Abstract
Background
Numerous cases of predator-induced polyphenisms, in which alternate phenotypes are produced in response to extrinsic stimuli, have been reported in aquatic taxa to date. The genus Daphnia (Branchiopoda, Cladocera) provides a model experimental system for the study of the developmental mechanisms and evolutionary processes associated with predator-induced polyphenisms. In D. pulex, juveniles form neckteeth in response to predatory kairomones released by Chaoborus larvae (Insecta, Diptera).
Results
Previous studies suggest that the timing of the sensitivity to kairomones in D. pulex can generally be divided into the embryonic and postembryonic developmental periods. We therefore examined which of the genes in the embryonic and first-instar juvenile stages exhibit different expression levels in the presence or absence of predator kairomones. Employing a candidate gene approach and identifying differentially-expressed genes revealed that the morphogenetic factors, Hox3, extradenticle and escargot, were up-regulated by kairomones in the postembryonic stage and may potentially be responsible for defense morph formation. In addition, the juvenile hormone pathway genes, JHAMT and Met, and the insulin signaling pathway genes, InR and IRS-1, were up-regulated in the first-instar stage. It is well known that these hormonal pathways are involved in physiological regulation following morphogenesis in many insect species. During the embryonic stage when morphotypes were determined, one of the novel genes identified by differential display was up-regulated, suggesting that this gene may be related to morphotype determination. Biological functions of the up-regulated genes are discussed in the context of defense morph formation.
Conclusions
It is suggested that, following the reception of kairomone signals, the identified genes are involved in a series of defensive phenotypic alterations and the production of a defensive phenotype.
Background
The ability to modulate development in the presence of predators is referred as "inducible defense" or "predator-induced polyphenism" [1]. Of the examples reported to date, the freshwater microcrustacean genus Daphnia, commonly called the waterflea, is considered to be a model case for elucidating the ecological and developmental underpinnings of this process [2]. D. pulex produces structures referred to as neckteeth on its head, primarily in the earlier instars (first to third instar), in the presence of predatory phantom midges (Chaoborus larvae) (Figure 1A, B, C). It is considered that this morphological change in daphnids occurs in response to being exposed to chemical cues referred to as "kairomones" released by predators [3]. Neckteeth effectively decrease the risk of predation because it is difficult for predators to capture Daphnia juveniles that have this outgrowth [3]. Several investigations have shown that D. pulex exhibits sensitivity to Chaoborus kairomones during embryonic development [4,5]. In addition, the additive effects of these chemical cues and sensitive phases of D. pulex embryos and juveniles have been elucidated [6].
Figure 1.
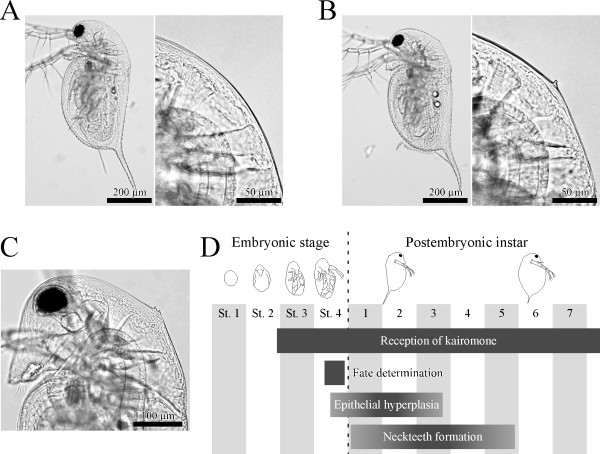
Chaoborus-induced defense morph formation in D. pulex. Chaoborus-induced defense morph formation in D. pulex. (A) A normal first-instar juvenile of Daphnia pulex (magnified view of occipital region: right). (B) A first-instar juvenile bearing neckteeth (magnified view of neckteeth: right). (C) A second-instar juvenile bearing larger neckteeth with two spines. (D) Schematic diagram of the developmental events in the process of the defense morph formation in D. pulex (based on [6]).
Predator-induced defenses most commonly involve morphological and behavioral changes, which allow them to escape from predators efficiently and contribute positively toward the overall ecological success of Daphnia [3,7]. The morphological changes in Daphnia that occur in response to the existence of predators are considered to be among the most remarkable examples of polyphenism, particularly since individuals with the same genetic background can express a variety of phenotypes in response to different environmental cues. During postembryonic development, neckteeth have been observed in the first instar, although the rate of induction in this instar is generally weak [6]. Induction rates were generally stronger in the second and third instars, before becoming weaker again in the fourth instar; neckteeth have not been reported in Daphnia after the fifth instar [8,9]. It is thought that Daphnia are most sensitive to kairomone signals during embryonic development [4], but exposure to the chemical cues after the first-instar juvenile stage is also required for the maintenance of these structures [6]. The morphogenesis of neckteeth appears to start during embryogenesis [10]. Although the developmental mechanisms by which multiple alternative phenotypes are produced have attracted considerable interest [11-19], little is known about the molecular basis underlying the developmental regulation of defense morph formation.
The waterflea Daphnia has come to be widely used as a model animal for a variety of reasons [20]. Specifically, since Daphnia is a keystone species in ponds and lakes [21], it has been used for decades as a standard organism for toxicity testing and its toxicological responses to environmental pollutants are well characterized [22,23]. Furthermore, Daphnia pulex is the first crustacean to have had its genome sequenced [23,24], and these new genomic data are likely to facilitate studies in the wide variety of research disciplines that employ Daphnia (e.g. [25,26]). Indeed, the availability of the draft Daphnia pulex genome sequence assembly and annotation v1.1 promotes the development of a new model system for ecological and evolutionary genomics [2,23,27].
In this study, several experiments using molecular tools were designed to elucidate the molecular mechanisms underlying defense morph formation in response to predator kairomones. Our previous study, which established the defense-morph induction by the exposure of dissected embryos to kairomone, revealed that kairomone reception during the late-embryonic stage (stages 3 and 4; the embryonic development is divided into 4 stages) is required for the future development of neckteeth, although the defensive traits only appear in postembryonic instars, particularly the second- and third-instars (Figure 1C) [6]. These findings suggest that, downstream of kairomone reception, the mechanisms responsible for fate determination and neckteeth development are activated in stage-4 embryos and first-instar juveniles, respectively (Figure 1D). In Daphnia, kairomone reception is thought to initiate a series of biological reactions; neuronal signals which are converted into endocrine signals that then subsequently induce changes in the expressions of morphogenetic factors and result in defense morph formation. The occurrence of these reactions has been inferred by recent studies on polyphenic development in insects [10,28-30]. Based on this working hypothesis, we searched the Daphnia genome database (wFleaBase) for genes affecting endocrine, morphogenetic and neuronal regulations, which are considered to be involved in the defense-morph formation. Then, using real-time quantitative RT-PCR, we sought to determine whether the observed mRNA expression levels of these gene sequences changed in response to kairomone treatment in stage-4 embryos and the first-instar juveniles. Furthermore, to identify any unknown genes involved in these processes, we also screened genes that had been differentially expressed in response to kairomone exposure using a differential display method (see Methods). Based on these results, the functions of these genes are discussed in terms of kairomone reception and defense morph formation.
Results and Discussion
Selection of candidate genes
In D. pulex defense morph formation, it is thought that morphogenetic factors such as those that have been identified in many arthropods are expressed downstream of physiological regulation [30,31]. The following 31 candidate genes were identified in the D. pulex genome: Hox genes [labial (lb), proboscipedia (pb), Hox3, Deformed (Dfd), Sex combs reduced (Scr), Antennapedia (Antp), Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B)], morphogenetic genes [Distal-less (Dll), aristaless (al), homothorax (hth), dachshund (dac), extradenticle (exd), escargot (esg), teashirt (tsh), epidermal growth factor receptor (EGFR1, 2), spitz (spi), decapentaplegic (dpp), wingless (wg) and hedgehog (hh)], endocrine genes [juvenile hormone acid methyltransferase (JHAMT), Methoprene-tolerant (Met), ultraspiracle (USP), ecdysone receptor (EcR), insulin-like receptor (InR), insulin receptor substrate-1 (IRS-1) and forkhead box O (FOXO)] and neuronal genes [tyramine beta-monooxygenase (TBM) and dopamine beta-monooxygenase (DBM)]. BLASTX searches http://blast.ncbi.nlm.nih.gov/Blast.cgi confirmed that the predicted D. pulex sequences are homologues of the candidate genes (Table 1, Additional file 1).
Table 1.
Expression profiles of investigated candidate genes
| Relative expressiona | |||
|---|---|---|---|
| Candidate gene (abbreviation) | Daphnia pulex gene ID | Embryo | Juvenile |
| Hox gene | |||
| labial (lb) | Dappu-36672 Dappu-97497 |
1.35 | 1.598 |
| proboscipedia (pb) | Dappu-44300 Dappu-97500 |
1.433 | 1.276 |
| Hox3 | Dappu-9456 | 1.312 | 1.9 |
| Deformed (Dfd) | Dappu-44270 Dappu-97505 |
1.35 | 1.541 |
| Sex combs reduced (Scr) | Dappu-37195 Dappu-44375 Dappu-97506 |
1.199 | 1.55 |
| Antennapedia (Antp) | Dappu-44334 Dappu-236216 |
1.254 | 1.248 |
| Ultrabithorax (Ubx) | Dappu-9277 Dappu-221891 |
1.253 | 1.601 |
| Abdominal-A (Abd-A) | Dappu-29076 | Not amplified | |
| Abdominal-B (Abd-B) | Dappu-29045 Dappu-97516 |
1.235 | 1.82 |
| Morphogenetic gene | |||
| Distal-less (Dll) | Dappu-9287 | 1.365 | 1.2 |
| aristaless (al) | Dappu-37455 | 1.212 | 1.597 |
| homothorax (hth) | Dappu-4560 Dappu-5009 |
1.1 | 1.539 |
| dachshund (dac) | Dappu-94521 Dappu-94522 Dappu-232746 Dappu-232754 |
1.05 | 1.591 |
| extradenticle (exd) | Dappu-219790 | 1.128 | 2.131 |
| escargot (esg) | Dappu-50534 | 0.7 | 1.951 |
| teashirt (tsh) | Dappu-99118 | 1.276 | 1.377 |
| epidermal growth factor receptor 1 (EGFR1) |
Not predicted | 1.193 | 1.505 |
| epidermal growth factor receptor 2 (EGFR2) |
Dappu-9119 Dappu-53919 |
1.174 | 1.67 |
| spitz (spi) | Dappu-271340 | 1.101 | 1.181 |
| decapentaplegic (dpp) | Dappu-40449 | 0.992 | 1.471 |
| wingless (wg) | Dappu-290640 | 1.306 | 1.616 |
| hedgehog (hh) | Dappu-290571 | 1.122 | 1.579 |
| Endocrine gene | |||
| juvenile hormone acid methyltransferase (JHAMT) | Dappu-300180 | 0.365 | 2.707 |
| Methoprene-tolerant (Met) | Dappu-247693 | 0.943 | 1.584 |
| ultraspiracle (USP) | Dappu-219609 | 0.775 | 1.354 |
| ecdysone receptor (EcR) | Dappu-319648 | 0.736 | 1.158 |
| insulin-like receptor (InR) | Dappu-270048 | 0.84 | 1.956 |
| insulin receptor substrate-1 (IRS-1) | Dappu-52188 Dappu-304473 |
0.825 | 1.81 |
| forkhead box O (FOXO) | Dappu-109005 | Not amplified | |
| Neuronal gene | |||
| tyramine beta-monooxygenase (TBM) | Dappu-1839 | 1.191 | 3.897 |
| dopamine beta-monooxygenase (DBM) | Dappu-62540 | 1.14 | 1.686 |
a Expression levels in the presence of the kairomones are displayed relative to control medium, measured by first real-time quantitative PCR. Values in bold indicate more than 1.5-fold increase.
See additional file 1 placing annotations of these genes.
Expression profiles of candidate genes
The relative expression levels of the candidate genes were quantified using real-time quantitative RT-PCR to examine whether these genes were differentially expressed after exposure to the predator kairomone in the embryonic stage (stage 4) and the postembryonic instar (first-instar). 18S ribosomal RNA, actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes were tested as reference genes for real-time qPCR. Since GAPDH was considered to be the most stable gene and its expression levels were closest to those of the candidate genes (data not shown), GAPDH was used as the reference gene. This result was consistent with a previous report of expression stability in D. magna following exposure to the drug, ibuprofen [32].
Real-time qPCR revealed that 21 of the 31 candidate genes were up-regulated in the presence of kairomones in first-instar juveniles. Conversely, none of the candidate genes were up-regulated by more than 1.5-fold at the embryonic stage 4 (Table 1, Additional file 1). Two genes (abd-A and FOXO) were not amplified by PCR, probably because the designed primer sites were inappropriate. Since a number of traits in first-instar juveniles change in response to defense morph formation (e.g. crest epithelial hyperplasia), the candidate genes were thought to be up-regulated in the preceding developmental stage (i.e. embryonic stage 4), although the expected results were not obtained. It is therefore suggested that the up-regulated genes are involved in neckteeth development, which is most pronounced at the second- and third-instar [6]. Furthermore, these results show that the candidate gene approach using a Daphnia genome database can be used for the analysis of the molecular mechanisms responsible for the defense morph formation.
To further clarify differences in the expression of the six candidate genes showing the most marked up-regulation (TBM, JHAMT, exd, InR, esg, Hox3) and the two gene candidates thought to be associated with JHAMT (Met) and InR (IRS-1), we reperformed the real-time qPCR using biological replicates to analyze the detailed expression profiles of kairomone-responsive genes whose functions were suspected of being involved in defense morph formation (Figure 2). Unfortunately, since the level of expression of TBM varied between trials (possibly due to low expression level), we excluded TBM from further analyses.
Figure 2.
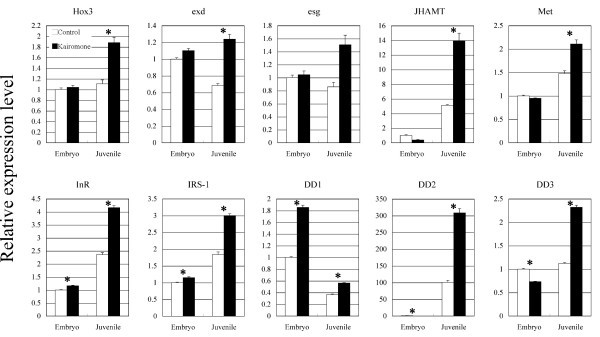
Relative expression levels of JHAMT, Met, InR, IRS-1, Hox3, exd, esg, DD1, DD2 and DD3 in stage 4 embryos and the first-instar juveniles treated with kairomone and control media, analyzed by real-time quantitative PCR. DD1 showed higher levels of expression after exposure to kairomone medium (black) than in control medium (white) during embryonic development. Other genes showed higher levels of expression after exposure to the kairomone medium than in the control during juvenile development. Y-axes indicate relative expression levels normalized by comparison with GAPDH expression (internal control gene). Technical triplicates were performed for all reactions. Bars indicate standard errors. Asterisks indicate significant differences (P < 0.05, based on [57]).
Endocrine genes (JHAMT, Met, InR and IRS-1)
Compared to when no kairomones were present, the expression levels of JHAMT, Met, InR and IRS-1 in first-instar juveniles exposed to kairomones increased by approximately 2.5-, 1.5-, 1.8- and 1.6-fold, respectively (Figure 2). JHAMT encodes the methyltransferase that mediates the final step of juvenile hormone synthesis [33], Met encodes a candidate receptor for juvenile hormone [34,35], InR encodes an insulin/insulin-like growth factor receptor, and IRS-1 encodes a downstream element that interacts directly with InR [36].
In addition to polyphenism [37-39], juvenile hormones (juvenoids) constitute a group of acyclic sesquiterpenoids that are key hormones in the regulation of a variety of physiological regulations in insect development and morphogenesis [28,40]. In crustaceans, methyl farnesoate (MF) is known to act as a juvenile hormone and plays important roles in the regulation of development [41]. For example, male production can be induced in female daphnids treated with MF [42]; however, little is known about other functions of MF in D. pulex. In addition to JH, the insulin-signaling pathway in many animals is also important for the regulation of a variety of developmental processes, including body-size and allometry controls [30,36,43,44]. It has been suggested that the crosstalk between the JH and insulin-signaling pathways is responsible for the expression of morphogenetic factors in the development of beetle horns [30]. Thus, it appears that physiological regulation by these endocrine factors may induce the expression of morphogenetic genes resulting in neckteeth formation in D. pulex.
Morphogenetic genes (Hox3, extradenticle, escargot)
Compared to conditions without kairomones, the expression levels of Hox3, extradenticle and escargot in first-instar juveniles increased by approximately 1.7-, 1.9- and 1.8-fold, respectively, in the presence of kairomones (Figure 2). Hox3 is a member of the Hox cluster and appears to have a typical Hox-like role in the centipede, whereas the insect Hox3 ortholog, zerknüllt (zen), has lost the function of specifying segmental identity during embryogenesis [31,45-47]. Although little is known about the functions of crustacean Hox3, expression in D. pulex has been reported in the nuchal area (where the neckteeth subsequently form) and in the mandibular mesoderm during the early- and mid-embryonic stages, respectively [48], suggesting a possible role in establishing the position of neckteeth development. Furthermore, although Daphnia neckteeth cannot be considered to be homologous to appendages, it is possible that the molecular mechanisms for appendage development are co-opted for neckteeth development.
Exd and esg respectively encode a homeobox transcription factor and a zinc finger transcription factor, and both are known to determine the proximal segmental identity of appendages (coxa and trochanter) in Drosophila melanogaster [49]. Furthermore, up-regulation of dac, a known selector gene for the femur and tibia in D. melanogaster, and Dll, which defines tarsus and pretarsus, did not produce as conspicuous a response as exd and esg in first-instar juveniles (1.6- and 1.2-fold, respectively) (Table 1, Additional file 1). This evidence showed that the genes responsible for the determination of proximal appendages were up-regulated in juveniles with neckteeth, implies that these genes might be co-opted for neckteeth formation. However, our results also showed that the expression level of al, a known selector gene for the most distal region of appendage [49], was also higher (Table 1, Additional file 1). This is probably because, in Drosophila, al is also expressed in the proximal regions of appendages [49]. It has recently been reported that Dll and al are both involved in the development of beetle horns, which are not homologous to appendages [29]. This is similar to the situation in the Daphnia neckteeth formation, except that the co-opted regions of appendages are different (proximal or distal). Indeed, further analyses of this hypothesis will provide us with insights, not only into defense morph formation in Daphnia, but also into the evolution of appendage morphology in arthropods.
Exploring novel genes by differential display
Next, differential display was performed to identify any novel genes that were related to neckteeth formation, but which were not included in the candidate gene approach. As a result, we obtained 22 fragments exhibiting differential expressions in response to kairomone exposure. To refine these results further, BLASTN searches were used to compare these fragments against wFleaBase and their coding sequences were predicted using a gene prediction software joined to wFleabase (Gnomon, Dappu v1.1 gene models, SNAP gene predictor). For the single fragment for which the functional sequence could not be predicted by the gene predictor, the full sequence was determined by rapid amplification of cDNA ends (RACE)-PCR. Of these 22 sequences, we performed real-time quantitative RT-PCR to confirm their responsiveness as described for the candidate genes above. As a result, three genes (DD1, DD2 and DD3) showed marked up-regulation in response to kairomone exposure (Figure 2, Table 2, Additional file 2).
Table 2.
Expression profiles of the genes obtained by differential display (DD)
| Relative expressiona | ||
|---|---|---|
| Sequence name | Embryo | Juvenile |
| DD1 | 1.35 | 1.598 |
| DD2 | 1.433 | 1.276 |
| DD3 | 1.312 | 1.9 |
a Expression levels in the presence of the kairomones are displayed relative to control medium, measured by first real-time quantitative PCR. Values in bold indicate more than 1.5-fold increase.
See additional file 2 placing annotations of these genes.
DD1
In stage 4 embryos, DD1 expression was up-regulated approximately 1.9-fold after exposure to kairomones (Figure 2); among all the genes examined in this study, this was the only gene that responded to kairomones in the embryonic stage. BLAST searches suggested that there were no genes homologous to DD1 in other crustaceans and insects. Motif searches using the InterPro database http://www.ebi.ac.uk/Tools/InterProScan/ revealed that DD1 has a signal peptide and a dopamine beta-monooxygenase N-terminal (DOMON) domain. DOMON domains are ubiquitous among plants and animals, and exist in a variety of proteins, including dopamine beta-monooxygenase, in which this domain was originally found [50]. In D. pulex, DD1 is thus considered to be a novel gene containing a DOMON domain. In addition to the aforementioned dopamine beta-monooxygenase (DBM), representative proteins containing a DOMON domain also include tyramine beta-monooxygenase (TBM), which is involved in the biosynthesis of biogenic amines [51]. However, the sequence and the domain structure of DD1 were completely different to those of DBM and TBM (data not shown), and the expression profiles of DD1 also did not correspond to those of DBM and TBM (Table 1, 2, Additional file 1, 2). Consequently, DD1 is considered to play a different role than either DBM or TBM. It is possible that DD1 is involved in kairmone reception and/or fate determination in the defense morph, because DD1 expression was initiated by the presence of kairomones at embryonic stage 4, which is considered to be a critical period for the reception of kairomones [4-6], before declining over the course of postembryonic development (Figure 2).
DD2, DD3
In postembryonic first-instars, expression levels of DD2 and DD3 in the presence of kairomones increased by approximately 3- and 2-fold, respectively (Figure 2). While DD2 showed extremely high homology to bacterial ribosomal RNA (Table 2, Additional file 2), the DD2 sequence in the D. pulex genome database was found to contain introns. Furthermore, the full DD2 sequence obtained by RACE-PCR had a 3' poly(A) tail, which is not typically present in bacterial transcripts and suggests that the identification of DD2 was not the result of contamination. Interestingly, in addition to kairomone responsiveness, the expression levels of DD2 were more than 100-fold higher in the first-instar juveniles than in the embryos (Figure 2). Based on these findings, it is possible that DD2 may have been acquired by horizontal transfer from bacteria.
DD3 exhibited similarity to growth and transformation-dependent protein (GTD-P) (Table 2, Additional file 2). Although GTD-P homologues have been identified in some arthropod species, little is known about its functions. However, it was reported that GTD-P was strongly expressed when a rat pheochromocytoma cell line (PC12) was exposed to nerve growth factor (NGF) [52]. As it was reported that PC12 cells exposed to NGF undergo proliferation, it is possible that GTD-P is involved in the cellular proliferation observed during defense morph formation in D. pulex.
Conclusion
In this study, we compared the gene expression profiles in the presence/absence of predator kairomones in D. pulex embryos and first-instar juveniles. Most of the differences in gene expression induced by the kairomone exposure were observed in postembryonic juveniles, while a single novel gene, DD1, was up-regulated in the embryonic stage. Taken together, a putative physiological and developmental cascade for the defense morph formation consisting of the following steps is suggested: 1) Kairomone reception by embryos, 2) Physiological changes through endocrine mechanisms including JH and insulin pathways, 3) Morphogenesis triggered by pattern formation genes (Figure 3).
Figure 3.
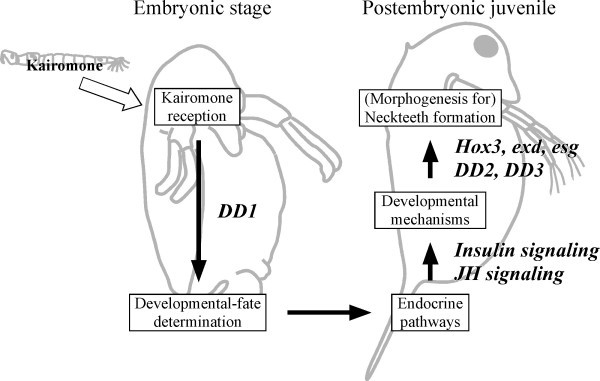
Schematic diagram showing the process of defense morph formation with the putative involved genes and biological pathways suggested by the present study. DD1 is thought to be involved in kairomone reception and/or fate determination during the embryonic stage. The other genes are considered to be involved in the morphogenesis of postembryonic juveniles.
Based on the results obtained in this study, many genes are thought to be involved in the D. pulex defense morph formation. Although we discussed about these genes exclusively in the developmental contexts, it is also possible that they contribute to the plasticity in life history traits such as depth selection [53]. Furthermore, we might underestimate gene expression levels in the kairmone-exposed first-instar juveniles because they were protected in mothers' brood chamber during their sensitive period. For these reasons, given that other genes not identified in this study may also be involved in the predator-induced polyphenisms, further analyses deserve to be undertaken to identify such genes. Furthermore, for the genes identified in this study, localization and functional analyses need to be performed to further clarify the mechanism of inducible defense in D. pulex.
Methods
Animals
The Daphnia pulex clone used in the experiments was provided by the National Institute for Environmental Studies (NIES), Tsukuba, Japan. The NIES clone, originally from Lake Kasumigaura in Japan, was reared in the laboratory at 20°C in aged tap water and fed unicellular green algae (Chlorella Industry Co. Ltd, Fukuoka, Japan) over generations. Using an established rearing method [54], populations of the Daphnia clone were maintained in 1 L beakers in a temperature- and photocycle-controlled incubator (20°C, 16-h light/8-h dark).
Fourth-instar Chaoborus flavicans larvae were collected from a pond at NIES and maintained in dechlorinated tap-water at a density of 10-15 larvae/L for more than 7 days in a temperature- and photocycle-controlled incubator on a diet of D. pulex.
Kairomone medium
After incubation with Chaoborus larvae, the water was filtered using a Whatman GF/C filter (Whatman, London, UK) to remove any daphnid juveniles and particles larger than approximately 1 μm, before being stored in plastic bottles at -20°C. For the experiments, the frozen water was thawed in an incubator (23°C) and used as a rearing medium for D. pulex (Chaoborus-conditioned medium). Dechlorinated tap-water, in which Chaoborus had never been reared, was filtered using a Whatman GF/C filter for use as control medium. The experimental and control media were changed every day.
Induction of defensive morph and total RNA extraction
Stage 4 embryos and postembryonic first instar juveniles of D. pulex were severally used to detect genes specifically expressed in response to the exposure to predator kairomones, with particular emphasis on the expression of those genes that manifested in defense morph formation. We focused on these developmental stages, because, after exposure to kairomones during the embryonic stages of development, fate determination into the defense morph should occur during embryonic stage 4 and morphogenesis of the crest and neckteeth should occur in the first instar (Figure 1B).
To obtain total RNA from stage 4 embryos, very early stage embryos (stage 1-2) were removed from the maternal brood chambers and treated with either the kairomone or control media. After incubation at 20°C for 24-48 h, stage 4 embryos were collected and total RNA was extracted from about 100 individuals for each treatment (kairomone or control), using RNAqueous®-Micro (Ambion, Austin, TX, USA). To obtain total RNA from first-instar juveniles, egg-bearing females were reared in the kairomone and control media (20 individuals/500 ml-beaker of adult female D. pulex: 1 × 106cells/ml of Chlorella for food). After one-week of rearing, newly-born juveniles were harvested and continuously incubated in the kairomone media. From the next day onward, only first-instar juveniles were collected everyday. Harvested individuals were then frozen in liquid nitrogen and preserved at -80°C. Total RNA was extracted from approximately 2,000 daphnia for each treatment (kairomone and control) using an SV Total RNA Isolation System (Promega, Madison, WI, USA). To ensure reproducibility, we conducted an additional series of induction experiments and repeated the RNA extraction procedure for quantitative RT-PCR. In addition, to control for any possible artifacts due to freezing of the kairomone medium or any elution from the plastic bottles used for sample storage, we also tested individuals that had been exposed to control medium that had been frozen and melted in the plastic bottles.
Candidate gene approach
Amino acid sequences of arthropod gene homologues that were thought to be involved in Daphnia defense morph formation (such as Hox genes, morphogenetic genes, endocrine genes and neuronal genes) were obtained using euGene's Arthropod genomes http://insects.eugenes.org/arthropods/, and aligned by the CLUSTALW program from GenomeNet http://align.genome.jp/. Amino acid alignments were made principally with the flour beetle (Tribolium castaneum), the pea aphid (Acyrthosiphon pisum), several species of Drosophila, and the tick (Ixodes scapularis). Based on the aligned sequences, conserved regions were identified and used to perform TBLASTN searches against the wFleaBase http://wfleabase.org/ to identify D. pulex homologues. Subsequently, the predicted coding sequences generated by gene prediction software on wFleaBase (Gnomon, Dappu v1.1 genes, SNAP gene predictor) were used as the sequences for the candidate genes (Table 1, Additional file 1). Furthermore, to improve reliability, these sequences were used as queries to perform BLASTX searches on the National Center for Biotechnology Information (NCBI) server http://blast.ncbi.nlm.nih.gov/Blast.cgi. These analyses helped us to confirm the true homologues of D. pulex (Table 1, Additional file 1).
Differential display
Differential display (DD) was employed to identify genes that were involved in defense morph formation, but which were not detected using the candidate gene approach. The DD assays were performed according to a previously-described method with slight modifications [19,55,56], using the total RNA of stage 4 embryos and first-instar juveniles exposed to the kairomone and control media. Briefly, first-strand cDNA was synthesized from DNase-treated total RNA (500 ng) using SuperScript III (Invitrogen, Carlsbad, CA) and oligo-dT anchor primer (5'-CCC GGA TCC T15 G-3'). The resultant cDNA samples were amplified by PCR in reaction mixtures (20 μl) containing 20 combinations of arbitrary 12-mers with a HindIII site (HindIII-1 to -20 primers, 5'-CGG GAA GCT TN12-3', where N is any base, 4 μM), the anchor primer (20 μM), and AmpliTaq Gold polymerase (0.5 units, Applied Biosystems, Inc., Foster City, CA). The PCR conditions were as follows: one cycle at 94°C for 5 min, followed by 40 cycles at 94°C for 30 sec, 40°C for 2 min and 72°C for 30 sec, and a final extension at 72°C for 5 min. The PCR products were separated on a non-denaturing 12% polyacrylamide gel and visualized by ethidium bromide staining to identify differential bands between the two experimental conditions. To ensure that the results were reproducible, duplicate PCRs and electrophoresis runs were performed.
Subcloning and sequencing
Differential cDNA bands were excised from the gels and re-amplified by PCR. Subsequently, the cDNA fragments were cloned into pGEM-T vector (Promega) according to the manufacturer's instructions before being sequenced with Big Dye terminator kit on a Model 3100 DNA sequencer (Applied Biosystems). To identify homologous sequences and to estimate gene function, similarity searches were performed with obtained sequences using the wFleaBase, NCBI BLAST database and the European Bioinformatics Institute (EBI) InterPro database http://www.ebi.ac.uk/Tools/InterProScan/.
Since the homologous gene was not found using the gene predictor for the obtained DD2 fragment, rapid amplification of cDNA ends (RACE)-PCR was performed using synthesized RACE primers (5'-CCGTTACTCTTTAGGAGGAGACCGCCCC-3' for the 5'-RACE and 5'-TAGGATAGGTGGGAGGCTTTGAAGCGGG-3' for the 3'-RACE) and the SMART RACE cDNA Amplification Kit (Clontech, Palo Alto, CA) to obtain the full-length sequence of DD2. The amplified cDNA fragments were cloned and sequenced as described above, and the full-length sequence thus obtained was subjected to an NCBI BLAST search.
Real-time quantitative RT-PCR
Among the identified genes obtained by the candidate gene approach and by the differential display, the relative expression levels associated with the two experimental conditions (kairomone and control) were quantified and compared using real-time quantitative RT-PCR. Total RNAs of stage 4 embryos and first-instar juveniles that had been exposed to the kairomone and control media were extracted and reverse-transcribed using the conditions described above, except that random hexamer primers were used instead of the oligo-dT primer. Relative quantification of cDNAs was performed using a SYBR Green I chemistry system and ABI Prism 7500 sequence detection system (Applied Biosystems). As endogenous controls of constitutive expression, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Accession No. FJ668125), actin (Accession no. AJ245732) and/or 18S ribosomal RNA (Accession no. AF014011) genes were used. Primers for both target and control genes were designed using Primer Express software (Applied Biosystems, see Additional file 3). Data acquisition and analysis were performed by ABI Prism 7500 SDS software ver. 2.0.1 (Applied Biosystems). The baseline and threshold for the Ct (cycle threshold) were set automatically. Each gene was tested in triplicate and standard errors were calculated by the relative standard curve method as described in User Bulletin 2 for the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Statistical analysis was performed based on [57].
Authors' contributions
HM, MI and TM designed the experiments; HM, MI, NS, YI, AI, HI, YO, SM, SK and RC performed the experiments; HM, MI, YI, AI, HI, SK and TM analyzed the data; HM, MI and TM wrote the paper. All authors have read and approved the final manuscript.
Supplementary Material
Table S1: Annotations and expression profiles of investigated candidate genes.
Table S2: Annotations and expression profiles of the genes obtained by differential display (DD).
Table S3: Protein ID and primers for real-time qPCR.
Contributor Information
Hitoshi Miyakawa, Email: h-miyakawa@ees.hokudai.ac.jp.
Maki Imai, Email: makiimai@ees.hokudai.ac.jp.
Naoki Sugimoto, Email: sugi@ees.hokudai.ac.jp.
Yuki Ishikawa, Email: yuki-i@ees.hokudai.ac.jp.
Asano Ishikawa, Email: asano@ees.hokudai.ac.jp.
Hidehiko Ishigaki, Email: daphnids@ees.hokudai.ac.jp.
Yasukazu Okada, Email: yasukazu@cc.okayama-u.ac.jp.
Satoshi Miyazaki, Email: ihsotas@vetmed.hokudai.ac.jp.
Shigeyuki Koshikawa, Email: skoshikawa@wisc.edu.
Richard Cornette, Email: cornette@affrc.go.jp.
Toru Miura, Email: miu@ees.hokudai.ac.jp.
Acknowledgements
We would like to thank J. Colbourne, B. D. Eads, R. Tollrian, C. Laforsch, L. De Meesterfor, S. Tochinai and M. Sakamoto for their valuable comments on the manuscript. We are also grateful to K. Takamura for his assistance with collecting Chaoborus larvae. Thanks are also due to Y. Naraki, S. Matsuzaki, C. Hiruta, M. Kawasaki and H. Gotoh for their support during the course of this study, specifically, their assistance with field sampling and laboratory experiments. This work was supported by Grants-in-Aid for Scientific Research (Nos. 18370007, 20033002 and 21677001) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Sequencing and certain sections of the analyses were performed at the DOE Joint Genome Institute under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231, Los Alamos National Laboratory under Contract No. W-7405-ENG-36 and in collaboration with the Daphnia Genomics Consortium (DGC). Additional analyses were performed by wFleaBase, developed at the Genome Informatics Lab of Indiana University with the support of Don Gilbert from the National Science Foundation and the National Institutes of Health. Coordination infrastructure for the DGC was provided by The Center for Genomics and Bioinformatics at Indiana University, which is supported in part by the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. Our work benefits from, and contributes to the Daphnia Genomics Consortium.
References
- Gilbert SF. Ecological developmental biology: developmental biology meets the real world. Dev Biol. 2001;233:1–12. doi: 10.1006/dbio.2001.0210. [DOI] [PubMed] [Google Scholar]
- Eads BD, Andrews J, Colbourne JK. Ecological genomics in Daphnia: stress responses and environmental sex determination. Heredity. 2008;100:184–190. doi: 10.1038/sj.hdy.6800999. [DOI] [PubMed] [Google Scholar]
- Tollrian R, Dodson SI. In: The Ecology and Evolution of Inducible Defenses. Tollrian R, Harvell CD, editor. Princeton, Princeton University Press; 1999. Inducible defenses in Cladocera: Constraints, costs, and multipredator environments; pp. 177–202. [Google Scholar]
- Krueger DA, Dodson SI. Embryological induction and predation ecology in Daphnia pulex. Limnol Oceanogr. 1981;26:219–223. doi: 10.4319/lo.1981.26.2.0219. [DOI] [Google Scholar]
- Parejko K. Embryology of Chaoborus-induced spines in Daphnia pulex. Hydrobiologia. 1992;231:77–84. doi: 10.1007/BF00006499. [DOI] [Google Scholar]
- Imai M, Naraki Y, Tochinai S, Miura T. Elaborate regulations of the predator-induced polyphenism in the water flea Daphnia pulex: kairomone-sensitive periods and life-history tradeoffs. J Exp Zool A Ecol Genet Physiol. pp. 788–795. [DOI] [PubMed]
- Gilbert SF. Developmental Biology. 8. Sunderland, Sinauer Associate; 2006. [Google Scholar]
- Tollrian R. Neckteeth formation in Daphnia pulex as an example of continuous phenotypic plasticity: morphological effects of Chaoborus kairomone concentration and their quantification. J Plankton Res. 1993;15:1309–1318. doi: 10.1093/plankt/15.11.1309. [DOI] [Google Scholar]
- Tollrian R. Chaoborus crystallinus predation on Daphnia pulex: can induced morphological changes balance effects of body size on vulnerability. Oecologia. 1995;101:151–155. doi: 10.1007/BF00317278. [DOI] [PubMed] [Google Scholar]
- Laforsch C, Tollrian R. Embryological aspects of inducible morphological defenses in Daphnia. J Morphol. 2004;262:701–707. doi: 10.1002/jmor.10270. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Development and evolution of adaptive polyphenisms. Evol Dev. 2003;5:9–18. doi: 10.1046/j.1525-142X.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental Plasticity and Evolution. New York, Oxford University Press; 2003. [Google Scholar]
- West-Eberhard MJ. Phenotypic accommodation: adaptive innovation due to developmental plasticity, with or without genetic change. Integr Comp Biol. 2003;43:970–970. [Google Scholar]
- West-Eberhard MJ. Phenotypic accommodation: adaptive innovation due to developmental plasticity. J Exp Zoolog B Mol Dev Evol. 2005;304:610–618. doi: 10.1002/jez.b.21071. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and the origin of species differences. Proc Natl Acad Sci USA. 2005;102(Suppl 1):6543–6549. doi: 10.1073/pnas.0501844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T. Proximate mechanisms and evolution of caste polyphenism in social insects: From sociality to genes. Ecol Res. 2004;19:141–148. doi: 10.1111/j.1440-1703.2003.00618.x. [DOI] [Google Scholar]
- Miura T. Developmental regulation of caste-specific characters in social-insect polyphenism. Evol Dev. 2005;7:122–129. doi: 10.1111/j.1525-142X.2005.05014.x. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nijhout HF. Evolution of a polyphenism by genetic accommodation. Science. 2006;311:650–652. doi: 10.1126/science.1118888. [DOI] [PubMed] [Google Scholar]
- Koshikawa S, Cornette R, Hojo M, Maekawa K, Matsumoto T, Miura T. Screening of genes expressed in developing mandibles during soldier differentiation in the termite Hodotermopsis sjostedti. FEBS Lett. 2005;579:1365–1370. doi: 10.1016/j.febslet.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Jenner RA, Wills MA. The choice of model organisms in evo-devo. Nat Rev Genet. 2007;8:311–319. doi: 10.1038/nrg2062. [DOI] [PubMed] [Google Scholar]
- Tessier AJ, Woodruff P. Cryptic trophic cascade along a gradient of lake size. Ecology. 2002;83:1263–1270. doi: 10.1890/0012-9658(2002)083[1263:CTCAAG]2.0.CO;2. [DOI] [Google Scholar]
- Nisbet RM, Muller EB, Lika K, Kooijman SALM. From molecules to ecosystems through dynamic energy budget models. Journal of Animal Ecology. 2000;69:913–926. doi: 10.1046/j.1365-2656.2000.00448.x. [DOI] [Google Scholar]
- Colbourne JK, Singan VR, Gilbert DG. wFleaBase: the Daphnia genome database. BMC Bioinformatics. 2005;6:45–45. doi: 10.1186/1471-2105-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu ME, Colbourne JK, Radivojac J, Lynch M. A microsatellite-based genetic linkage map of the waterflea, Daphnia pulex: On the prospect of crustacean genomics. Genomics. 2006;88:415–430. doi: 10.1016/j.ygeno.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Hebert PD. Habitat-dependent hybrid parentage and differential introgression between neighboringly sympatric Daphnia species. Proc Natl Acad Sci USA. 1993;90:7079–7083. doi: 10.1073/pnas.90.15.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarczyk M. Predator-induced diapause in Daphnia magna may require two chemical cues. Oecologia. 1999;119:159–165. doi: 10.1007/s004420050772. [DOI] [PubMed] [Google Scholar]
- Wade MJ. The co-evolutionary genetics of ecological communities. Nat Rev Genet. 2007;8:185–195. doi: 10.1038/nrg2031. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Control mechanisms of polyphenic development in insects - In polyphenic development, environmental factors alter same aspects of development in an orderly and predictable way. Bioscience. 1999;49:181–192. doi: 10.2307/1313508. [DOI] [Google Scholar]
- Moczek AP, Nagy LM. Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. Evol Dev. 2005;7:175–185. doi: 10.1111/j.1525-142X.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Emlen DJ, Szafran Q, Corley LS, Dworkin I. Insulin signaling and limb-patterning: candidate pathways for the origin and evolutionary diversification of beetle 'horns'. Heredity. 2006;97:179–191. doi: 10.1038/sj.hdy.6800868. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity. 2. Malden, Blackwell Publishing; 2005. [Google Scholar]
- Heckmann LH, Connon R, Hutchinson TH, Maund SJ, Sibly RM, Callaghan A. Expression of target and reference genes in Daphnia magna exposed to ibuprofen. BMC Genomics. 2006;7:175. doi: 10.1186/1471-2164-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T, Itoyama K. Juvenile hormone acid methyltransferase: a key regulatory enzyme for insect metamorphosis. Proc Natl Acad Sci USA. 2003;100:11986–11991. doi: 10.1073/pnas.2134232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Robinson GE, Palli SR. Identification and characterization of a juvenile hormone response element and its binding proteins. J Biol Chem. 2007;282:37605–37617. doi: 10.1074/jbc.M704595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM. Juvenile hormone action: a 2007 perspective. J Insect Physiol. 2008;54:895–901. doi: 10.1016/j.jinsphys.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. The control of growth. Development. 2003;130:5863–5867. doi: 10.1242/dev.00902. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Wheeler DE. Juvenile hormone and the physiological basis of insect polymorphisms. Q Rev Biol. 1982;57:109–133. doi: 10.1086/412671. [DOI] [Google Scholar]
- Nijhout HF. Developmental perspectives on evolution of butterfly mimicry. Bioscience. 1994;44:148–157. doi: 10.2307/1312251. [DOI] [Google Scholar]
- Hartfelder K, Emlen DJ. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editor. Vol. 3. Oxford, Elsevier; 2005. Endocrine control of insect polyphenism; pp. 651–703. full_text. [Google Scholar]
- Truman JW, Hiruma K, Allee JP, Macwhinnie SG, Champlin DT, Riddiford LM. Juvenile hormone is required to couple imaginal disc formation with nutrition in insects. Science. 2006;312:1385–1388. doi: 10.1126/science.1123652. [DOI] [PubMed] [Google Scholar]
- Laufer H, Ahl JSB, Sagi A. The role of juvenile hormones in crustacean reproduction. Am Zool. 1993;33:365–374. [Google Scholar]
- Tatarazako N, Oda S, Watanabe H, Morita M, Iguchi T. Juvenile hormone agonists affect the occurrence of male Daphnia. Chemosphere. 2003;53:827–833. doi: 10.1016/S0045-6535(03)00761-6. [DOI] [PubMed] [Google Scholar]
- Shingleton AW, Das J, Vinicius L, Stern DL. The temporal requirements for insulin signaling during development in Drosophila. PLoS Biol. 2005;3:e289. doi: 10.1371/journal.pbio.0030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Falciani F, Hausdorf B, Schroder R, Akam M, Tautz D, Denell R, Brown S. Class 3 Hox genes in insects and the origin of zen. Proc Natl Acad Sci USA. 1996;93:8479–8484. doi: 10.1073/pnas.93.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. Exploring the myriapod body plan: expression patterns of the ten Hox genes in a centipede. Development. 2002;129:1225–1238. doi: 10.1242/dev.129.5.1225. [DOI] [PubMed] [Google Scholar]
- Panfilio KA, Liu PZ, Akam M, Kaufman TC. Oncopeltus fasciatus zen is essential for serosal tissue function in katatrepsis. Dev Biol. 2006;292:226–243. doi: 10.1016/j.ydbio.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Papillon D, Telford MJ. Evolution of Hox3 and ftz in arthropods: insights from the crustacean Daphnia pulex. Dev Genes Evol. 2007;217:315–322. doi: 10.1007/s00427-007-0141-8. [DOI] [PubMed] [Google Scholar]
- Kojima T. The mechanism of Drosophila leg development along the proximodistal axis. Dev Growth Differ. 2004;46:115–129. doi: 10.1111/j.1440-169X.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- Aravind L. DOMON: an ancient extracellular domain in dopamine beta-monooxygenase and other proteins. Trends Biochem Sci. 2001;26:524–526. doi: 10.1016/S0968-0004(01)01924-7. [DOI] [PubMed] [Google Scholar]
- Hess CR, McGuirl MM, Klinman JP. Mechanism of the insect enzyme, tyramine beta-monooxygenase, reveals differences from the mammalian enzyme, dopamine beta-monooxygenase. J Biol Chem. 2008;283:3042–3049. doi: 10.1074/jbc.M705911200. [DOI] [PubMed] [Google Scholar]
- Marek L, Levresse V, Amura C, Zentrich E, Van Putten V, Nemenoff RA, Heasley LE. Multiple signaling conduits regulate global differentiation-specific gene expression in PC12 cells. J Cell Physiol. 2004;201:459–469. doi: 10.1002/jcp.20087. [DOI] [PubMed] [Google Scholar]
- De Meester L, Dawidowicz P, Van Gool E, Loose CJ. In: The Ecology and Evolution of Inducible Defenses. Tollrian R, Harvell CD, editor. Princeton, Princeton University Press; 1999. Ecology and evolution of predator-induced behavior of zooplankton: depth selection behavior and diel vertical migration; pp. 160–176. [Google Scholar]
- Hanazato T. Induction of helmet development by a Chaoborus factor in Daphnia ambigua during juvenile stages. J Plankton Res. 1990;12:1287–1294. doi: 10.1093/plankt/12.6.1287. [DOI] [Google Scholar]
- Linskens MH, Feng J, Andrews WH, Enlow BE, Saati SM, Tonkin LA, Funk WD, Villeponteau B. Cataloging altered gene expression in young and senescent cells using enhanced differential display. Nucleic Acids Res. 1995;23:3244–3251. doi: 10.1093/nar/23.16.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Kamikouchi A, Sawata M, Takeuchi H, Natori S, Kubo T, Matsumoto T. Soldier caste-specific gene expression in the mandibular glands of Hodotermopsis japonica (Isoptera: termopsidae) Proc Natl Acad Sci USA. 1999;96:13874–13879. doi: 10.1073/pnas.96.24.13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming G, Fidler F, Vaux DL. Error bars in experimental biology. J Cell Biol. 2007;177:7–11. doi: 10.1083/jcb.200611141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Annotations and expression profiles of investigated candidate genes.
Table S2: Annotations and expression profiles of the genes obtained by differential display (DD).
Table S3: Protein ID and primers for real-time qPCR.


