Abstract
MyD88, IRAK4 and IRAK2 are critical signaling mediators of the TLR/IL1-R superfamily. Here we report the crystal structure of the MyD88: IRAK4: IRAK2 death domain (DD) complex, which surprisingly reveals a left-handed helical oligomer that consists of 6 MyD88, 4 IRAK4 and 4 IRAK2 DDs. Assembly of this helical signaling tower is hierarchical, in which MyD88 recruits IRAK4 and the MyD88: IRAK4 complex recruits the IRAK4 substrates IRAK2 or the related IRAK1. Formation of these Myddosome complexes brings the kinase domains of IRAKs into proximity for phosphorylation and activation. Composite binding sites are required for recruitment of the individual DDs in the complex, which are confirmed by mutagenesis and previously identified signaling mutations. Specificities in Myddosome formation are dictated by both molecular complementarity and correspondence of surface electrostatics. The MyD88: IRAK4: IRAK2 complex provides a template for Toll signaling in Drosophila and an elegant mechanism for versatile assembly and regulation of DD complexes in signal transduction.
Toll-like receptors (TLRs) and receptors for pro-inflammatory cytokines IL-1 and IL-18 share a common TIR domain in their intracellular region and belong to the TLR/IL1-R superfamily 1,2. TLRs recognize pathogen-associated molecular patterns (PAMPs) to initiate protective immune responses 3,4. The molecular pathways for these receptors are complex and their dysregulation is associated with many human diseases in the immune system such as inflammatory disorders 5,6, autoimmune diseases 7–10 and allergy 11 as well as diseases beyond the immune system such as cancer 12, insulin resistance 13, atherosclerosis 14,15, and painful neuropathy 16.
Signal transduction is initiated by the approximation of the receptor TIR domains upon binding of PAMPs and cytokines 1. This leads to the recruitment of intracellular TIR-containing adaptors such as MyD88, Mal/TIRAP, TRIF and TRAM 17. MyD88 is critical for signaling responses of IL-1, IL-18, and all TLRs except TLR33,17–19. In addition to its C-terminal TIR domain, MyD88 contains an N-terminal death domain (DD) and a short intermediate domain (ID). Through the DD, MyD88 interacts with IRAKs, including IRAK1, IRAK2, IRAK4 and IRAK-M, which are characterized by an N-terminal DD and a C-terminal Ser/Thr kinase or kinase-like domain 3,20,21. Eventually, the ensuing pathway activates transcription factors NF-κB, AP-1, and IRFs to elicit anti-pathogen responses and inflammation 3,17,22. Targeted deletions in mice have identified IRAK1, IRAK2 and IRAK4 as essential positive players and IRAK-M as a critical negative regulator in the pathway 23–26. In humans, inherited MyD88 and IRAK4 deficiencies cause recurrent, often life-threatening infections by pyogenic bacteria 27,28.
Structure determination
To elucidate the molecular basis of DD-mediated TLR/IL-1R signaling, we co-expressed DDs of MyD88 and IRAKs. MyD88 DD interacted with IRAK4 DD to form a binary Myddosome complex that eluted from a gel filtration column with a molecular mass of 135.4 kDa (0.5% error) as measured by multi-angle light scattering (MALS) (Supplementary Fig. 1). Furthermore, a ternary Myddosome complex that also contains IRAK2 DD were formed and eluted with an apparent molecular mass of ~180 kDa.
We crystallized the MyD88: IRAK4: IRAK2 DD complex and determined its structure at 3.4 Å resolution using multiple heavy atom derivatives that included Au, Hg, Pt, Se and Ta cluster (Supplementary Table 1, Supplementary Fig. 2). Because the IRAK4 DD structure is known 29, we used it as a model to identify 14 DD structures in the experimental electron density map. We then distinguished the DD molecules as MyD88, IRAK4 and IRAK2 using the Se positions (Supplementary Table 2). The final atomic model comprises 6 MyD88, 4 IRAK4, and 4 IRAK2 DD molecules. The observed stoichiometries in the crystal agree well with the measured and the apparent molecular masses of the binary MyD88: IRAK4 and the ternary MyD88: IRAK4: IRAK2 complexes (Supplementary Discussion 1), the gross staining intensities of the DDs in the complexes on SDS-PAGE (Supplementary Fig. 3) and a previous study showing the 6:4 stoichiometry when the MyD88: IRAK4 complex became slightly “stripped” under a mild mass spectrometry condition 30.
Overall structure of the complex
The ternary Myddosome complex forms a tower-shaped structure of about 110 Å in height and about 70 Å in diameter (Fig. 1a). It contains approximately four layers, with MyD88 at the bottom two layers, IRAK4 in the middle layer and IRAK2 at the top layer. The center of the MyD88 layers has a sizable cavity (Supplementary Fig. 4), in agreement with the prominent depression in the EM structure of the MyD88: IRAK4 complex 30.
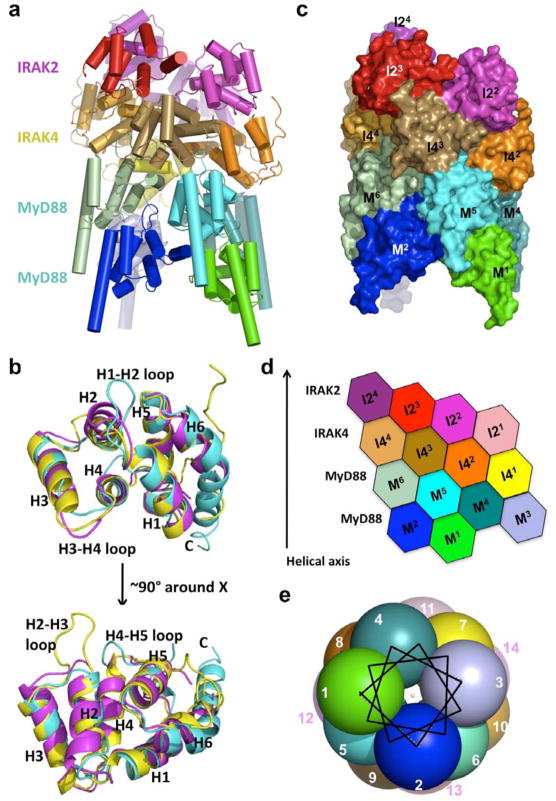
Figure 1. Structure of the ternary Myddosome complex.
a, A ribbon diagram of the structure, with the 6 MyD88 molecules in cold colors, the 4 IRAK4 molecules in earth-tone colors and the 4 IRAK2 molecules in warm colors. b, Superposition of MyD88 DD (cyan), IRAK4 DD (yellow) and IRAK2 DD (magenta). Helices H1 to H6 and the H1-H2, H2-H3 and H4-H5 loops are labeled. c, Surface diagram of the complex with each subunit labeled using the same color coding as in a. M: MyD88; I4: IRAK4; I2: IRAK2. d, Planar arrangement of the complex. e, The helical symmetry is shown in a helical wheel representation with each ball representing a molecule and looking down the helical axis. Each molecule is labeled as an integer sequentially from M1 to I24.
The six helical bundle structures 31 of MyD88, IRAK4 and IRAK2 DDs containing helices H1 to H6 each have their distinct features (Fig. 1b) with MyD88 being the most dissimilar (Supplementary Table 3). MyD88 has a short H3 and an extraordinarily long H6 from residue 99 to the end of the construct at residue 120, which includes part of the intermediate domain (ID, residues 110–154). MyD88s, a splice variant of MyD88 that does not contain the ID, did not interact with IRAK4 nor activate NF-κB 32,33. Our structure suggests that the DD boundary of MyD88 does not end until about residue 120 and deletion of the MyD88 ID would result in a truncated DD. For the loop regions, MyD88 has the longest H1-H2 loop, shortest H3-H4 loop and the longest H4-H5 loop, and IRAK4 has the longest H2-H3 loop. The MyD88 structure explains the disruptive phenotypes of mutations ΔE52 and L93P in children suffering from life-threatening pyogenic bacterial infections 28 (Supplementary Fig. 5).
The ternary complex forms a helical oligomer
To our surprise, the MyD88: IRAK4: IRAK2 complex is a single stranded left-handed helix of DDs, starting from the six molecules of MyD88 (M1–6), continuing with the four molecules of IRAK4 (I41–4) and ending with the four molecules of IRAK2 (I21–4) (Fig. 1c, 1d). In the helical oligomer, the adjacent individual DD molecules are related by a rotation of 98 ± 2 ° and a translation along the helical axis of about 6 Å. This approximate helical symmetry provides a quasi-equivalent environment for each DD molecule in the ternary complex. The DD molecules complete almost four turns, with approximately 3.7 DD molecules per turn.
If we cut open the structure from the side and lay the molecules flat, the locations of the DDs form a staggered hexagonal pattern (Fig. 1d). Each molecule has maximally six immediate neighboring DDs in this two-dimensional representation. This remains true even in the rolled up three-dimensional structure, because a given DD has limited contacts with DDs beyond the immediate vicinity. The helical assembly can also be represented in a similar helical wheel used to show amphipathic α-helices in protein structures (Fig. 1e). The nature of a helical assembly dictates that molecules at either ends of the helix are less ordered or perhaps exhibit less occupancy than the central molecules. This can be shown by the temperature factor distribution in the ternary complex (Supplementary Fig. 6).
Composite binding sites and interaction specificity
The interactions in the helical assembly can be classified into three types. Type I and type II mostly mediate interactions between the layers while type III represents contacts between adjacent DD molecules in the helical spiral (Fig. 2a–2e, Supplementary Fig. 7). Superposition of the three type I interaction pairs, MyD88: MyD88, MyD88: IRAK4, and IRAK4: IRAK2, showed that they are highly similar to one another (Supplementary Fig. 8). Likewise, the three kinds of type II interaction pairs and the five kinds of type III interaction pairs superimpose well (Supplementary Fig. 8). The type I interaction is formed by H2 and H3 on one DD and H1 and H4 region on the other DD. The type II interaction is formed from two opposite edges of the DDs, centered around the H4-H5 loop on one DD and around the H1-H2 loop on the other DD. The type III interaction is formed between H3 of one DD and an edge of the other DD composed of the H1-H2 and H3-H4 loops. The type I and type II interactions are more extensive in buried surface areas than the type III interaction (Supplementary Fig. 7).
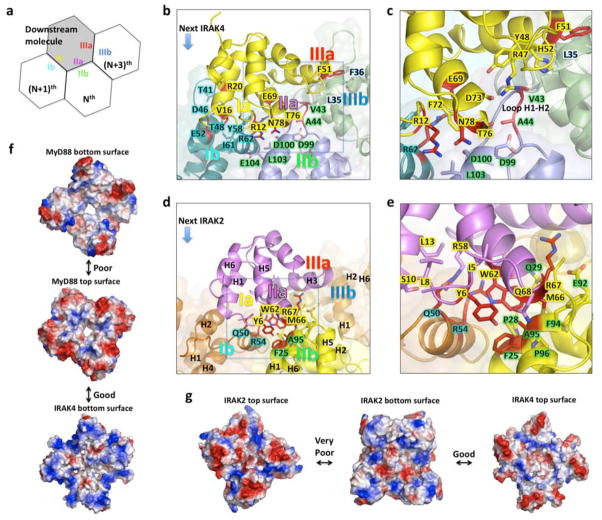
Figure 2. Composite interactions and specificity in the ternary complex.
a, Schematic representation of a composite binding site. The Nth, (N+1)th, and (N+3)th molecules provide type IIb, Ib and IIIb surfaces to interact with the type IIa, Ia and IIIa surfaces of a downstream molecule, respectively. b, A composite binding site for IRAK4 (yellow) formed from three MyD88 molecules (M3, M4, and M6) through type I, II and III interactions. c: Enlargement of the square in b. Note that the unique H1-H2 loop of MyD88 is critical for Myddosome assembly. d, A composite binding site for IRAK2 (purple) formed from three IRAK4 molecules (I41, I42, and I44) through type I, II, and III interactions. e, Enlargement of the square in d. f, Good charge complementarity between IRAK4 and MyD88 and poor charge complementarity between the top and bottom surfaces of MyD88. g, Good charge complementarity between IRAK2 and IRAK4 and very poor charge complementarity between the top and bottom surfaces of IRAK2.
During signaling, MyD88 molecules are first recruited to activated receptors and oligomerized. This then forms the “seeds” for inducing assembly of the complex. Recruitment of a downstream molecule requires direct interaction with the composite binding site of 3 molecules via all three types of interactions, type II with an Nth molecule, type I with an (N+1)th molecule and type III with an (N+3)th molecule (Fig. 2a). For example, the first IRAK4 molecule (I41) interacts with three MyD88 molecules (M3, M4 and M6) simultaneously during its recruitment (Fig. 2b, 2c), explaining the requirement of MyD88 oligomerization in signal-dependent IRAK4 recruitment. We confirmed the importance of all three sites in this IRAK4: MyD88 interaction using structure-based mutagenesis, which identified R12, V16, R20, E69, T76 and N78 of IRAK4 (Supplementary Fig. 9a, 9b), and V43, A44, E52, Y58, I61 and R62 of MyD88 (Supplementary Fig. 9a, 9c) as critical residues. The importance of MyD88 residues E52 and Y58 is consistent with previous mutational studies of MyD88 in IRAK4 recruitment and NF-κB signaling 34. Similarly, recruitment of the first IRAK2 molecule would require interaction with the composite binding site of three IRAK4 molecules (I41, I42, I44) through type I, II, and III interfaces (Supplementary Fig. 9a, 9d, 9e). The importance of these sites in the IRAK2: IRAK4 interaction were also confirmed by structure-based mutagenesis, which identified Y6, W62, M66 and R67 of IRAK2 (Supplementary Fig. 9d) and F25, Q50, F51, R54, and A95 of IRAK4 (Supplementary Fig. 9e) as critical residues.
The IRAK4: MyD88 and the IRAK2: IRAK4 interactions are quite different, providing the specificity in Myddosome formation. At the MyD88: IRAK4 interface, the uniquely long H1-H2 loop of MyD88, including residues V43 and A44, interacts with R47, Y48, H52, D73, and T76 of IRAK4 (Fig. 2b, 2c, Supplementary Fig. 7). There is no such corresponding interaction between IRAK2 and IRAK4. At the IRAK2: IRAK4 interface, there is a reciprocal interdigitation of the surfaces. Y6, W62 and M66 of IRAK2 insert into a region surrounded by F25, P28, Q29, Q50, R54, E92, F94, A95 and P96 of IRAK4. Conversely, Q50 and R54 of IRAK4 insert into a region surrounded by I5, Y6, L8, S10, L13, R58, E59 and W62 of IRAK2 (Fig. 2d, 2e, Supplementary Fig. 7). The equivalent inserted residues at the IRAK4: MyD88 interface are R12, F72 and T76 of IRAK4 and Y58 and R62 of MyD88, which are not only different but also much less interdigitating in the interaction.
The specificity in Myddosome formation may also be elucidated from analysis on the surface charge and shape complementarity between the layers of DD molecules. Inspection of the top and bottom surfaces of MyD88 layers suggested that the charge complementarity between these surfaces is weak (Fig. 2f). In addition, the shape complementarity scores (Sc) 35 between MyD88 DDs (such as the type II and type III interactions) are in the range of 0.3 and 0.4, the lowest among all the interactions in the complex (Supplementary Fig. 7). The relatively poor charge and shape complementarity may explain the observed variable stoichiometry of MyD88 in the binary Myddosome complex 30 (Supplementary Discussion 1).
In contrast to the weak complementarity between the top and bottom surfaces of MyD88, the bottom surface of IRAK4 matches well with the top surface of MyD88 both in charge complementarity (Fig. 2f) and in shape complementarity as shown in the Sc scores (Supplementary Fig. 7). The top surface of IRAK4 has good charge complementarity (Fig. 2g) and shape complementarity (Supplementary Fig. 7) with the bottom surface of IRAK2, resulting in specific recruitment of IRAK2. Because of the incompatibility between the top and bottom surfaces of IRAK2, only one layer of IRAK2 can assemble into the helical oligomer (Fig. 2g).
Hierarchical and sequential assembly
Our reconstitution experiments suggest the presence of hierarchy in formation of the helical signaling tower in the Myddosome (Supplementary Fig. 10). MyD88 DD and IRAK4 DD formed a stable complex while IRAK2 DD did not form a stable complex with either MyD88 or IRAK4 alone. Instead, IRAK2 DD interacts with the MyD88: IRAK4 complex. These data support the concept that IRAK4 recruitment to MyD88 is an event upstream to IRAK2 recruitment in the TLR/IL-1R signaling pathways. IRAK1 DD did not express in any of our reconstitution experiments; however previous data showed that IRAK4 and IRAK1 did not directly associate and that addition of MyD88 permitted assembly of a complex containing both IRAKs 32,36. In the ternary complex, it has been shown that IRAK4 auto-phosphorylation occurs first and that IRAK4 activation is critical for IRAK1 and IRAK2 phosphorylation and activation 36. In IRAK4-deficient mice, neither IRAK1 nor IRAK2 could be recruited to the TLR signaling complexes 24. While IRAK4 is absolutely required for MyD88-dependent signaling 24, IRAK1 and IRAK2 play somewhat redundant roles 25. Our sequence alignment showed that IRAK2 residues critical for interaction with the MyD88: IRAK4 complex are conserved in IRAK1 (Supplementary Fig. 9a), confirming the biological observation.
It has been shown that in solution, MyD88 DD can form oligomers under a high protein concentration in a reversible manner 30 while IRAK4 DD is a monomer in solution. This indicates that when brought into proximity during TLR/IL-1R signaling, the weak MyD88 oligomer would be stabilized and acts as an initial platform for IRAK4 recruitment and subsequently IRAK1 and IRAK2 recruitment (Fig. 3a). In turn, IRAK4 recruitment further stabilizes the MyD88: IRAK4 complex, a notion supported by our mutagenesis results. The IRAK4 residue F25 participates in both type II interaction with IRAK2 and type III interaction with its neighboring IRAK4. Unexpectedly, the F25D mutation not only impaired the ability of IRAK4 to pull down IRAK2, but also weakened the ability of IRAK4 to pull down MyD88 (Supplementary Fig. 9e), suggesting that a stabilized type III interaction among IRAK4 DDs is important for Myddosome assembly. A most possible mode of DD assembly during signaling is a sequential assembly starting from clustered MyD88 then to IRAK4, then to IRAK2. The composite binding sites provided by the initial segments of the helical oligomer would provide a platform for concerted recruitment of IRAK4 and IRAK2 DDs, in a highly cooperative “chain” reaction (Fig. 3a, Supplementary Discussion 2).
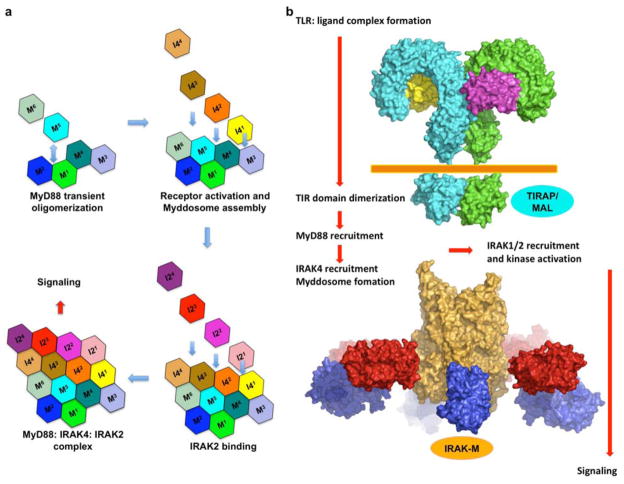
Figure 3. Model of sequential assembly upon TLR/IL1-R signaling.
a, Upon ligand binding, TLR/IL-1Rs, including the cytoplasmic TIR domains, are dimerized or oligomerized. This results in the recruitment of other TIR containing adaptor proteins. In this case, MyD88 is recruited to the receptor complex and the death domain is oligomerized. In the presence of IRAK4, the death domain of IRAK4 (I41) can be recruited to the oligomerized MyD88 DDs through the three interfaces (M3, M4, M6) and quickly forms the binary Myddosome complex. Downstream kinases (IRAK2 in this case, but IRAK1 as well) can then be recruited in a similar fashion and signaling is triggered. b, A model of the TLR receptor signaling complex that recruits the MyD88: IRAK4: IRAK2 complex with proteins drawn in scale. TLRs: cyan and green (PDB code 3FXI for the extracellular domain of TLR4 and PDB code 2J67 for the TIR domain of TLR10). MD2: yellow and magenta (PDB code 3FXI in complex TLR4). Orange: the MyD88: IRAK4: IRAK2 complex. Red: the IRAK4 kinase domain (PDB code 2NRU). Blue: the IRAK2 kinase domain using that of IRAK4.
If we place the ternary Myddosome complex next to the TIR domains of TLR/IL1-Rs with the C-terminal helices of MyD88 pointing towards the cellular membrane, the C-terminal TIR domains of MyD88 molecules would be poised to interact with clustered receptor TIR domains and other TIR-containing adaptors such as TIRAP/MAL (Fig. 3b). Given that 4 MyD88 molecules are minimally required to form one layer in the helical assembly, it is likely that dimerization of TLR/IL1-Rs per se is not efficient for inducing signaling. It has been shown that TLRs can be recruited to lipid rafts upon stimulation 37, which would favor higher order receptor oligomerization to facilitate Myddosome assembly. The clustered IRAK4 kinase domains can then undergo auto-phosphorylation. In the crystal structure of the ternary complex, IRAK4 DDs and IRAK2 DDs pack closely against each other. The C-terminal tail of IRAK4 DD points toward the IRAK2 DD layer, while the C-terminal tail of IRAK2 DD points toward the IRAK4 DD layer. This arrangement would then bring the kinase domains of IRAK4 to the proximity of IRAK1 or IRAK2 to allow efficient phosphorylation by IRAK4 (Fig. 3b). These observations explain why a splice variant of IRAK1 that does not have an intact kinase domain 38 and two splice isoforms of IRAK2 that lack functional DDs 39 are negatively regulators in TLR/IL-1R signaling. It has been shown that once phosphorylated, IRAK1 or IRAK2 leaves the Myddosome and interacts with TRAF6 to elicit Lys63-linked polyubiquitination and downstream signaling 40,41. We speculate that IRAK-M, a negative regulator of the TLR pathway, builds next to the IRAK2 layer of the ternary complex to prevent IRAK2 or IRAK1 dissociation and signaling.
Implications on formation of the dMyD88: Tube: Pelle complex
The mammalian TLRs are orthologues of the Drosophila Toll receptor, which is activated by the endogenous protein ligand Spätzle in response to microbial stimuli in immunity and spatial cues during embryonic development 42. Intracellular signaling of Toll is mediated by dMyD88, Tube, and the kinase Pelle 43. A recent study suggested that Tube, although not having a kinase domain, is a homologue for the mammalian IRAK444. Structural homology search using DALI 45 supports this conclusion because IRAK4 DD is most similar to Tube DD while IRAK2 DD is most similar to Pelle DD (Supplementary Table 4). Characterization of the DD interactions among these three Drosophila signaling proteins have also shown an unexpected parallel in the order of the assembly; formation of the dMyD88: Tube binary complex is the most stable (KD = 1.2 nM) and Pelle interacts much more preferentially with the dMyD88: Tube complex (KD = 51 nM) than with Tube alone (KD = ~0.5 μM) 46.
The organizational similarity prompted us to model the structure of the Drosophila Myddosome. Unlike the oligomeric MyD88: IRAK4: IRAK2 complex, the dMyD88: Tube: Pelle ternary complex in solution is 1: 1: 146. Superposition of the known structure of the Tube: Pelle DD complex 47 onto a pair of type II IRAK4: IRAK2 interaction showed that they superimpose well with a slight orientational difference (Fig. 4a), suggesting a conservation of this interaction between Drosophila and mammals. Superposition of Tube DD with IRAK4 DD in a type II IRAK4: MyD88 interaction pair created a Tube: MyD88 complex that simulates the Tube: dMyD88 interaction. Combining this model with the structure of the Tube: Pelle complex created an elongated molecule with MyD88 and Pelle at either side of Tube (Fig. 4b). The shape of the ternary complex model is similar to the kidney shaped outline of the dMyD88: Tube: Pelle complex derived from small angle X-ray scattering (SAXS) 46.
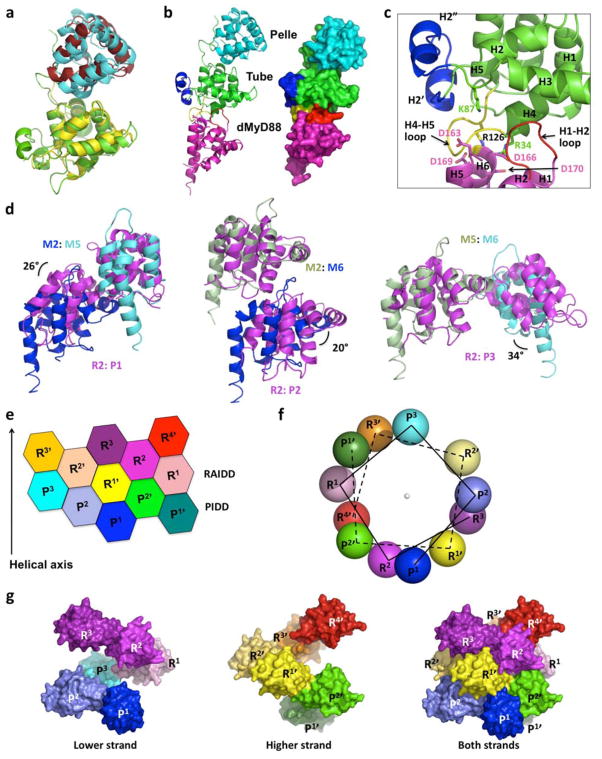
Figure 4. Common architecture in Drosophila Toll signaling and DD assembly in general.
a, Superposition of Tube: Pelle (green and cyan) and IRAK4: IRAK2 (yellow and brick red) complexes. b, A model of the dMyD88: Tube: Pelle complex in both ribbon and surface representations. Pelle is colored in cyan. Tube is colored in green except that the H2-H3 loop insertion (H2′ and H2″) is in blue and the H4-H5 loop is in yellow. The model of dMyD88, obtained based on the MyD88 DD structure but without modeling the insertion in the H1-H2 loop, is colored in magenta except that the H1-H2 loop is in red. c, Enlargement of the dMyD88: Tube interface. Residues important for dMyD88: Tube complex formation are shown in sticks. Note that the real H1-H2 loop of dMyD88 is 6 residues longer than that in the model. d, Superposition of the type I, II, and III interactions in the MyD88: IRAK4: IRAK2 and the PIDD: RAIDD complexes. The resultant angular differences are labeled. e, Planar arrangement of the PIDD: RAIDD complex. R: RAIDD; P: PIDD. Subunits in one strand of the double helix are labeled as R and P while those in the other strand are labeled R′ and P′. f. The helical symmetry is shown in a helical wheel representation with each ball representing a molecule and looking down the helical axis. Note that two helical strands are present in the complex. g, Surface representations of the two helical strands in the PIDD: RAIDD complex. The lower strand is about 5 Å lower than the higher strand, but they can be superimposed well. The lower strand (left) plus the higher strand (middle) equals the PIDD: RAIDD complex (right).
The validity of the ternary complex model is supported by published mutations, R34E, K87E and R126E of Tube and D113K, D163K, D166K, and D169K/D170K of dMyD88, that prevented interaction between Tube and dMyD8848. R34, K87, and R126 reside on the side of Tube modeled to interact with dMyD88, while D163, D166, and D169/D170 (equivalent to G97, D100, and L103/E104 of MyD88) are located on the surface of MyD88 modeled to interact with Tube (Fig. 4b, 4c). There is no equivalent of D113 of dMyD88 in MyD88 because it is located in a 6-residue insertion in the already long H1-H2 loop, which directly interacts with Tube in the model. As in the IRAK4: MyD88 complex (Fig. 2f), there may also be charge complementarity in this Tube: dMyDD interface.
Notably, the structure of the Tube: Pelle complex shows a large surface burial of about 1,000 Å2 to allow 1:1 complex formation. This is provided by the C-terminal loop extension of Tube to enlarge the type II interface 47. A similar situation may also apply to the Tube: dMyD88 interaction; Tube has a 5-residue insertion in the H4-H5 loop and a long insertion in the H2-H3 loop that forms a pair of helices (H2′ and H2″) in addition to the insertion in the H1-H2 loop of dMyD88 (Fig. 4b, 4c), all of which may increase the interaction surface and strength for the 1:1 complex formation. There is no direct interaction between dMyD88 and Pelle in our model of the complex, which is consistent with earlier biochemical studies 46. It appears that a conformational change or a loss of entropy at the C-terminal loop extension of Tube may have been induced upon dMyD88 interaction to enhance the ability of the dMyD88: Tube complex to interact with Pelle. In summary, Drosophila and mammals use a similar three-player system with different stoichiometry in Toll or TLR/IL-1R signaling. It is possible that upon oligomerization of dMyD88 by activated Toll, an oligomeric form of the dMyD88: Tube: Pelle complex may be formed to allow auto-activation of the Pelle kinase. The apparent, higher complexity of the MyD88: IRAK4: IRAK2 complex may represent a more cooperatively controlled signaling system to accommodate the more complicated biology in mammals.
Common helical oligomers for DD assembly
Comparison with the PIDD: RAIDD complex that forms the core of the PIDDosome for caspase-2 activation 49, surprisingly showed that the three types of interfaces in the Myddosome are similar to those observed in the PIDD: RAIDD complex despite the different apparent oligomerization schemes (Fig. 4d). Superposition of analogous interactions showed that the type I, II and III interactions differ by rotational differences of 25.8°, 19.9° and 34°, respectively. The conservation of the three types of interactions may be indicative of evolution of primordial DD interaction pairs as well as general match of shape concavity and convexity in these interfaces as dictated by the six helical bundle fold.
The MyD88: IRAK4: IRAK2 complex structure prompted us to re-inspect the structure of the PIDD: RAIDD complex. We discovered that the PIDD: RAIDD complex may be seen as a double stranded left-handed helical oligomer (Fig. 4e–4g). One strand has three PIDD DD and three RAIDD DD molecules and the other strand contains two PIDD DDs and four RAIDD DDs, leading to the 5: 7 stoichiometry of the PIDD DD: RAIDD DD complex. The relationship between a double-stranded and a single-stranded oligomer appears to be quite simple: when the two-dimensional representation is rolled up with one notch down, the arrangement turns from a double-stranded oligomer to a single-stranded oligomer (Supplementary Fig. 11). This versatility explains how the PIDD: RAIDD complex and the MyD88: IRAK4: IRAK2 complex use similar interaction surfaces to create structures of different helical symmetries and stoichiometries.
A significant fraction of proteins in all kingdoms exists in the form of helical oligomers, which have been studied extensively by electron microscopy (EM) 50. However, helical symmetry is rarely known in signal transduction. In the case of the MyD88: IRAK4: IRAK2 complex, the relative small size prevents detailed studies by EM and its tendency to be heterogeneous makes crystallization difficult. Yet helical symmetry seems to be especially suitable for regulating the pathway and the strength in signal transduction because complexes with helical symmetries can be evolved to accommodate variable number of binding partners with specificity. In addition, assembly of such complexes is sensitive to the number of receptors that are activated and the degree of their aggregation so that a threshold for eliciting complex formation can be set for the signaling processes. Many more signaling proteins may function as helical oligomers, including other DD superfamily members such as CARD, Pyrin and DED.
METHODS SUMMARY
The DDs of human MyD88, IRAK4 and IRAK2 were co-expressed in E. coli and co- purified by Ni-NTA affinity resin followed by gel filtration chromatography. Crystals grew at 20°C at a protein concentration of about 1 mg/ml using 50 mM Tris-HCl at pH 8.0, 100–250 mM MgCl2, and 8–15% ethanol.
Methods
Protein Expression, Purification, and Crystallization
The DDs of human MyD88 (residues 20–117), IRAK4 (residues 4–106), and IRAK2 (residue 1–112) were co-expressed in E. coli. Both MyD88 and IRAK4 DDs were fused to a C-terminal His-tag. The MyD88: IRAK4: IRAK2 complex was purified by Ni-NTA affinity resin and Superdex 200 gel filtration chromatography. The protein concentration for crystallization is about 1 mg/ml. Crystals appeared at 20 °C in hanging drops in conditions of 50 mM Tris at pH 8.0, 100–250 mM MgCl2, and 8–15 % ethanol. The MyD88: IRAK4 complex was co-expressed in E. coli with the exact same constructs except that IRAK2 gene was not included. The complex was purified with the same method described above. For mutagenesis, all tagged or non-tagged proteins were expressed similarly. In order to obtain sufficient amount of soluble MyD88 for analysis, a longer MyD88 construct containing residues 20–154 (MyD88-long) was used for expression. In order to distinguish the IRAK2 band from the MyD88-long and IRAK4 bands on SDS-PAGE, the non-tag version of IRAK2 was fused to two ubiquitins at the N-terminus.
Mutational Analysis of Complex Formation in Vitro
Site-directed mutagenesis was performed using the Quikchange kit. All mutations were done on the tagged protein and were confirmed by DNA sequencing. For the co-expression constructs and the His-tagged constructs, the expressed proteins were mixed with the resin and subjected to three times of wash with 60 mM immidazole. The resin was then mixed with non-tagged protein lysates at room temperature for 1 hr, and then washed again with three times of 60 mM immidazole. The samples were eluted with 200 mM immidazole and subjected to SDS-PAGE. The gels were stained with Coomassie blue. The complex formation was determined by the ability of the mutant proteins to pull-down other DDs.
Structure Determination and Model Building
Diffraction data sets were collected at the X29 and X25 beamlines of NSLS and the 24-ID-E beamline of APS, and processed using HKL200051. To obtain initial phases, extensive heavy atom screenings were done with gold, mercury, platinum and tantalum compounds and selenomethione substituted crystals were obtained. Phases for initial model building were obtained by multiple isomorphous replacement using the program SOLVE 52, with one gold, two mercury, one platinum, one selenium, and one tantalum derivatives. The electron density was modified and extended by the program RESOLVE 52. With heavy atom positions and the expected structural homology between the three DDs, six MyD88, four IRAK4, and four IRAK2 molecules can be identified and built accurately. There is one ternary Myddosome complex per crystallographic asymmetric unit with a solvent content of ~72 %. The models were subjected to repeated rounds of building with Coot 53 and refinement with CNS 54. The model was also subjected to TLS refinement by the program Phenix 55. Residues 94 to 112 of IRAK2 were not visible in the electron density maps. The structure was analyzed using the CCP4 suite 56, the ProtorP server 57, and the Dali server 45. All figures were made using PyMOL 58.
Supplementary Material
Acknowledgments
We thank Drs. Kanagalaghatta Rajashankar, Igor Kourinov and Narayanasami Sukumar for data collection at the NE-CAT of APS and Dr. Xiaojing Ma for help with the manuscript. This work was supported by NIH (HW), theCancer Research Institute (SCL and YCL), and the American Heart Association (YCL).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions H.W. initiated the project idea. S.C.L., and Y.C.L. designed and performed experiments. S.C.L. and H.W. interpreted data and wrote the manuscript.
Author Information The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 3MOP. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol Rev. 2008;226:10–18. doi: 10.1111/j.1600-065X.2008.00701.x. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 5.Verstak B, Hertzog P, Mansell A. Toll-like receptor signalling and the clinical benefits that lie within. Inflamm Res. 2007;56:1–10. doi: 10.1007/s00011-007-6093-7. [DOI] [PubMed] [Google Scholar]
- 6.Marx J. Biomedicine. Puzzling out the pains in the gut. Science. 2007;315:33–35. doi: 10.1126/science.315.5808.33. [DOI] [PubMed] [Google Scholar]
- 7.O’Neill LA. Primer: Toll-like receptor signaling pathways--what do rheumatologists need to know? Nat Clin Pract Rheumatol. 2008;4:319–327. doi: 10.1038/ncprheum0802. [DOI] [PubMed] [Google Scholar]
- 8.Marta M, Meier UC, Lobell A. Regulation of autoimmune encephalomyelitis by toll-like receptors. Autoimmun Rev. 2009;8:506–509. doi: 10.1016/j.autrev.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Lande R, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 10.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 11.Horner AA, Raz E. Do microbes influence the pathogenesis of allergic diseases? Building the case for Toll-like receptor ligands. Curr Opin Immunol. 2003;15:614–619. doi: 10.1016/j.coi.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Chen R, et al. Cancers take their Toll--the function and regulation of Toll-like receptors in cancer cells. Oncogene. 2008;27:225–233. doi: 10.1038/sj.onc.1210907. [DOI] [PubMed] [Google Scholar]
- 13.Dasu MR, Devaraj S, Park S, Jialal I. Increased Toll-like Receptor activation and TLR ligands in Recently Diagnosed Type 2 diabetes Subjects. Diabetes Care. 2010;33:861–868. doi: 10.2337/dc09-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart CR, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol. 2010;11 (2):155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Dekker WK, Cheng C, Pasterkamp G, Duckers HJ. Toll like receptor 4 in atherosclerosis and plaque destabilization. Atherosclerosis. 2010;209:314–320. doi: 10.1016/j.atherosclerosis.2009.09.075. [DOI] [PubMed] [Google Scholar]
- 16.Romero-Sandoval EA, Horvath RJ, DeLeo JA. Neuroimmune interactions and pain: focus on glial-modulating targets. Curr Opin Investig Drugs. 2008;9:726–734. [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 18.Dinarello CA. Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int Rev Immunol. 1998;16:457–99. doi: 10.3109/08830189809043005. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, et al. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 20.Dunne A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003;2003:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 21.Bowie A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal generators for pro-inflammatory interleukins and microbial products. J Leukoc Biol. 2000;67:508–514. doi: 10.1002/jlb.67.4.508. [DOI] [PubMed] [Google Scholar]
- 22.Beutler B, et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi K, et al. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki N, et al. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature. 2002;416:750–756. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 25.Kawagoe T, et al. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 26.Wan Y, et al. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control. J Biol Chem. 2009;284:10367–10375. doi: 10.1074/jbc.M807822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 28.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasker MV, Gajjar MM, Nair SK. Cutting edge: Molecular structure of the IL-1R-associated kinase-4 death domain and its implications for TLR signaling. J Immunol. 2005;175:4175–4179. doi: 10.4049/jimmunol.175.7.4175. [DOI] [PubMed] [Google Scholar]
- 30.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park HH, et al. The Death Domain Superfamily in Intracellular Signaling of Apoptosis and Inflammation. Ann Rev Immunology. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns K, et al. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janssens S, et al. MyD88S, a splice variant of MyD88, differentially modulates NF-kappaB- and AP-1-dependent gene expression. FEBS Lett. 2003;548:103–107. doi: 10.1016/s0014-5793(03)00747-6. [DOI] [PubMed] [Google Scholar]
- 34.Loiarro M, et al. Identification of critical residues of the MyD88 death domain involved in the recruitment of downstream kinases. J Biol Chem. 2009;284:28093–28103. doi: 10.1074/jbc.M109.004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci U S A. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szabo G, Dolganiuc A, Dai Q, Pruett SB. TLR4, ethanol, and lipid rafts: a new mechanism of ethanol action with implications for other receptor-mediated effects. J Immunol. 2007;178:1243–1249. doi: 10.4049/jimmunol.178.3.1243. [DOI] [PubMed] [Google Scholar]
- 38.Rao N, Nguyen S, Ngo K, Fung-Leung WP. A novel splice variant of interleukin-1 receptor (IL-1R)-associated kinase 1 plays a negative regulatory role in Toll/IL-1R-induced inflammatory signaling. Mol Cell Biol. 2005;25:6521–6532. doi: 10.1128/MCB.25.15.6521-6532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardy MP, O’Neill LA. The murine IRAK2 gene encodes four alternatively spliced isoforms, two of which are inhibitory. J Biol Chem. 2004;279:27699–27708. doi: 10.1074/jbc.M403068200. [DOI] [PubMed] [Google Scholar]
- 40.Conze DB, et al. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao Z, et al. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 42.Belvin MP, Anderson KV. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. 1996;12:393–394. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 43.Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci U S A. 2002;99:12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towb P, Huaiyu S, Wasserman SA. Tube Is an IRAK-4 Homolog in a Toll Pathway Adapted for Development and Immunity. J Innate Immun. 2009;1:309–321. doi: 10.1159/000200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 46.Moncrieffe MC, Grossmann JG, Gay NJ. Assembly of oligomeric death domain complexes during Toll receptor signaling. J Biol Chem. 2008;283:33447–33454. doi: 10.1074/jbc.M805427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao T, Towb P, Wasserman SA, Sprang SR. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun H, et al. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. Embo J. 2004;23:100–110. doi: 10.1038/sj.emboj.7600033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egelman EH. Single-particle reconstruction from EM images of helical filaments. Curr Opin Struct Biol. 2007;17 (5):556–561. doi: 10.1016/j.sbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11 (Pt 1):49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60 (Pt 12 Pt 1):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58 (Pt 11):1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 56.Collaborative Computational Project. Number 4., The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 57.Reynolds C, Damerell D, Jones S. ProtorP: a protein-protein interaction analysis server. Bioinformatics. 2009;25 (3):413–4. doi: 10.1093/bioinformatics/btn584. [DOI] [PubMed] [Google Scholar]
- 58.Delano WL. The PyMol Molecular Graphics System. 2002 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.