Abstract
Ribonucleases are evoking medical interest because of their intrinsic cytotoxic activity. Most notably, ranpirnase, which is an amphibian ribonuclease, is in advanced clinical trials as a chemotherapeutic agent for the treatment of cancer. Here, we describe a strategy to create a novel antiviral agent based on bovine pancreatic ribonuclease (RNase A), a mammalian homologue of ranpirnase. Specifically, we have linked the N- and C-termini of RNase A with an amino acid sequence that is recognized and cleaved by human immunodeficiency virus (HIV) protease. This linkage obstructs the active site, forming an HIV-specific RNase A zymogen. Cleavage by HIV-1 protease increases ribonucleolytic activity by 50-fold. By relying on the proper function of HIV-1 protease, rather than its inhibition, our approach will not engender known mechanisms of resistance. Thus, we report an initial step toward a new class of agents for the treatment of HIV/AIDS.
Introduction
Since the first cases of human immunodeficiency virus (HIV) were described in the early 1980s,1 much has been done to improve survival. Four classes of antiretroviral drugs are now in clinical use, with protease inhibitors and two types of reverse-transcriptase inhibitors comprising the three major classes.2 In addition, the first fusion inhibitor was approved in 2003.3,4 A combination of these chemotherapeutic agents, known as highly active antiretroviral therapy, or HAART, has led to a significant decline in the morbidity and mortality of HIV patients.5,6 Although the number of deaths among persons with AIDS in the United States declined substantially during the late 1990s, the rate of decline has since decreased significantly. Declines in the numbers of new cases have also leveled off.7
This slowing of progress against HIV/AIDS, combined with several major problems with HIV/AIDS therapy, continues to motivate the design of new drugs.8,9 Side effects, especially disturbances in lipid metabolism, are an increasing problem as patients are living longer and developing cardiac disease.2,10 Rapid viral replication and the high error rate of reverse transcriptase have led to high rates of resistance to HAART,5 with about 10% of newly acquired infections in the United States and Europe being resistant to at least one of the three major drug classes.11 The rapid development of resistance necessitates strict patient compliance with therapy. Finally, AIDS remains a chronic disease due to the existence of latent HIV infection in memory T cells and other cells that are not pharmacologically accessible.12,13
One strategy for slowing viral progression and eliminating latent viral infection is to kill those cells that are infected with HIV. Although cytotoxic cancer drugs could be used, there is a risk of serious side effects that would compound the side effects of HAART.14 To circumvent this problem, toxins that are specific for HIV-infected cells have been designed. One approach has been to use toxins that will be activated by the HIV-1 protease (HIV PR), a virally encoded protein that cleaves the viral polyprotein late in the life cycle of HIV.15 In one case, an HIV PR-activated variant of caspase-3 was constructed and its ability to kill HIV-infected cells was demonstrated.16 In another case, diphtheria toxin was modified such that a degradation signal was removed by cleavage with HIV PR.17 These “pro-drug” strategies have the potential to reduce the problems with both side effects and drug resistance.
Bovine pancreatic ribonuclease (RNase A) has been studied intensely by biochemists for decades.18,19 Recently, interest in RNase A has resurged with the discovery of the facinating biological properties of ranpirnase, angiogenin, and bovine seminal ribonuclease, which are homologues of RNase A.20–25 For example, ranpirnase26 is toxic to cancer cells in vitro and in vivo in a manner that is dependent on its catalytic activity,27 and is currently in Phase IIIb clinical trials for the treatment of malignant mesothelioma.28 In addition, RNase A and other ribonucleases are known to inhibit HIV replication.29,30
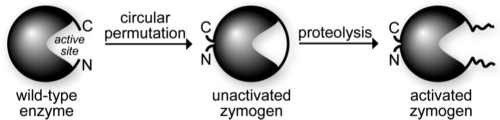
Here, we describe the creation of an HIV-specific zymogen from RNase A. Inspired by the design of zymogens specific for malaria and hepatitis C,31,32 we sought to exploit the activity of HIV PR to activate a cytotoxin. The N- and C-termini of RNase A were joined by circular permutation, thereby occluding the active site with an amino acid sequence containing a cleavage site for HIV PR (Fig. 1). In the presence of HIV PR, this sequence is cleaved and the catalytic activity is unmasked. It is our hope that because a chemotherapeutic strategy based on a ribonuclease zymogen would rely on catalysis by HIV PR rather than merely an affinity for that enzyme, the development of resistance would be unlikely.
FIG. 1.
Design of an HIV-specific ribonuclease A zymogen. Circular permutation was used to block access to the active site until activation by HIV PR.
Materials and Methods
Materials
Escherichia coli strain BL21(DE3) was from Novagen (Madison, WI). K-562 cells,33 which are human erythroleukemia cells, were obtained from American Type Culture Collection (Manassas, VA). HIV PR expression vector pET-HIVPR34 was a kind gift from J. Tang (Oklahoma Medical Research Foundation).
Enzymes were obtained from Promega (Madison, WI). Protein purification columns and resin were from Amersham Biosciences (Piscataway, NJ). Synthetic oligonucleotides, including the ribonuclease substrate 6-FAM-dArU(dA)2-6-TAMRA,35 were from Integrated DNA Technologies (Coralville, IA). Poly(cytidylic acid) [poly(C)] was from Sigma–Aldrich (St. Louis, MO) and was precipitated with ethanol before use to remove short RNA fragments. [methyl-3H]Thymidine was from Perkin–Elmer (Boston, MA). MES buffer (Sigma-Aldrich, St. Louis, MO) was purified by an-ion-exchange chromatography to remove any contaminating oligo(vinylsulfonic acid) (OVS).36 All other chemicals were of commercial grade or better, and were used without further purifications.
Phosphate-buffered saline (PBS) contained (in 1 liter) NaCl (8.0 g), KCl (2.0 g), Na2HPO4 ∙ 7H2O (1.15 g), and KH2PO4 (2.0 g), and had a pH of 7.4.
Zymogen expression and purification
Plasmids used to direct the production of HIV RNase A zymogens were derived from the plasmid pET22b(+)/19N.31 The region of the plasmid that encoded the linker was replaced with DNA encoding the sequence GSTATIMMQRGNAG (Zymogen 1) or GGSTATIMMQRGNAG (Zymogen 2) using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Zymogens were expressed and purified as described previously.32 Zymogens were ∼95% pure according to SDS–PAGE, and zymogen identity was confirmed by MALDI-TOF (Table 1).
Table 1.
Biochemical Properties of Unactivated Ribonuclease A Zymogens
| |
|
|
|
m/zd |
|
|---|---|---|---|---|---|
| Ribonuclease | Tma(°C) | Kdb(nM) | IC50c(μM) | Expected | Observed |
| Wild type | 64e | 44 × 10−6f | >25 | 13,682 | 13,692 |
| Zymogen 1 | 46 | 150 ± 10 | >25 | 15,165 | 15,142 |
| Zymogen 2 | 49 | — | >25 | 15,222 | 15,208 |
Values of Tm were determined in PBS by UV spectroscopy.
Value of Kd (± SE) was determined for the complex with hRI at (23 ± 2)°C.
Values of IC50 are for the incorporation of [methyl-3H]thymidine into the DNA of K-562 cells.
Values of m/z were determined by MALDI-TOF mass spectrometry.
From Rutkoski et al.42
From Lee et al.53
Protease expression and purification
The substitutions Q7K/L33I/L63I37 and C67A/C95A38 were made with the Quikchange mutagenesis kit to improve protease stability. The resulting plasmid, pET-HIVPRV, was transformed into E. coli BL21(DE3) cells for expression. Transforming, plating, and subculturing were done in Luria–Bertani medium containing glucose (1% w/v) to reduce leaky expression of HIV PR, which is toxic to E. coli.39 Protease expression was induced in Terrific Broth containing isopropyl-β-d-thiogalactoside (1 mM). The protease was purified from inclusion bodies by methods described previously,40 with the following modifications. Following two passes through a French pressure cell (>16,000 psi), inclusion bodies were collected by centrifugation at 16,000 × g for 45 min, washed with extraction buffer, resuspended in extraction buffer, and again collected by centrifugation. Ion-exchange columns were poured from resin and run by gravity at (23 ± 2)°C.
Zymogen cleavage
Zymogens were activated by mixing them with 0.04–0.2 M equivalents of HIV PR in reaction buffer, which was 100 mM sodium acetate buffer, pH 5.0, containing NaCl (100 mM), EDTA (1 mM), glycerol (5% v/v), and PEG-8000 (0.1% w/v), and incubating the resulting solution at 37°C for 1 h. Activation was stopped by dilution into gel-loading buffer to a final composition of 50 mM Tris–HCl, pH 6.8, containing DTT (100 mM), SDS (0.2% w/v), glycerol (10% v/v), and bromophenol blue (0.75 mM), or by dilution (by 100-fold) into 0.10 M MES–NaOH, pH 6.0, containing NaCl (0.10 M). Following dilution, reaction mixtures were placed on ice. SDS-PAGE in the presence of DTT was used to assess zymogen cleavage (Fig. 2).
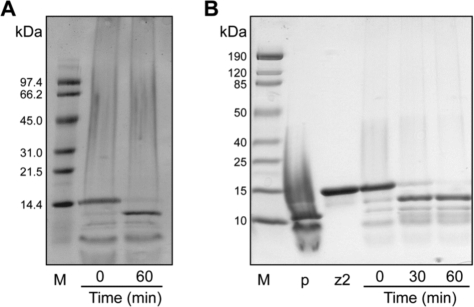
FIG. 2.
Activation of zymogens by HIV PR. Activation at 37°C was monitored by SDS–PAGE in the presence of DTT. (A) Activation of zymogen 1 after addition of HIV PR at a 1:5 (protease:zymogen) molar ratio. (B) Activation of zymogen 2 after addition of HIV PR at a 1:25 (protease:zymogen) molar ratio. M is the protein molecular weight marker, p is HIV PR, and z2 is zymogen 2.
Ribonucleolytic activity
The ability of zymogens to catalyze the cleavage of the fluorogenic substrate 6-FAM-dArU(dA)2-6-TAMRA, which exhibits a 180-fold increase in fluorescence (excitation at 493 nm; emission at 515 nm) upon cleavage,35 was assessed. Assays were carried out at (23 ± 2)°C in 2.0 ml of 0.10 M MES–NaOH (OVS-free), pH 6.0, containing NaCl (0.10 M), substrate (20 nM), and zymogen (5 pM–1 nM). Values of kcat/KM were obtained with Eq. (1):
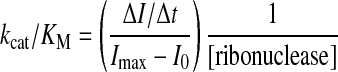 |
(1) |
where ∆I/∆t represents the initial reaction velocity, Imax is the fluorescence following complete cleavage of the substrate by excess RNase A, and I0 is the fluorescence prior to addition of ribonuclease.
In addition, the ability of zymogens to cleave poly(C) (ɛ268nm = 6200 M−1 cm−1 per nucleotide) was monitored by the change in UV absorption, which increases upon cleavage (∆ɛ250 nm = 2380 M−1 cm−1). Assays were carried out at (23 ± 2)°C in 0.10 M MES–NaOH (OVS free), pH 6.0, containing NaCl (0.10 M), poly(C) (10 μM–1.5 mM), and zymogen (20 nM–1 μM). Initial velocity data were used to calculate values of kcat, KM, and kcat/KM with the program Prism 4 for Macintosh (GraphPad Software, San Diego, CA).
Conformational stability
The value of Tm, which is the temperature at the midpoint of the thermal transition between the folded and unfolded states, of each zymogen was determined by monitoring its UV absorption at 287 nm. Zymogen solutions (∼25 μM in PBS) were heated incrementally (0.15°C/min from 25 to 75°C). Data were collected and analyzed with the program THERMAL from Varian Analytical Instruments (Walnut Creek, CA).
Inhibition by the ribonuclease inhibitor protein
Binding of unactivated zymogen 1 to the ribonuclease inhibitor protein was assessed via a microplate assay as described previously.41
Cytotoxic activity
K-562 cells were grown in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). The effect of zymogens on K-562 proliferation was measured by the incorporation of [methyl-3H]thymidine as described previously.42
Results
Design of HIV-specific zymogen
The p2/NC recognition sequence of HIV PR (TATIM/MQRGN) was chosen because it is cleaved initially and efficiently in vitro and in vivo.43,44 It is desirable for a zymogen to contain a linker that is long enough to allow for the flexibility necessary to be cleaved by the protease, but also short enough to prevent indiscriminate ribonucleolytic activity prior to activation by the specific protease. We designed two HIV-specific RNase zymogens, one with a 14-residue linker (zymogen 1) and another with a 15-residue linker (zymogen 2). The 10-residue cleavage sequence was extended by a linker designed for solubility and flexibility, containing glycine and serine residues.
Circular permutation was done to produce new N- and C-termini at residues 89 and 88 of RNase A (Fig. 1). These two residues were linked by a new disulfide bond.31 In addition, residues 4 and 118 (RNase A numbering) were replaced with cysteine residues and linked by a disulfide bond in order to improve stability.45
Zymogen activation
Zymogens 1 and 2 were incubated with protease in a 1:25 to 1:5 (PR:zymogen) ratio at 37°C. Activation was monitored by SDS–PAGE. Following reduction with DTT, zymogen fragments of 10,548 Da and 4635 Da are formed from zymogen 1. The formation of a 10.5-kDa fragment from a 15.2-Da fragment can be followed by SDS–PAGE. (The 4.6-kDa fragment runs off the gel and was not observed.) Both zymogens 1 and 2 were more than 95% cleaved by HIV PR after an incubation of 60 min at 37°C (Fig. 2). Control incubations of zymogens 1 and 2 without protease for 1 h did not result in any cleavage (data not shown). Native RNase A has been shown not to be a substrate of HIV PR.46
The ability of zymogens to cleave two RNA substrates was measured (Tables 2 and 3). The shorter substrate was a fluorogenic tetranucleotide that increases in fluorescence upon cleavage (Table 2).35 The catalytic efficiency (kcat/KM) of zymogen 1 increases from 2.9 × 105 to 1.4 × 107 M−1 s−1 upon activation, an increase of 48-fold. The kcat/KM value of the activated zymogen is nearly that of wild-type RNase A. For zymogen 2, the increase is more modest, from 8.7 × 105 to 7.6 × 106 M−1 s−1, or 9-fold.
Table 2.
Kinetic Parameters of Ribonuclease A Zymogens with an Oligonucleotide Substratea
| Ribonuclease | (kcat/KM)unactivated103 M−1s−1 | (kcat/KM)activated106 M−1s−1 | 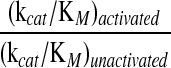 |
|---|---|---|---|
| Wild type | N/Ab | 52 ± 4c | N/A |
| Zymogen 1 | 290 ± 40 | 14 ± 1 | 48 |
| Zymogen 2 | 870 ± 220 | 7.6 ± 5 | 9 |
Values of Kcat/KM (± SE) were determined for catalysis of 6-FAM-dArU(dA)2-6-TAMRA cleavage at 25°C in 0.10 M MES–NaOH buffer (OVS free), pH 6.0, containing 0.10 M NaCl.
N/A, not applicable.
From Rutkoski et al.42
Table 3.
Kinetic Parameters of Ribonuclease A Zymogens with a Polynucleotide Substratea
| Ribonuclease | (kcat)unactivateds−1 | (kcat)activateds−1 | (KM)unactivated10−6M | (KM)activated10−6M | (kcat/KM)unactivated103M−1s−1 | (kcat/KM)activated103M−1s−1 | 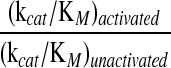 |
|---|---|---|---|---|---|---|---|
| Wild type | N/A | 280 ± 29 | N/Ac | 3.3 ± 0.2 | N/A | 83 ± 7 | N/A |
| Zymogen 1 | 3.2 ± 0.1 | 38 ± 2 | 120 ± 16 | 60 ± 10 | 27 ± 3 | 0.65 ± 0.1 | 24 |
Values of kcat, KM, and kcat/Km (± SE) were determined for catalysis of poly(C) cleavage at 25°C in 0.10 M MES–NaOH buffer (OVS free), pH 6.0, containing 0.10 M NaCl. Initial velocity data were used to calculate values of kcat, KM, and kcat/KM with the program Prism 4 (GraphPad Software, San Diego, CA).
From Johnson et al.32
N/A, not applicable.
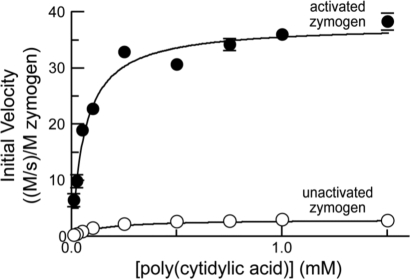
This sensitive assay for ribonucleolytic activity might not be an accurate reflection of the ability of ribonucleases to cleave therapeutically relevant substrates.47 Hence, we measured the ribonucleolytic activity of zymogen 1 with a longer polynucleotide substrate, poly(C) (Fig. 3 and Table 3). This assay also allows for delineation of the contributions of the kcat and KM values to catalytic efficiency. Upon activation, the kcat value increased from 3.2 to 38 s−1. The KM value decreased from 1.2 × 10−4 to 6 × 10−5 M, resulting in a change in kcat/KM of 24-fold, from 2.7 × 104 to 6.5 × 105 M−1 s−1.
FIG. 3.
Enzyme kinetics of unactivated zymogen 1 (○; 1.0 μM) and activated zymogen 1 (•; 20 nM). Initial velocity data (±SE) at increasing amounts of poly(C) are normalized for zymogen concentration. Data were used to determine values of kcat, KM, and kcat/KM (Table 3).
Conformational stability of zymogens
The ability of a cytotoxic ribonuclease to retain its conformation at physiological temperatures is an important factor in determining its potential as a chemotherapeutic agent.45 Zymogens 1 and 2 are stable above physiological temperature (37°C). Both, however, are less stable than wild-type RNase A (Table 1).
Binding of ribonuclease inhibitor protein
The ability of a ribonuclease to evade binding by the cytoplasmic ribonuclease inhibitor protein (RI), which binds RNase A with femtomolar affinity, is essential for its cytotoxicity.42,48,49 The active site of the ribonuclease comprises a large portion of the buried surface area in the ribonuclease–inhibitor complex.50,51 We would expect that the unactivated RNase A zymogen would have diminished affinity for RI, and indeed the Kd value for the ribonuclease–RI complex with the unactivated zymogen is >106-fold greater than that for wild type RNase A (Table 1).
Cytotoxicity of unactivated zymogen
A cytotoxic therapeutic agent must have little or no toxicity to healthy cells—those that are not infected with HIV. To assess this attribute, unactivated zymogens were assayed for their ability to inhibit the proliferation of K-562 cells, which are human cells that are not infected with HIV. Neither zymogen 1 nor zymogen 2 was toxic to K-562 cells up to a concentration of 38 μM (Table 1).
Discussion
We have created two HIV-specific RNase A zymogens. Their activation by HIV PR is 95% complete following a 1-h incubation with a substoichiometeric amount of HIV PR (Fig. 2). One factor that determines the efficiency of this cleavage reaction is the ability of the protease to bind to the zymogen. HIV PR and zymogens 1 and 2 are highly cationic proteins, having theoretical isoelectric points of pI = 9.3, 8.8, and 8.8, respectively.52 Coulombic repulsion between HIV PR and a zymogen could diminish the efficiency of activation, a problem that could be overcome by additional mutagenesis.
The two zymogens differ as catalysts of RNA cleavage. The ribonucleolytic activity of zymogens 1 and 2 toward a tetranucleotide substrate was measured before and after activation with HIV PR (Tables 2 and 3). Prior to activation, both zymogens had low catalytic efficiency (105 M−1 s−1), with zymogen 2 having 3-fold higher activity than zymogen 1. This difference is likely due to the longer length of the linker of zymogen 2, which could alter the ability of substrates to access the active site prior to activation. Following activation, zymogen 1 regains a catalytic efficiency that is close to that of wild-type RNase A, suggesting that the cleaved linker no longer obscures the active site. Zymogen 2, however, does not regain as much activity, which suggests that the longer linker continues to interact with the active site after activation.
The use of a longer RNA substrate revealed additional information. Using poly(C) as a substrate, unactivated zymogen 1 has a KM value that is 36-fold higher than that of wild-type RNase A, confirming that the linker inhibits the binding of RNA substrates (Table 3). Yet, the kcat value of the unactivated zymogen is also decreased by 88-fold, suggesting that the linker also prevents the turnover of RNA substrates by disturbing active-site residues.32 Upon activation of zymogen 1 with HIV PR, the KM value decreases by 2-fold and the kcat value increases by 10-fold. Neither value was similar to that of wild-type RNase A, suggesting that, after cleavage, the linker continues to interfere with the binding and turnover of a longer RNA substrate.
The development of a therapy that kills HIV-infected cells is highly attractive. Such a therapy could eradicate completely the viral reservoir in patients, a challenge that remains in the era of HAART.13 Drugs that are used to kill cancer cells could be used for this purpose but would result in the side effects typical of cancer chemotherapy, including bone marrow suppression. Any potential for decreasing white blood cells in a patient with a weakened immune system should be avoided. As an alternative, our strategy seeks the selective destruction of HIV-infected cells.
Ribonuclease zymogens have other desirable attributes. Ribonucleases that evade the endogenous ribonuclease inhibitor protein are known to be potent cytotoxins.49 In contrast, we have shown that ribonuclease zymogens have no observable toxicity toward a human cell line that does not contain HIV PR (Table 1). This absence of cytotoxicity suggests that a drug based on a ribonuclease zymogen could have a high therapeutic index. Another attractive feature is the potential difficulty in the development of viral resistance to a ribonuclease zymogen. Rapid viral replication and the high error rate of reverse transcriptase have led to the rapid development of resistance to reverse transcriptase inhibitors and protease inhibitors, small-molecule drugs that rely on binding to these enzymes. It is reasonable to anticipate that resistance to a drug that requires the catalytic activity of HIV PR toward a native substrate would be significantly more difficult for a virus to develop. Moreover, resistance derived from a protease variant with altered specificity could be countered by replacing the linker sequence with one that is recognized by the protease variant. Finally, we note that a chemotherapeutic agent based on a ribonuclease zymogen would not be used concurrently with protease inhibitor therapy, which would likely diminish zymogen activation and hence efficacy.
In conclusion, we have described the creation of an HIV-specific RNase A zymogen that increases in catalytic efficiency by approximately 50-fold upon activation with HIV PR, is stable at physiological temperature, and is not toxic to uninfected cells. These data establish a proof of principle for a new class of agents for the treatment of HIV/AIDS. We anticipate testing our strategy with human cells containing HIV PR.
Acknowledgments
We are grateful to J.F. May and S.M. Fuchs for assistance with cloning, L.D. Lavis for the diethylfluorescein derivatives, used in the microplate assay, and M.N. Levine, S.M. Fuchs, K.A. Dickson, and R.J. Johnson for contributive discussions. This work was supported by Grants 51670 (Bill & Melinda Gates Foundation) and CA073808 (NIH). R.F.T. was supported by a Wisconsin Distinguished Rath Graduate Fellowship. The University of Wisconsin–Madison Biophysics Instrumentation Facility was established with grants BIR-9512577 (NSF) and RR13790 (NIH). The Keck Center for Chemical Genomics was established with a grant from the W.M. Keck Foundation.
Disclosure Statement
No competing financial interests exist.
References
- 1.Hymes KB. Cheung T. Greene JB, et al. Kaposi's sarcoma in homosexual men—a report of eight cases. Lancet. 1981;2:598–600. doi: 10.1016/s0140-6736(81)92740-9. [DOI] [PubMed] [Google Scholar]
- 2.Carr A. Toxicity of antiretroviral therapy and implications for drug development. Nat Rev Drug Discov. 2003;2:624–634. doi: 10.1038/nrd1151. [DOI] [PubMed] [Google Scholar]
- 3.Matthews T. Salgo M. Greenberg M. Chung J. DeMasi R. Bolognesi D. Enfuvirtide: The first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 4.Hanson K. Hicks C. New antiretroviral drugs. Curr HIV/AIDS Rep. 2006;3:93–101. doi: 10.1007/s11904-006-0024-z. [DOI] [PubMed] [Google Scholar]
- 5.Barbaro G. Scozzafava A. Mastrolorenzo A. Supuran CT. Highly active antiretroviral therapy: Current state of the art, new agents and their pharmacological interactions useful for improving therapeutic outcome. Curr Pharm Des. 2005;11:1805–1843. doi: 10.2174/1381612053764869. [DOI] [PubMed] [Google Scholar]
- 6.Temesgen Z. Warnke D. Kasten MJ. Current status of antiretroviral therapy. Expert Opin Pharmacother. 2006;7:1541–1554. doi: 10.1517/14656566.7.12.1541. [DOI] [PubMed] [Google Scholar]
- 7.Kellerman S. Begley E. Boyett B. Clark H. Schulden J. Changes in HIV and AIDS in the United States: Entering the third decade. Curr Infect Dis Rep. 2005;7:138–143. doi: 10.1007/s11908-005-0074-1. [DOI] [PubMed] [Google Scholar]
- 8.Weiss RA. HIV and AIDS: Looking ahead. Nat Med. 2003;9:887–891. doi: 10.1038/nm0703-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flexner C. HIV drug development: The next 25 years. Nat Rev Drug Discov. 2007;6:959–966. doi: 10.1038/nrd2336. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal L. Lu X. Jin Q. Alkhatib G. Anti-HIV therapy: Current and future directions. Curr Pharm Des. 2006;12:2031–2055. doi: 10.2174/138161206777442100. [DOI] [PubMed] [Google Scholar]
- 11.Shafer RW. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin Microbiol Rev. 2002;15:247–277. doi: 10.1128/CMR.15.2.247-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon V. Ho DD. HIV-1 dynamics in vivo: Implications for therapy. Nat Rev Microbiol. 2003;1:181–190. doi: 10.1038/nrmicro772. [DOI] [PubMed] [Google Scholar]
- 13.Yang QE. Human immunodeficiency virus reservoir might be actively eradicated as residual malignant cells by cytotoxic chemotherapy. Med Hypotheses. 2004;62:358–363. doi: 10.1016/j.mehy.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Ravot E. Lisziewicz J. Lori F. New uses for old drugs in HIV infection: The role of hydroxyurea, cyclosporin and thalidomide. Drugs. 1999;58:953–963. doi: 10.2165/00003495-199958060-00001. [DOI] [PubMed] [Google Scholar]
- 15.Kohl NE. Emini EA. Schleif WA, et al. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vocero-Akbani AM. Heyden NV. Lissy NA. Ratner L. Dowdy SF. Killing HIV-infected cells by transduction with an HIV protease-activated caspase-3 protein. Nat Med. 1999;5:29–33. doi: 10.1038/4710. [DOI] [PubMed] [Google Scholar]
- 17.Falnes PO. Welker R. Krausslich HG. Olsnes S. Toxins that are activated by HIV type-1 protease through removal of a signal for degradation by the N-end-rule pathway. Biochem J. 1999;343(Pt 1):199–207. [PMC free article] [PubMed] [Google Scholar]
- 18.Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 19.Marshall GR. Feng JA. Kuster DJ. Back to the future: Ribonuclease A. Biopolymers. 2008;90:259–277. doi: 10.1002/bip.20845. [DOI] [PubMed] [Google Scholar]
- 20.D'Alessio G, editor; Riordan JF, editor. Ribonucleases: Structures, Functions. Academic Press; New York: 1997. [Google Scholar]
- 21.Matousek J. Ribonucleases and their antitumor activity. Comp Biochem Physiol. 2001;129C:175–191. doi: 10.1016/s1532-0456(01)90202-9. [DOI] [PubMed] [Google Scholar]
- 22.Leland PA. Raines RT. Cancer chemotherapy—ribonucleases to the rescue. Chem Biol. 2001;8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarov AA. Ilinskaya ON. Cytotoxic ribonucleases: Molecular weapons and their targets. FEBS Lett. 2003;540:15–20. doi: 10.1016/s0014-5793(03)00225-4. [DOI] [PubMed] [Google Scholar]
- 24.Benito A. Ribó M. Vilanova M. On the track of antitumor ribonucleases. Mol Biosyst. 2005;1:294–302. doi: 10.1039/b502847g. [DOI] [PubMed] [Google Scholar]
- 25.Arnold U. Ulbrich-Hofmann R. Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol Lett. 2006;28:1615–1622. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 26.Lee JE. Raines RT. Ribonucleases as novel chemotherapeutics: The ranpirnase example. BioDrugs. 2008;22:53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ardelt W. Mikulski SM. Shogen K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. J Biol Chem. 1991;266:245–251. [PubMed] [Google Scholar]
- 28.Pavlakis N. Vogelzang NJ. Ranpirnase—an antitumour ribonuclease: Its potential role in malignant mesothelioma. Expert Opin Biol Ther. 2006;6:391–399. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- 29.Youle RJ. Wu YN. Mikulski SM, et al. RNase inhibition of human immunodeficiency virus infection of H9 cells. Proc Natl Acad Sci USA. 1994;91:6012–6106. doi: 10.1073/pnas.91.13.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedoya VI. Boasso A. Hardy AW. Rybak S. Shearer GM. Rugeles MT. Ribonucleases in HIV type 1 inhibition: Effect of recombinant RNases on infection of primary T cells and immune activation-induced RNase gene and protein expression. AIDS Res Hum Retroviruses. 2006;22:897–907. doi: 10.1089/aid.2006.22.897. [DOI] [PubMed] [Google Scholar]
- 31.Plainkum P. Fuchs SM. Wiyakrutta S. Raines RT. Creation of a zymogen. Nat Struct Biol. 2003;10:115–119. doi: 10.1038/nsb884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson RJ. Lin SR. Raines RT. A ribonuclease zymogen activated by the NS3 protease of the hepatitis C virus. FEBS J. 2006;273:5457–5465. doi: 10.1111/j.1742-4658.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- 33.Lozzio CB. Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 34.Ido E. Han HP. Kezdy FJ. Tang J. Kinetic studies of human immunodeficiency virus type 1 protease and its active-site hydrogen bond mutant A28S. J Biol Chem. 1991;266:24359–24366. [PubMed] [Google Scholar]
- 35.Kelemen BR. Klink TA. Behlke MA. Eubanks SR. Leland PA. Raines RT. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999;27:3696–3701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith BD. Soellner MB. Raines RT. Potent inhibition of ribonuclease A by oligo(vinylsulfonic acid) J Biol Chem. 2003;278:20934–20938. doi: 10.1074/jbc.M301852200. [DOI] [PubMed] [Google Scholar]
- 37.Mildner AM. Rothrock DJ. Leone JW, et al. The HIV-1 protease as enzyme and substrate: Mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry. 1994;33:9405–9413. doi: 10.1021/bi00198a005. [DOI] [PubMed] [Google Scholar]
- 38.Davis DA. Dorsey K. Wingfield PT, et al. Regulation of HIV-1 protease activity through cysteine modification. Biochemistry. 1996;35:2482–2488. doi: 10.1021/bi951525k. [DOI] [PubMed] [Google Scholar]
- 39.Pan SH. Malcolm BA. Reduced background expression and improved plasmid stability with pET vectors in BL21 (DE3) Biotechniques. 2000;29:1234–1238. doi: 10.2144/00296st03. [DOI] [PubMed] [Google Scholar]
- 40.Todd MJ. Semo N. Freire E. The structural stability of the HIV-1 protease. J Mol Biol. 1998;283:475–488. doi: 10.1006/jmbi.1998.2090. [DOI] [PubMed] [Google Scholar]
- 41.Lavis LD. Rutkoski TJ. Raines RT. Tuning the pKa of fluorescein to optimize binding assays. Anal Chem. 2007;79:6775–6782. doi: 10.1021/ac070907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutkoski TJ. Kurten EL. Mitchell JC. Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J Mol Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Pettit SC. Moody MD. Wehbie RS, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettit SC. Lindquist JN. Kaplan AH. Swanstrom R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2:66. doi: 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klink TA. Raines RT. Conformational stability is a determinant of ribonuclease A cytotoxicity. J Biol Chem. 2000;275:17463–17467. doi: 10.1074/jbc.M001132200. [DOI] [PubMed] [Google Scholar]
- 46.Hui JO. Tomasselli AG. Zurcher-Neely HA. Heinrikson RL. Ribonuclease A as a substrate of the protease from human immunodeficiency virus-1. J Biol Chem. 1990;265:21386–21389. [PubMed] [Google Scholar]
- 47.Suhasini AN. Sirdeshmukh R. Transfer RNA cleavages by onconase reveal unusual cleavage sites. J Biol Chem. 2006;281:12201–12209. doi: 10.1074/jbc.M504488200. [DOI] [PubMed] [Google Scholar]
- 48.Leland PA. Schultz LW. Kim B-M. Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc Natl Acad Sci USA. 1998;98:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutkoski TJ. Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr Pharm Biotechnol. 2008;9:185–199. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobe B. Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 51.Johnson RJ. McCoy JG. Bingman CA. Phillips GN., Jr Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007;367:434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjellqvist B. Basse B. Olsen E. Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–539. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- 53.Lee FS. Shapiro R. Vallee BL. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]