.1016/0041-0101(94)90409-x. [DOI] [PubMed] [Google Scholar]
- 14.Hines HB, Brueggemann EE. J. Chromatogr. A. 1994;670:199–208. [Google Scholar]
- 15.Krutchinsky AN, Zhang W, Chait BT. J. Am. Soc. Mass. Spectrom. 2000;11:493–504. doi: 10.1016/S1044-0305(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 16.Rubina AY, Dyukova VI, Dementieva EI, Stomakhin AA, Nesmeyanov VA, Grishin EV, Zasedatelev AS. Anal. Biochem. 2005;340:317–329. doi: 10.1016/j.ab.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 17.Gabai R, Segev L, Joselevich E. J. Am. Chem. Soc. 2005;127:11390–11398. doi: 10.1021/ja051642v. [DOI] [PubMed] [Google Scholar]
- 18.Barattin R, Voyer N. Chem. Commun. 2008:1513–1532. doi: 10.1039/b614328h. [DOI] [PubMed] [Google Scholar]
- 19.Kienberger F, Ebner A, Gruber HJ, Hinterdorfer P. Acc. Chem. Res. 2006;39:29–36. doi: 10.1021/ar050084m. [DOI] [PubMed] [Google Scholar]
- 20.Stroh C, Wang H, Bash R, Ashcroft B, Nelson J, Gruber H, Lohr D, Lindsay SM, Hinterdorfer P. Proc. Nat. Acad. Sci. U.S.A. 2004;101:12503–12507. doi: 10.1073/pnas.0403538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riener CK, Stroh CM, Ebner A, Klampfl C, Gall AA, Romanin C, Lyubchenko YL, Hinterdorfer P, Gruber HJ. Anal. Chim. Acta. 2003;479:59–75. [Google Scholar]
- 22.Yam CM, Xiao Z, Gu J, Boutet S, Cai C. J. Am. Chem. Soc. 2003;125:7498–7499. doi: 10.1021/ja0350435. [DOI] [PubMed] [Google Scholar]
- 23.Berquand A, Xia N, Castner DG, Clare BH, Abbott NL, Dupres V, Adriaensen Y, Dufrene YF. Langmuir. 2005;21:5517–5523. doi: 10.1021/la050162e. [DOI] [PubMed] [Google Scholar]
- 24.Nandivada H, Jiang XW, Lahann J. Adv. Mat. 2007;19:2197–2208. [Google Scholar]
- 25.Moses JE, Moorhouse AD. Chem. Soc. Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 26.Kolb HC, Sharpless KB. Drug Discovery Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 27.Bain CD, Whitesides GM. Science. 1988;240:62–63. doi: 10.1126/science.240.4848.62. [DOI] [PubMed] [Google Scholar]
- 28.Hartmuth C, Kolb MGF, Barry Sharpless K. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Ulman A. Chem. Rev. 1996;96:1533–1554. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 30.Cleveland JP, Manne S, Bocek D, Hansma PK. Rev. Sci. Instrum. 1993;64:403–405. [Google Scholar]
- 31.Evans E. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 32.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 33.Novotny J, Bruccoleri RE, Saul FA. Biochem. 1989;28:4735–4749. doi: 10.1021/bi00437a034. [DOI] [PubMed] [Google Scholar]