Abstract
Cancer patients are subject to multiple competing risks of death and may die from causes other than the cancer diagnosed. The probability of not dying from the cancer diagnosed, which is one of the patients’ main concerns, is sometimes called the “personal cure” rate. Two approaches of modeling competing-risk survival data, namely the cause-specific hazards approach and the mixture model approach, have been used to model competing-risk survival data. In this article, we first show the connection and differences between crude cause-specific survival in the presence of other causes and net survival in the absence of other causes. The mixture survival model is extended to population-based grouped survival data to estimate the personal cure rate. Using the colorectal cancer survival data from the Surveillance, Epidemiology and End Results (SEER) Program, we estimate the probabilities of dying from colorectal cancer, heart disease, and other causes by age at diagnosis, race and American Joint Committee on Cancer (AJCC) stage.
Keywords: Competing risks, grouped survival data, mixture model, personal cure, SEER Program
1 Introduction
Net survival, i.e., survival in the absence of other causes, is a measure of excess mortality due to cancer. This is a hypothetical measure of survival if all causes of death other than cancer of interest were to be eliminated. Net survival is a desirable measure to evaluate the progress of cancer treatment and control efforts since the interpretation as excess mortality due to cancer is not a ected by changes in mortality due to other diseases.1 However, net survival does not represent the actual survival patterns observed in a cohort of cancer patients. Comorbidity for cancer patients may limit treatment options and increase the risk of death from other causes. Usually comorbidity from competing causes increases with advancing age and is greater for patients in poor health. Thus, net survival may not be an ideal measure for assessing the impact of a cancer diagnosis in the presence of multiple competing risks. From another perspective, crude cause-specific probabilities of death provide measures of cause-specific mortality in the presence of causes of death in addition to cancer and reflect mortality patterns actually observed among patients.2 Thus, they are appropriate measures when the focus is on inference and comparison of cause-specific failures under a variety of conditions. Such probabilities can be used to weigh the risks and benefits of various treatment options, particularly for patients diagnosed at older ages when comorbidity is high.
There has been considerable progress against cancer due to improvements in treatment, and the dissemination of early diagnosis and screening. Thus, successfully treated cancer patients may die from a cause other than the diagnosed cancer, which is called “personal cure.” The corresponding proportion of dying from causes other than the diagnosed cancer is defined as the personal cure rate. Gordon3 originally applied the mixture model to estimate the personal cure rate using breast cancer data from a clinical trial. To assess mortality from cancer at the population level, we extend the mixture model for competing-risk survival to population-based cancer survival data in order to calculate crude probabilities of dying from cancer and other competing risks. The rest of the paper is organized as follows: In Section 2, we first describe survival data with competing risks and review the mixture model for continuous survival data. Next, we extend the mixture model to grouped survival data, describe the estimation method and discuss the connection between net survival and crude survival. In Section 3, we apply the mixture model to colorectal cancer survival data to calculate the probabilities of dying from three competing causes of death. We discuss the potential use and limitations of the mixture model in the Discussion section.
2 The mixture model for grouped survival data with competing risks
We consider a patient subject to K mutually exclusive competing risks of death and assume that the primary outcome is a random pair (D, T), where D takes a value from the set {1, 2, ..., K} indicating cause of death and T is a non-negative random variable representing time to death. Let z denote a vector of covariates. The cause-specific hazard rate, defined as the probability of dying from cause k alone in [t, t + dt) in the presence of all acting risks, given T ≥ t,4 is given by
Let and . The survival function for T is
David and Moescheberger5 consider the observed death time T as the minimum of K latent death times, i.e., T = min(T1,···, TK). When the competing risks T1, . . . , TK, are independent, Sk(t|z) can be interpreted as the net survival function from cause k in the absence of other causes and hk(t|z) is the net hazard.6 Net survival can be estimated by the Kaplan-Meier method or the actuarial method by treating the other causes of deaths as censored.
The crude cumulative probability or cumulative incidence function (CIF) of dying from cause k in the presence of other causes is
| (1) |
The functions is also called sub-distribution function for cause k, k = 1, ..., K. Let πk(z) = P(D = k|z) be the probability of dying from cause k and Qk(t|z) = P(T > t|D = k, z) be the conditional crude survival function. Then .
2.1 Review of mixture model for competing risk data
The mixture model7 specifies the death probabilities πk(z) and the crude survival P(T > t|D = k, z), k = 1, ..., K. It does not require the independence assumption among competing risks. The cause of death D follows a multinomial distribution with probabilities P1(z), ..., PK(z) with
| (2) |
where μk is a scalar constant and γk is a vector of regression coefficients. Because , we set μK = 0 and γK = 0 for identifiability purpose. We also assume cause D = 1 is death due to cancer of interest. The personal cure rate is then calculated as 1 − P1(z).
The functions Qk(t|z) = P(T > t|D = k, z), k = 1, ..., K are called crude cause-specific survival functions.1 Larson and Dinse7 use a proportional hazards (PH) model for Qk(t|z):
| (3) |
where , qk(t) is the baseline hazard function and βk is a vector of regression coefficients. The overall survival function can also be expressed as
| (4) |
It can be shown that Sk(t|z) ≥ Qk(t|z) (see Appendix I) for all t unless Pj(z) = 0 for j ≠ k. The inequality implies that, under the independent competing-risks assumption, the net survival function in the absence of other causes is always greater than the crude cause-specific survival function in the presence of other causes. Hence, the Kaplan-Meier and actuarial estimates always overestimate the conditional crude cause-specific survival probability.
2.2 The mixture model for grouped survival data
Survival times from population-based cancer registries are usually grouped into annual or monthly intervals, Ij = (tj−1, tj] for j = 1, ..., J, where t0 = 0 and tJ = τ denote the beginning and end of follow-up, respectively. For the cohort with covariates z, let njz be the number of people alive at the beginning of interval Ij, dkjz be the number of people who die from cause k, k = 1, ..., K and let ljz be the number of people lost to follow-up in the interval. For simplicity of notation, we omit the subscript z and denote the observed data as . The total number of people who die during interval Ij is .
The probability of dying from cause k during the interval Ij is
| (5) |
Because some people are lost to follow-up during the interval, a widely used technique is to adjust the person-years at risk as and the resulting estimate is called the actuarial estimate. Gail6 showed that the actuarial estimate is a good approximation of the maximum likelihood estimate (MLE) of S(t) under the assumption that time when lost to follow-up and time of death from competing risks are independent. The actuarial estimate can also be justified by assuming that time when lost to follow-up is uniform in interval Ij. Then the number of people who are censored at time tj, i.e., T > tj, is
Let θ0 = (μk, γk, k = 1, ..., K) be the parameters in the logistic model (2) for the cause of death and let θk be the parameters for the crude survival functions Qk(t), k = 1, ..., K. The likelihood function for observed competing-risk survival data is
where θ = (θ0, θ1, ..., θK). As the Newton-Raphson method requires the calculation of a complex Hessian matrix, the Expectation-Maximization (EM) algorithm is used to find the MLEs of θ.
The complete data are (, dkj, ckj, k = 1, ..., K, j = 1, ..., J), where ckj is the number of people censored at time tj who would ultimately die from cause k. Using Equation (4), the loglikelihood for the complete data is log , where
The E-step assigns the censored observations, i.e., the people who are lost to follow-up or who are still alive at the end of the study, into one of the K causes of death according to their conditional probabilities P(D = k|z, t > tj). The expected number of deaths due to causes k in interval Ij for those censored people is
| (6) |
The M-step involves maximizing the loglikelihood functions and , k = 1, ..., K.
The MLEs of the parameters in πk(z) can be obtained by multinomial logistic regression, and the estimation of the parameters in Qk(t|z) depends on the model specifications for Qk(t|z). The popular models include the Weibull and Gompertz models and the semi-parametric proportional hazards model. For arbitrary interval-censored survival data, various methods are proposed by Finkelstein10, Pan and Chappell11 and Goetghebeur and Ryan12. For grouped survival data, we follow Prentice and Gloeckler13 and write the loglikelihood as
where pkj(z) = Qk(tj|z)/Qk(tj−1|z) and . Let , j = 1, ..., J. Then,
| (7) |
The estimates of θk = (αk1, . . . , αkJ, βk) can be obtained by SAS PROC LOGISTIC.
Several factors complicate variance estimation for the parameter estimates. First the dimension of parameters is large for the semiparametric model. Second variance estimates do not come from the EM algorithm as a byproduct. Several approaches to calculate the observed information matrix in an EM context have been proposed.14,15 But, these approaches involve tedious algebra and are analytically intractable. Another variance estimator, which is simple to compute and also turns out to have good small sample properties, is based on multiple imputation.12,16 First the expected numbers of deaths due to cause k in interval Ij, ckj, j = 1, ..., J, are imputed M times using a multinomial distribution with conditional probabilities given in (6), after the final step of the EM algorithm is completed. Then for each imputed “complete” data set, a point estimate and a variance estimate vm of θ are calculated, m = 1, ..., M. Let . The variance estimate for the MLE is given by
| (8) |
This is a weighted sum of the within-imputation variance and the between-imputation variance.
2.3 Estimating net survival from the mixture model
The direct output from the mixture model consists of death probabilities from different causes and the conditional crude survival function. Under the assumption of independent competing risks, the net survival Sk(t|z) can be derived from output from the mixture model. We assume that hk(t|z) = wkj(z)h(t|z) for t ∈ Ij, where the weights wkj(z) are constants for each interval Ij and covariate z. From (1), we have
hence
| (9) |
Because
the net survival function Sk(tj|z) can be estimated by
| (10) |
where wkl is given by (9). The variance estimates for crude survival Qk(t|z) and net survival Sk(tj|z) are given in Appendix II. The relationship between Sk(t|z) and ST (t|z) and implies that the net survival functions can be calculated as a by-product of the mixture survival model under the independent competing risk assumption.
3 Application
The Surveillance, Epidemiology, and End Results (SEER) Program of the National Cancer Institute is an authoritative source of information on cancer incidence and survival in the United States (http://www.seer.cancer.gov). SEER currently collects and publishes cancer incidence and survival data from population-based cancer registries covering approximately 26 percent of the US population. SEER coverage includes 23 percent of African Americans, 40 percent of Hispanics, 42 percent of American Indians and Alaska Natives, 53 percent of Asians, and 70 percent of Hawaiian/Pacific Islanders. The SEER Program began collecting data on cancer cases in 1973 in the states of Connecticut, Iowa, New Mexico, Utah, and Hawaii and the metropolitan areas of Detroit and San Francisco-Oakland. In 1974-1975, the metropolitan area of Atlanta and the 13-county Seattle-Puget Sound area were added. These original 9 regions are referred to as the SEER 9 registries, covering 10% of the US population.
Colorectal cancer is the third most common cancer and the third leading cause of cancer-related mortality in the United States. Over the past decade, colorectal cancer incidence and mortality rates have modestly decreased or remained level. The most recent estimates from the American Cancer Society show that there are 106,100 new cases of colon cancer, 40,870 new cases of rectal cancer and 49,920 deaths from colorectal cancer for the year 2009. If diagnosed early and treated successfully, colorectal cancer can be cured. In fact, many colorectal cancer patients live long enough to die ultimately from other causes, most commonly from heart disease.
Because of the long history of the SEER 9 registries, we use the colorectal cancer survival data for illustration. We consider three competing risks (K = 3), namely, colorectal cancer death (D = 1), heart disease death (D = 2) and death due to other causes (D = 3). Here, we use the mixture model to estimate probabilities of dying from different causes given a patient's age at diagnosis, race and the American Joint Committee on Cancer (AJCC) stage. There are remarkable differences between racial and ethnic groups in both incidence and mortality. The mortality rate from colorectal cancer for African Americans is higher than that for whites (American Cancer Society, 2008). To confirm this conclusion, we also test the difference in probabilities of dying from multiple causes between whites and African Americans.
The SEER data we consider consist of 199,715 colorectal cancer cases diagnosed from 1975 to 2002. The end of followup is December, 2003 and the maximum followup time is 28 years. The survival data are then stratified by single age (50, 51,...,99, 100+), race (white, black) and AJCC stage (I, II, III, IV). The average age at diagnosis is 64, and 92% are whites and 8% are African Americans. The percentages of cancer stages I-IV are 20%, 33%, 25% and 22%, respectively.
In the first analysis, we fit separate mixture models for each combination of race and AJCC stage in order to estimate the probabilities of dying from different causes. Age at diagnosis is used as a covariate in equations (2) and (7). To account for the possible quadratic effect of age, the square of age is also included as a covariate. Figure 1 plots the observed and modeled probabilities of dying from colorectal cancer with respect to age at diagnosis. The observed probabilities are calculated as the proportion dying from colorectal cancer for each single age group . The modeled probabilities are the estimates in Equation (2). For example, less than 40% of the patients diagnosed with Stage I colorectal cancer actually die from colorectal cancer. We see that the modeled probabilities fit the observed probabilities reasonably well. The probabilities of colorectal cancer death increase with AJCC stage. This makes sense as a diagnosis of more advanced colorectal cancer implies higher probability of death. Overall, the probability of colorectal cancer death decreases in older patients except for very old ages. Figure 1 also shows that the probabilities of dying from colorectal cancer are much lower for whites than blacks in the same cancer stage.
Figure 1.
Observed and modeled probabilities of dying from colorectal cancer by age at diagnosis *
Figure 2 plots the nonparametric and modeled cumulative probabilities of death due to colorectal cancer within 5 years of cancer diagnosis by age at diagnosis. The nonparametric estimate17 is calculated as
where is the actuarial survival. The modeled probabilities are the estimates of . We see that the 5-year probabilities of colorectal cancer death do not change much with age at diagnosis for stage I, and the corresponding probabilities increase slightly with age at diagnosis for Stage III and IV. This shows that patients with more severe diagnosis are more likely to die from cancer.
Figure 2.
Nonparametric and modeled cumulative probabilities of death due to colorectal cancer within 5 years of diagnosis *
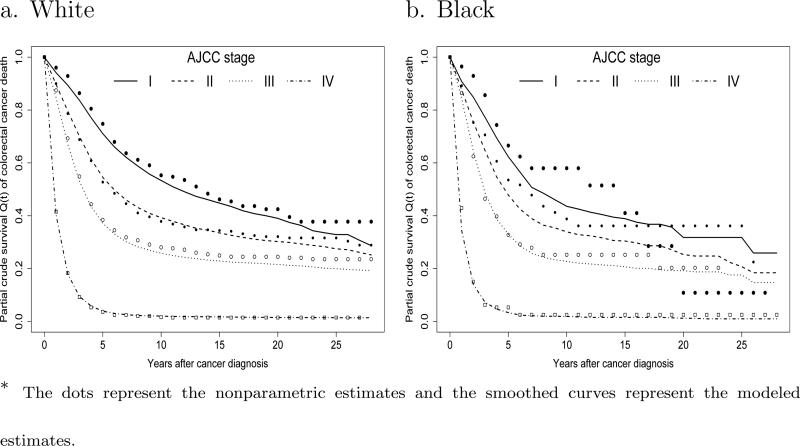
One may be interested in the actual survival pattern after diagnosis. As an example, we show the nonparametric and modeled conditional crude cause-specific survival estimates in the presence of competing risks for patients diagnosed at age 70 in Figure 3. We see that white patients have slightly higher crude survival rates than blacks. Figure 3 also shows that for stage IV cancer the conditional cause-specific survival rates are all less than 5% after 5 years of diagnosis. This implies that for patients diagnosed with stage IV cancer at age 70, 95% of cancer deaths would occur within 5 years after diagnosis.
Figure 3.
Conditional crude survival estimates Q1(t) of colorectal cancer death in the presence of competing risks for patients diagnosed at age 70 *
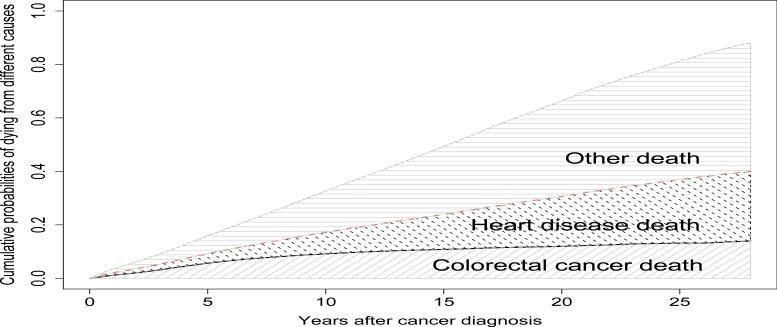
The unconditional cumulative probability of death due to cancer versus other causes is useful to describe the experience of individual patients. For example, Figure 4 shows the cumulative probabilities of death due to three causes for white patients diagnosed with stage I cancer at age 70. The personal cure rate is about 90%. The cumulative colorectal cancer death probability levels off after about 10 years from diagnosis, but the probability of death due to other causes still increases. So if a patient does not die from colorectal cancer within 10 years, it is very likely that he will die from another cause.
Figure 4.
Cumulative cause-specific probabilities of death for white patients diagnosed at age 70 with Stage I colorectal cancer
The analysis above can be used to describe the survival patterns experienced by cancer patients. In contrast, a statistical model presents a great advantage when some form of inference is required. As we see from Figures 1 and 2, differences exist in survival patterns between racial and ethnic groups. In the second analysis, we perform a formal test to examine the effect of race on probabilities of dying from different causes for each cancer stage. The covariate z of interest is the indicator of being black. In logistic model (2), the parameters exp(γ1) and exp(γ2) represent the odds ratios of being blacks on the probabilities of dying from cancer and heart disease, respectively. For example, exp(γ1) > 1 means that blacks have higher probability of dying from colorectal cancer than whites. The estimates of odds ratio and the 95% confidence intervals (CI) are shown in Table 1. For stages I and II, blacks have a significantly higher risk of dying from colorectal cancer than whites. For Stage III, whites and blacks have similar cause-specific probabilities. For Stage IV, blacks have a lower probability of dying from cancer, but a higher probability of dying from heart disease.
Table 1.
Odds ratios of race on the probabilities of dying from different causes
| Stage | Parameter | Cause of death | Estimate | 95% CI | p-value |
|---|---|---|---|---|---|
| I | exp(γ1) | Cancer | 1.322 | (1.187, 1.472) | <0.001 |
| |
exp(γ2) |
Heart Disease |
1.202 |
(1.047, 1.381) |
0.009 |
| II | exp(γ1) | Cancer | 1.229 | (1.138, 1.328) | <0.001 |
| |
exp(γ2) |
Heart Disease |
1.046 |
(0.954, 1.147) |
0.335 |
| III | exp(γ1) | Cancer | 1.060 | (0.968, 1.161) | 0.212 |
| |
exp(γ2) |
Heart Disease |
1.017 |
(0.896, 1.154) |
0.797 |
| IV | exp(γ1) | Cancer | 0.807 | (0.729, 0.894) | <0.001 |
| exp(γ2) | Heart Disease | 1.223 | (1.037, 1.441) | 0.017 |
By using a logistic model for probabilities of dying from different causes, the mixture model implicitly assumes that hazards from different causes are constant after the end of the follow-up. Usually, hazards due to causes other than cancer will increase remarkably as people get older, while the hazard due to cancer death may remain similar with respect to age. Misspecification of the logistic model might yield biased estimates. It is also necessary to have a sufficiently long follow-up time to observe most deaths and their corresponding causes, so that death probabilities can be modeled reliably.
As shown in Figures 1 and 2, the mixture model provides a reasonably good fit to the observed data. However, one can argue that most of the patterns and probabilities can be easily obtained by smoothing the raw data. For example, a multinomial logistic model with splines can be used to estimate πk, the probabilities of dying from different causes. Here, we are trying to model the probabilities πk and crude survival functions Qk(t) simultaneously. This provides a complete picture of survival patterns after cancer diagnosis.
4 Discussion
In this article, we apply the mixture model to grouped survival data with competing risks from population-based cancer registries. This model can be used to estimate probabilities of death due to different competing causes. The personal cure rate is helpful to describe the survival experience after cancer diagnosis. This model can also be applied to data from clinical trials. For example, one can compare the probabilities of different types of failures to evaluate the risk and benefit of two treatment options. Physicians can determine the appropriate treatments for cancer patients based on their comorbidities and prognosis.
An alternative approach to competing-risk data is to model the net survival functions5,13. To ensure identifiability, the competing risks are assumed to be independent. The cancer patients, especially in their old ages, have higher comorbidity problems than the general cancer-free population. The mixture model assumes that the process of loss to follow-up is independent of the competing risks of death, but it does not require independence among the K competing risks. Another advantage of using the mixture model is that the net survival function can be derived as a by-product.
Acknowledgements
The research of Dr. Yu was carried out in part at the Information Management Services, Inc. and was supported in part by the contract with the National Cancer Institute and by the Intramural Research Program of the National Institute on Aging.
Appendix I. Proof of Sk(t|z)≥Qk(t|z)
Based on equation (3), we have . Because and from (1),
Thus
Appendix II. Variances for Qk(t|z) and Sk(t|z)
The variances of Qk(t|z) and Sk(t|z) can be derived from the covariance matrix of θ in (8) using the delta method. Let θk = (αk1, ..., αkJ, βk) be the parameters in θk(t|z) and V(θk) be the covariance matrix of θk. Because
the variance of Qk(tj|z) can be calculated as
where γkj = (log pk1(z), ..., log pkj(z), z log Qk(tj|z)).
Let . Then . Assuming the parameters (θk, μk, γk), k = 1, ..., K, are functionally independent, we have
| (11) |
where the partial derivatives of are:
The variance of ST(tj|z) can be calculated as:
where V(θ) is the variance of .
The variance of Sk(tj|z) is given by , where
Based on Equation (10), . We have
where is given in (11) and
Note that for i ≠ k.
References
- 1.Cronin A, Feuer EJ. Cumulative cause-specific mortality for cancer patients in the presence of other causes: a crude analogue of relative survival. Statistics in Medicine. 2000;19:1729–1740. doi: 10.1002/1097-0258(20000715)19:13<1729::aid-sim484>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 2.Shairer C, Mink PJ, Carroll L, Devesa SS. Probabilities of death from breast cancer and other causes among female breast cancer patients. Journal of the National Cancer Institute. 2004;96:1311–1321. doi: 10.1093/jnci/djh253. [DOI] [PubMed] [Google Scholar]
- 3.Gordon NH. Application of the theory of finite mixtures for the estimation of ‘cure’ rates of treated cancer patients. Statistics in Medicine. 1990;9:397–407. doi: 10.1002/sim.4780090411. [DOI] [PubMed] [Google Scholar]
- 4.Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data. 2nd ed. Wiley; New York: 2002. [Google Scholar]
- 5.David HA, Moeschberger ML. The Theory of Competing Risks. Griffin; London: 1978. [Google Scholar]
- 6.Gail M. A review and critique of some models used in competing risk analysis. Biometrics. 1975;31:209–222. [PubMed] [Google Scholar]
- 7.Larson MG, Dinse GE. A mixture model for the regression analysis of competing risks data. Applied Statistics. 1985;34:201–211. [Google Scholar]
- 8.Cutler SJ, Ederer F. Maximum utilization of the life table method in analyzing survival. Journal of Chronical Disease. 1958;8:699–712. doi: 10.1016/0021-9681(58)90126-7. [DOI] [PubMed] [Google Scholar]
- 9.Chiang CL. Competing risks in mortality analysis. Annual Review of Public Health. 1991;12:281–307. doi: 10.1146/annurev.pu.12.050191.001433. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein DM. A proportional hazards model for interval-censored failure time data. Biometrics. 1986;42:845–854. [PubMed] [Google Scholar]
- 11.Pan W, Chappell R. Estimation in the Cox proportional hazards model with left-truncated and interval-censored data. Biometrics. 2002;58:64–70. doi: 10.1111/j.0006-341x.2002.00064.x. [DOI] [PubMed] [Google Scholar]
- 12.Goetghebeur E, Ryan L. Semiparametric regression analysis of interval-censored data. Biometrics. 2000;56:1139–1144. doi: 10.1111/j.0006-341x.2000.01139.x. [DOI] [PubMed] [Google Scholar]
- 13.Prentice RL, Gloeckler LA. Regression analysis of grouped survival data with application to breast cancer data. Biometrics. 1978;34:57–67. [PubMed] [Google Scholar]
- 14.Louis TA. Finding the observed information matrix when using the EM algorithm. Journal of the Royal Statistical Society Series B. 1982;44:226–233. [Google Scholar]
- 15.Meng XL, Rubin DB. Using EM to obtain asymptotic variance-covariance matrices: the SEM algorithm. Journal of the American Statistical Association. 1991;86:899–909. [Google Scholar]
- 16.Rubin DB, Schenker N. Multiple imputation in health-care data bases: An overview and some applications. Statistics in Medicine. 1991;10:585–598. doi: 10.1002/sim.4780100410. [DOI] [PubMed] [Google Scholar]
- 17.Gaynor JJ, Feuer EJ, Tan CC, Wu DH, Little CR, Straus DJ, Clarkson BD, Brennan MF. On the use of cause-specific failure and conditional failure probabilities: examples from clinical oncology data. Journal of the American Statistical Association. 1993;88:400–409. [Google Scholar]