Summary
Opisthorchis viverrini is an important helminth pathogen of humans that is endemic in Thailand and Laos. Adult flukes reside within host bile ducts and feed on epithelial tissue and blood cells. Chronic opisthorchiasis is associated with severe hepatobiliary diseases such as cholangiocarcinoma. Here we report that adult O. viverrini secrete two major cysteine proteases: cathepsin F (Ov-CF-1) and cathepsin B1 (Ov-CB-1). Ov-CF-1 is secreted as an inactive zymogen that auto-catalytically processes and activates to a mature enzyme at pH 4.5 via an intermolecular cleavage at the prosegment-mature domain junction. Ov-CB-1 is also secreted as a zymogen but, in contrast to Ov-CF-1, is fully active against peptide and macromolecular substrates despite retaining the N-terminal prosegment. The active Ov-CB-1 zymogen was capable of trans-activating Ov-CF-1 by proteolytic removal of its prosegment at pH 5.5, a pH at which the Ov-CF-1 zymogen cannot auto-catalytically activate. Both cathepsins hydrolyse human haemoglobin but their combined action more efficiently degrades haemoglobin to smaller peptides than each enzyme alone. Ov-CF-1 degraded extracellular matrix proteins more effectively than Ov-CB-1 at physiological pH. We propose that Ov-CB-1 regulates Ov-CF-1 activity and that both enzymes work together to degrade host tissue contributing to the development of liver fluke-associated cholangiocarcinoma.
Introduction
The helminth parasite Opisthorchis viverrini is an important food-borne pathogen of humans. The disease caused by this parasite, opisthorchiasis or liver fluke disease, is endemic to mainland Southeast Asia, predominantly Northeast Thailand, Laos, Cambodia and central Vietnam where at least 79.8 million people live at risk of infection (Jongsuksuntigul and Imsomboon, 2003; Keiser and Utzinger, 2005; Sripa et al., 2007, 2008; Hotez et al., 2008). Humans become infected by eating raw or undercooked fish containing the infective stage (metacercariae) which excyst in the gut and migrate via the ampulla of Vater into the bile ducts of the liver where they develop and become sexually mature adult flukes. While many O. viverrini infections are asymptomatic, chronic disease is associated with a range of hepatobiliary complications, including inflammation, epithelial desquamation, goblet cell metaplasia, adenomatous hyperplasia and periportal/periductal fibrosis (Kaewpitoon et al., 2008). The most serious symptoms, however, are cholangitis, cholecystitis and the development of bile duct cancer (cholangiocarcinoma; CCA) (Chai et al., 2005; Sripa et al., 2007). CCA is highly prevalent throughout East Asia where there is a strong link between infection with this parasite and human cancer (Sripa et al., 2007). O. viverrini is one of only three metazoan pathogens of humans that is considered a Group 1 carcinogen by the World Health Organization's International Agency for Research on Cancer (Parkin, 2006; Sripa et al., 2007; Bouvard et al., 2009).
Several factors may enhance the development of CCA including mechanical damage, prolonged immunopathological damage and the continual action of parasite-secreted molecules (Kaewpitoon et al., 2008; Sripa and Pairojkul, 2008). It has been demonstrated that proteins secreted by adult O. viverrini induce proliferation of cells in culture suggesting that the parasites liberate carcinogenic molecules (Thuwajit et al., 2004; Smout et al., 2009). Amongst these secretions are proteases that may contribute to the pathologies associated with O. viverrini–induced hepatobiliary abnormalities by degrading macromolecules and damaging cells of the bile duct wall (e.g., Suttiprapa et al., 2009; Pinlaor et al., 2009).
In the current study, we report on the two major cysteine proteases secreted by adult O. viverrini, a cathepsin F (Ov-CF-1), the discovery of which we reported earlier (Pinlaor et al., 2009), and a newly discovered cathepsin B1 (Ov-CB-1). Ov-CF-1 is secreted as an inactive precursor enzyme, or zymogen, that auto-catalytically processes to a fully active mature enzyme at low pH (pH 4.5) via a specific inter-molecular cleavage at the juncture between the N-terminal prosegment and mature protease domain. Ov-CB-1 is also secreted as a zymogen but in contrast to Ov-CF-1 and despite retaining its N-terminal prosegment, this is fully active and, therefore, does not need undergo processing for activation. The active Ov-CB-1 zymogen can trans-activate the Ov-CF-1 zymogen by proteolytic removal of its prosegment at pH 5.5, a pH at which Ov-CF-1 cannot autocatalytically activate. Analysis of the substrate specificity of the proteases using fluorogenic peptides and the physiologically relevant substrate haemoglobin demonstrated that Ov-CF-1 and Ov-CB-1 exhibit overlapping and distinct specificities for peptide bonds. While the two proteases can hydrolyse human haemoglobin to small peptides this is far more effective when both Ov-CF-1 and Ov-CB-1 function in concert. Ov-CF-1 and Ov-CB-1 degrade extracellular matrix proteins fibronectin and laminin at near physiological pH, although Ov-CF-1 exhibited greater activity against these substrates. Our observations suggest that Ov-CF-1 and Ov-CB-1 are primary tissue-degrading proteases secreted by adult O. viverrini and that the hydrolytic activity of Ov-CF-1 is regulated by trans-processing by Ov-CB-1, presumably in the low pH milieu of the trematode gut.
Experimental Procedures
Materials
Z-Phe-Arg-NHMec, Z-Leu-Arg-NHMec, Z-Pro-Arg-NHMec, Z-Arg-Arg-NHMec and Z-Val-Val-Arg-NHMec were obtained from Bachem (St. Helens, UK). E-64, DTT, EDTA, trypsin (proteomics grade), fibronectin and laminin were obtained from Sigma-Aldrich (Sydney, Australia). Restriction enzymes were obtained from New England Biolabs (UK) Ltd. (Hitchin, UK). Primers were obtained from Sigma-Genosys (Pampisford, UK). The pPIC ZαA vector and Pichia pastoris strain X33 were obtained from Invitrogen Corp. (San Diego, CA, USA). Ni-NTA agarose and columns were obtained from Qiagen (Crawley, UK). Pre-cast NuPage 4-12 % Bis-Tris gels and pre-stained molecular weight markers were purchased from Invitrogen (Australia).
O. viverrini RNA extraction and RT-PCR
O. viverrini metacercariae were obtained by digesting the flesh of naturally infected cyprinoid fish (collected from an endemic area of Khon Kaen province, Thailand) with pepsin. About 100 metacercariae of O. viverrini were used to infect hamsters, Mesocricetus auratas, by stomach intubation as previously described (Pinlaor et al. 2004) using procedures approved by the Animal Ethics Committee of Khon Kaen University. Hamsters were euthanized at either three weeks or six weeks post-infection from which the 3 week-old juvenile flukes or adult O. viverrini respectively were recovered by perfusing the bile ducts with phosphate-buffered saline (PBS), pH 7.2. Eggs of O. viverrini were recovered from tissue culture medium where they had been discharged from adult worms (Suttiprapa et al. 2008). Total RNA was prepared from O. viverrini eggs, metacercariae, 3 week-old juveniles and adult flukes using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Contaminating genomic DNA was removed by treatment with DNAse I (Promega). Reverse transcription was performed with 1 μg total RNA using the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas). Aliquots of the resulting cDNA from each life-cycle stage (200 ng) were subjected to PCR amplification under the following conditions: 94°C for 1 minute, 55°C for 1 minute and 72°C for 2 minutes with a final extension at 72°C for 10 minutes. A total of 35 cycles were performed. The following gene-specific primers were used: Ov-CB forward 5′-GGAACAATGGCCTCACTGTC and reverse 5′-CCGCAGTGACTTCATCTTCA-3′ and Ov-CF-1 forward 5′-TCGGACCAGTATTGGACCAAG-3′ and reverse 5′-TACGCTGGAAAGCACACAACG-3′. RT-PCR amplification of constitutively expressed O. viverrini β-actin was performed as a positive control. PCR products were separated by 0.8 % agarose gel electrophoresis and stained with ethidium bromide.
Expression and purification of recombinant O. viverrini peptidases in yeast
Recombinant O. viverrini procathepsin F (Ov-CF-1) was produced in yeast as previously described (Pinlaor et al., 2009). Two O. viverrini expressed sequence tag (EST) clones encoding cathepsin B-like sequences (EST identifier OVAE615 designated Ov-CB-1 and EST identifier OVAE532 designated Ov-CB-2) that were previously identified by Laha et al., (2007) were used to design specific primers for PCR. Using the forward primer 5′-GCGCGCGAATTCGGAGAACTTGAAGATGTA and reverse primer 5′-GCGCGCGCGGCCGCTCCTTTCTCACCCCAGTC both procathepsin B coding sequences immediately downstream of the predicted N-terminal signal peptides were amplified from the lambda TriplEx2 plasmids using Taq polymerase (Invitrogen). Cycling conditions were: 95°C for 1 minute, 60°C for 1 minute and 72°C for 2 minutes with a final extension at 72°C for 10 minutes. A total of 35 cycles were performed. PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions and sequenced at the Bioservice Unit (Bangkok, Thailand) to ensure congruence with the original cDNA. Inserts were digested from plasmid preparations with NotI and EcoRI restriction enzymes and inserted in-frame with the yeast alpha-factor at the NotI/EcoRI site of Pichia pastoris expression vector pPIC ZαA (Invitrogen). Constructs were linearized with SacI and the digestion products employed to transform competent X33 P. pastoris cells using the Pichia EasyComp™ Kit (Invitrogen) according to the manufacturer's instructions.
P. pastoris yeast transformants were cultured in 500 ml BMGY broth, buffered to pH 8.0, in 5 L baffled flasks at 30°C until an OD600 of 2-6 was reached (Collins et al., 2004). Cells were harvested by centrifugation at 2000 × g for 5 min and protein expression induced by resuspending in 100 ml BMMY broth, buffered at pH 6.0 containing 1% methanol (Dowd et al., 1997). Recombinant proteins were affinity purified from yeast using Ni-NTA-agarose (Qiagen). Recombinant propeptidases were dialysed against phosphate buffered saline (PBS) and stored at −20°C.
Autocatalytic Processing and Activation of Ov-CB-1 and Ov-CF-1
To determine whether the recombinant Ov-CB-1 and Ov-CF-1 proenzymes were capable of autocatalytic activation, 20 μl of each enzyme (100 μg) was added to 100 μl activation buffer (0.1 M sodium acetate, pH 4.5 or 5.5; 0.1 M sodium phosphate, pH 6.5 each containing 1 mM DTT and 1 mM EDTA) and incubated for up to 6 h at 37°C. Aliquots of 10 μl were removed at various times and transferred into tubes containing 1 μl of 1 mM E-64 to halt the enzymatic reaction. After separation on 4 – 12 % Bis-Tris NuPage gels, auto-activated Ov-CB-1 and Ov-CF-1 proteins were transferred to polyvinylidene fluoride (PVDF) immobilon-P membranes (Millipore) at 120 mA for 45 min. The membranes were washed with distilled water and stained with 0.025 % Coomassie Brilliant Blue R-250 in 40 % methanol, 10 % acetic acid. Selected protein bands were subjected to 5 cycles of N-terminal (Edman) sequencing using an Applied Biosystems 494 Procise Protein Sequencing System at the Australian Proteome Analysis Facility (Sydney, Australia).
The trans-processing of Ov-CF-1 was carried out by mixing 50 μg of the purified recombinant with 5.0 μg of recombinant Ov-CB-1. The mixtures were incubated in 0.1 M sodium acetate (pH 5.5) containing 1 mM EDTA and 1 mM DTT for 1 h at 37°C, and samples were removed at various time points for analysis on 4 – 12 % Bis-Tris NuPage gels. Following SDS-PAGE, Coomassie blue-stained protein bands corresponding to Ov-CF-1 that had been trans-processed by Ov-CB-1 were excised and analysed by mass spectrometry as described (Robinson et al., 2008b; Robinson et al., 2009). Briefly, individual gel bands were cut into smaller pieces (approximately 1 mm2) and reduced and alkylated with 5 mM tributylphosphine and 20 mM acrylamide (Sigma) in 100 mM NH4HCO3 for 90 min. The excised sections were in-gel digested with trypsin (Sigma Proteomics grade) and the peptides solubilised with 2 % formic acid (Sigma) prior to analysis by nano liquid chromatography electrospray ionisation tandem mass spectrometry (nanoLC-ESI-MS/MS) using a Tempo nanoLC system (Applied Biosystems) with a C18 column (Vydac) coupled to a QSTAR Elite QqTOF mass spectrometer running in IDA mode (Applied Biosystems). Peak list files generated by the Protein Pilot v1.0 software (Applied Biosystems) using default parameters were exported to a local PEAKS (Bioinformatics Solutions Inc.) search engine and employed as queries to search a custom-made database containing only Ov-CF-1 and Ov-CB-1 protein sequences. The present analysis was performed to identify peptides released from the N-terminal of trans-processed Ov-CF-1 following digestion with trypsin. Accordingly, the enzyme specificity of the PEAKS search engine was set to “no enzyme” in order to match peptides generated by Ov-CB-1 cleavage at the N-terminal and by trypsin cleavage at the C-terminal (cuts after Arg/Lys). Propionamide (acrylamide) modification of cysteines was used as a fixed parameter and oxidation of methionines was set as a variable protein modification. The mass tolerance was set at 0.1 Da for both precursor and fragment ions and only one missed cleavage was allowed. Only high-scoring (> 60 %) peptides were considered to be significant.
Enzyme assays with fluorogenic peptide substrates
Assays to monitor auto-activation and trans-processing of the cathepsin zymogens and activity of the mature enzymes were performed as previously described (Stack et al., 2008). Briefly, initial rates of hydrolysis of the fluorogenic dipeptide substrates were monitored by the release of the fluorogenic leaving group, NHMec, at an excitation wavelength of 380 nm and an emission wavelength of 460 nm using a Bio-Tek KC4 microfluorometer. Ov-CF-1 and Ov-CB-1 (0.1 nM) were incubated in a range of 100 mM buffers: glycine-HCl (pH 2.0 – 3.0), formate (pH 3.0 – 4.0), sodium acetate (pH 4.0 – 5.5), sodium phosphate (pH 5.5 – 8.0), and sodium borate (pH 8.0 – 9.0), each containing 1 mM DTT and 1 mM EDTA in the presence of 2 μM substrate.
Preparation of red blood cell lysates and Hb digestion assays
Human red blood cells were washed three times by resuspending 0.25 ml of whole blood in 5 ml PBS and centrifugation at 5000 rpm. The supernatant with the buffy coat was removed each time. After the final wash, the cells were lysed to release haemoglobin (Hb) by adding 1 ml ice-cold distilled H2O for 10 min, after which the suspension was centrifuged at 15000 rpm to remove insoluble material (Brady et al., 1999). To remove any free amino acids or low molecular mass material, Hb was dialysed twice against 1.5 L phosphate-buffered saline (PBS), pH 7.3, for 3 h using a dialysis membrane with a 3000 Da molecular mass cut-off (Sigma, Australia). Hb was quantified using an extinction coefficient of 125 000 M-1cm-1 at 414 nm (Gabay and Ginsburg, 1993) and was in good agreement with the total protein in lysates measured by the Lowry method (Lowry et al., 1951) using BSA as standard.
Hb (1.8 nmoles) was incubated with either Ov-CB-1 or Ov-CF-1 as well as both enzymes together (0.2 nM) in 100 mM sodium acetate (pH 4.0) containing 1 mM DTT for 6 h at 37°C. Control reactions contained no enzyme. The reactions were stopped at 0, 15, 30, 60, 90, 120, 240 or 360 min by addition of 1 μl 1 mM E-64 to the tube. Aliquots were analysed on 4 - 12 % NuPage gels under reducing conditions. Gels were visualised by staining with Flamingo fluorescent protein stain (Bio-Rad) and images of the gels documented using the PharosFX laser imaging system (Bio-Rad).
Analysis of Hb hydrolysis by nanoLC-ESI-MS/MS
Hb digests (15 min samples) were spun at 13,000 rpm for 15 min to remove particulates and were concentrated to a final volume of 15 μl using a Concentrator 5301 (Eppendorf). Peptides were analysed by nanoLC-ESI-MS/MS using a Tempo nanoLC system (Applied Biosystems) with a C18 column (Vydac) coupled to a QSTAR Elite QqTOF mass spectrometer running in IDA mode (Applied Biosystems). Peak list files generated by the Protein Pilot v1.0 software (Applied Biosystems) were exported to local MASCOT (Matrix Science) and PEAKs (Bioinformatics Solutions Inc.) search engines for protein database searching. MS/MS data was used to search 3239079 entries in the MSDB (20060809) database using MASCOT whereas PEAKs software was used to search a custom-made database containing only human Hb-alpha and Hb-beta sequences. The enzyme specificity was set to “no enzyme” and propionamide (acrylamide) modification of cysteines was used as a fixed parameter and oxidation of methionines was set as a variable protein modification. The mass tolerance was set at 100 ppm for precursor ions and 0.2 Da for fragment ions. Only one missed cleavage was allowed. For MASCOT searches, matches with a MOWSE score > 70 were considered to be significant (Robinson and Connolly, 2005; Robinson et al., 2007); and matched peptides achieving a score > 60 % were accepted during PEAKs searches (Robinson et al., 2009). The matching peptides were then mapped onto the primary amino acid sequences of human Hb-alpha and Hb-beta to identify Ov-CB-1 and Ov-CF-1 cleavage sites and to plot P1-P4 preference for each enzyme.
Digestion of extracellular matrix (ECM) proteins by Ov-CB-1 and Ov-CF-1
Fibronectin and laminin (both dissolved in distilled water at 1 mg/ml) were dialysed for two days against 0.1 M sodium acetate (pH 4.5) or PBS (pH 6.5). Digestion reactions contained 2.0 nM of dialysed ECM protein substrates, 1 mM DTT and 1 mM EDTA and 0.2 nM activated peptidase in a final volume of 100 μl of one of the above buffers. Reactions were performed for 3 h at 37 °C, after which they were stopped by addition of E-64 to 10 μM. ECM protein digests were analyzed on reducing 4 – 12 % NuPage gels and visualised by staining with Flamingo fluorescent stain (Bio-Rad).
Results
Autocatalytic activation of Ov-CF-1
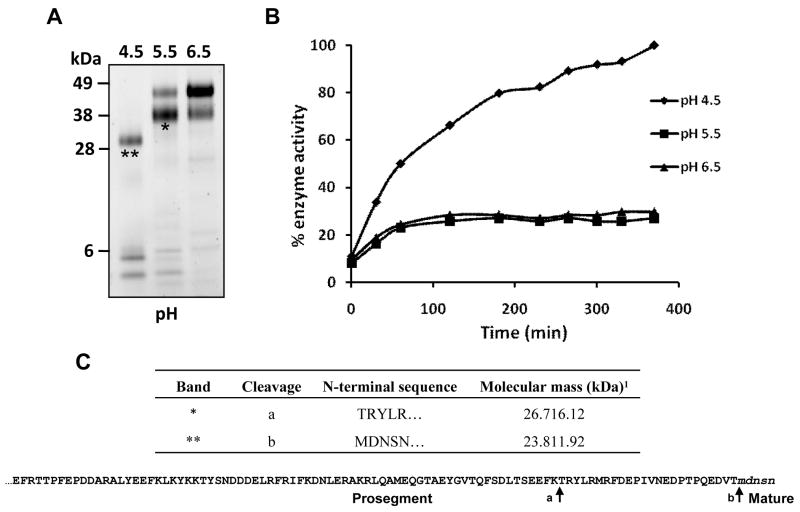
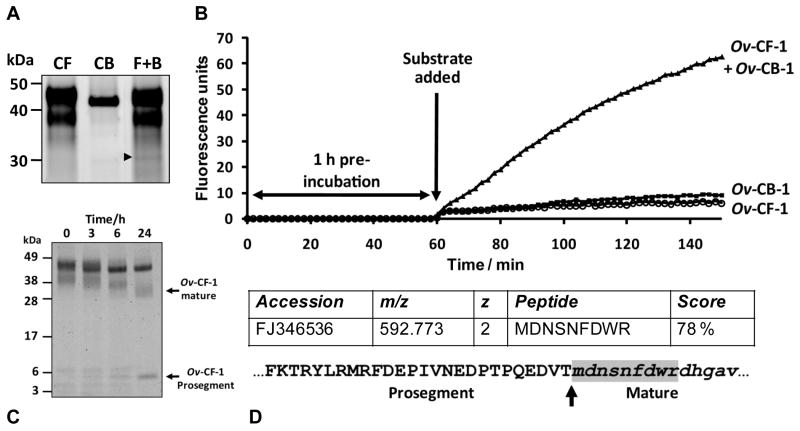
We reported previously the expression of Ov-CF-1 in the yeast P. pastoris and isolation by affinity chromatography (Pinlaor et al, 2009). In that study we found that the Ov-CF-1 proenzyme in the yeast medium was present as two major protein bands migrating at 41 kDa and 47 kDa as a result of differential addition of N-linked glycans. Moreover, the recombinant Ov-CF-1 zymogen was unable to auto-catalytically activate under standard conditions (pH 4.5 for 3 h) that result in auto-activation of other trematode cathepsins (Stack et al., 2008). However, we have now determined that the Ov-CF-1 zymogen can autocatalytically process upon prolonged incubation (6 h) at pH 4.5 by inter-molecular cleavage and removal of the prosegment to release a fully mature and active enzyme (Figure 1). Analysis of the in vitro auto-activation process by 4 -12 % SDS-PAGE shows that following 6 h incubation at pH 5.5 and pH 6.5, the 47 kDa and 41 kDa bands were evident but the 41 kDa band was much more prominent at pH 5.5. N-terminal sequencing showed that the 41 kDa species (pH 5.5) represented an intermediate Ov-CF-1 that had undergone partial prosegment removal via cleavage at Phe-26-Lys-25↓Thr-24, leaving 24 residues of the prosegment still attached to the mature enzyme. In contrast, when incubated at pH 4.5, the 47 kDa and 41 kDa bands were hydrolytically reduced to a single major band of ∼ 30 kDa. N-terminal sequencing confirmed that the 30 kDa band represents the mature enzyme with no prosegment attached generated by cleavage at Val-2-Thr-1 ↓ Met1. This was the same cleavage site observed when Ov-CF-1 was trans-activated in vitro upon addition of F. hepatica cathepsin L1 (Pinlaor et al., 2009). Peptides representing products of the cleaved prosegment were observed below the 6 kDa molecular mass standard (see Figure 1).
Figure 1.
Auto-activation of Ov-CF-1 at pH 4.5. (A) Purified recombinant Ov-CF-1 (50 μg) was incubated in either 0.1 M sodium acetate (pH 4.5 or 5.5) or 0.1 M sodium phosphate (pH 6.5) for 6 h. Aliquots of the reaction mixtures were removed after 6 h, the hydrolysis stopped by addition of E-64 on ice, and analyzed on 4-12 % Bis-Tris NuPage gels (Invitrogen). At pH 6.5, Ov-CF-1 migrated as a major band of 47 kDa representing the unprocessed zymogen (Pinlaor et al., 2009). Following incubation at pH 5.5, the majority of the enzyme migrated as an intermediate band with a molecular mass of 41 kDa. At pH 4.5, Ov-CF-1 had been fully processed and migrated as a single band at 30 kDa with low molecular mass prosegment peptides (< 6 kDa) clearly visible. (B) The Ov-CF-1 auto-activation reactions shown in (A) were assayed for peptidolytic activity against the fluorogenic dipeptide substrate Z-Leu-Arg-NHMec (measured by monitoring the release of the fluorogenic leaving group (-NHMec) over 360 min (6 h) at 37°C. (C) N-terminal sequences obtained for the Ov-CF-1 samples marked with asterisks in (A). The cleavage sites (arrows) identified by N-terminal sequencing are also mapped onto the primary amino acid sequence of the Ov-CF-1 prosegment (bottom). The EF found at the N-terminal was introduced by the EcoRI cloning site used in the pPIC ZαA expression vector. 1Theoretical molecular mass of the Ov-CF-1 polypeptides calculated by Compute pI/MW.
The rate of formation of the active mature enzyme from the inactive zymogen was monitored between pH 4.5 – 6.5 by performing the auto-catalytic reaction in the presence of the fluorogenic substrate Z-Leu-Arg-NHMec and monitoring the release of -NHMec over time. The rate of hydrolysis of Z-Leu-Arg-NHMec, and hence the rate of activation from proOv-CF-1 to mature Ov-CF-1 increased with time and occurred much more rapidly at pH 4.5 than at either pH 5.5 or 6.5 indicating that auto-catalytic activation occurs much more efficiently in an acidic environment. Interestingly, when Ov-CF-1 was incubated at pH 5.5 or pH 6.5, the enzyme showed only modest activity against Z-Leu-Arg-NHMec even after prolonged incubation periods of up to 6 h. This indicates that the 24-residue section of the C-terminal end of the Ov-CF-1 prosegment that remained attached to the mature domain of the 41 kDa intermediate at pH 5.5 inhibits optimal processing of the zymogen.
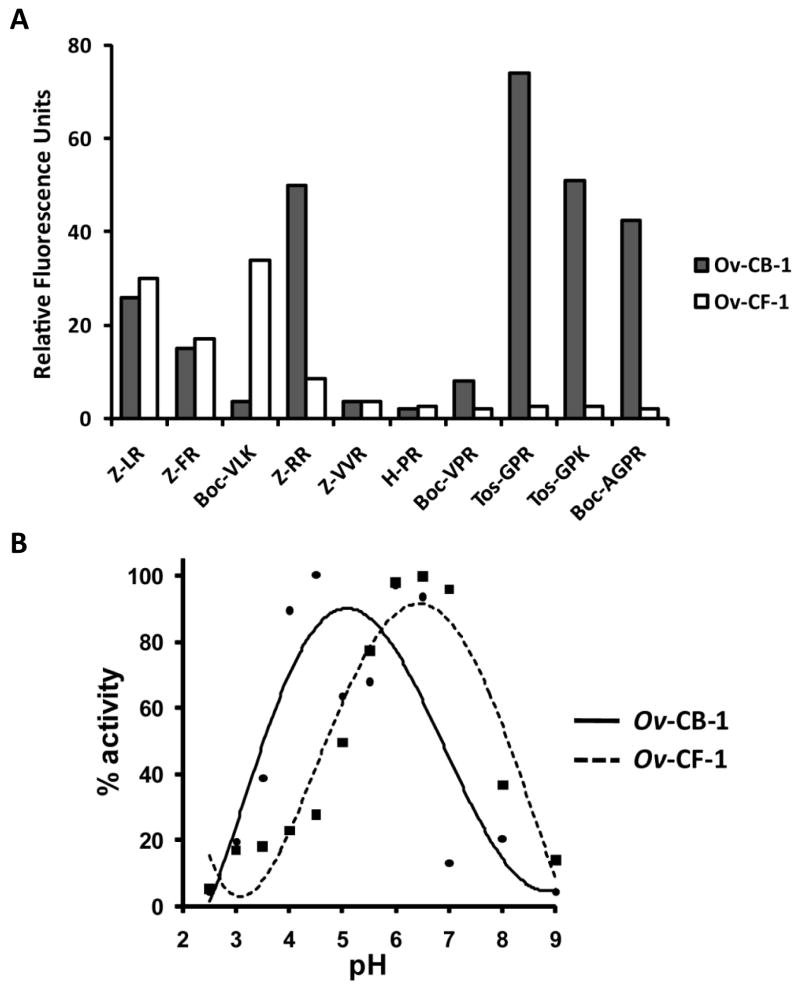
The fully activated Ov-CF-1 cleaved fluorogenic -NHMec substrates over a wide pH range - pH 3.5 - 8.5 with an optimum at pH 6.5 – (Figure 4B) with the preference: Val-Leu-Arg > Leu-Arg > Phe-Arg > Arg-Arg with little or no activity against Pro-Arg or Pro-Lys substrates (Figure 4A). Collectively, the data show that autocatalytic cleavage of the proOv-CF-1 at Val-2-Thr-1 ↓ Met1 generates a 30 kDa mature active enzyme capable of cleaving small and large substrates.
Figure 4.
Activity of Ov-CB-1 and Ov-CF-1 against a panel of diagnostic fluorescent peptides. (A) The relative activity of Ov-CB-1 and Ov-CF-1 (enzymes were pre-incubated in 0.1 M sodium acetate, pH 4.5 for 6 h) against a range of fluorescent substrates was determined by measured by monitoring the release of the fluorogenic leaving group (-NHMec) over 1 h at pH 5.5. (B) Initial rates of hydrolysis of Z-Leu-Arg-NHMec by Ov-CB-1 and Ov-CF-1 were measured over 1 h min at 37°C in a variety of buffers (in the range pH 2-10).
Characterisation and expression of the Ov-CB-1 and Ov-CB-2 transcripts
The full length Ov-CB-1 (OvAE615 clone) and Ov-CB-2 (OvAE532 clone) transcripts were isolated from an adult O. viverrini cDNA library (Laha et al., 2007). For Ov-CB-1, the resulting 1102 bp cDNA (GenBank accession number GQ303560) comprised a 1011 bp open reading frame encoding a 337 amino acid protein. The encoded protein contained a 16-residue N-terminal signal peptide predicted by the SignalP algorithm (Bendtsen et al., 2004). The 1053 bp Ov-CB-2 cDNA (GenBank GQ303559) comprised a 939 bp open reading frame encoding a 313 amino acid protein. The deduced Ov-CB-2 protein contained a putative 22-residue N-terminal signal peptide predicted by SignalP. The molecular mass of the Ov-CB-1 and Ov-CB-2 zymogens (without the predicted N-terminal signal peptides) were calculated as 36.0 kDa and 32.6 kDa with theoretical pI values of 5.28 and 5.95, respectively. The conceptually translated cathepsin B cDNAs (sharing ∼ 62 % amino acid sequence identity) showed identity to cathepsin B proteases from other pathogenic trematodes including Clonorchis sinensis (86 %), Trichobilharzia regenti (52 %) and Schistosoma japonicum (51 %). Primary sequence alignments showed that both Ov-CB-1 and Ov-CB-2 contained the conserved active site dyad residues Cys108 and His277 as well as conserved catalytic Gln102 and Asn297 residues. The predicted mature cathepsin B domain of Ov-CB-1 (molecular mass 28.7 kDa; theoretical pI 5.51) and Ov-CB-2 (molecular mass 25.6 kDa; theoretical pI 5.59) contained two putative N-linked glycosylation sites: Asn126 and Asn226.
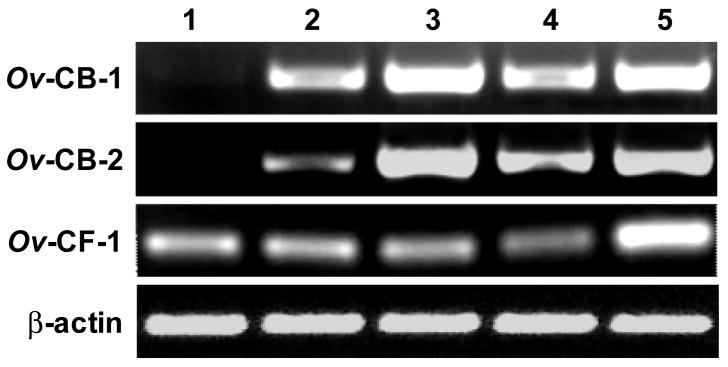
The relative levels of Ov-CB-1, Ov-CB-2 and Ov-CF-1 gene expression in O. viverrini eggs, metacercariae, immature worms and mature adults were determined using RT-PCR (Figure 2). Ov-CF-1 was constitutively expressed at similar levels throughout the life-cycle stages analysed. In contrast, Ov-CB-1 and Ov-CB-2 showed little or no expression in O. viverrini eggs but were co-expressed with Ov-CF-1 at similar levels in the other developmental stages tested.
Figure 2.
RT-PCR analysis of Ov-CB-1, Ov-CB-2 and Ov-CF-1 transcripts. RT-PCR analysis of Ov-CB-1, Ov-CB-2 and Ov-CF-1 expression in (1) O. viverrini eggs, (2) metacercariae, (3) immature worms, (4) adult worms, and (5) an adult worm cDNA library (5). Amplification of constitutively expressed O. viverrini β-actin was used as a control transcript.
The un-processed Ov-CB-1 zymogen exhibits full enzymatic activity
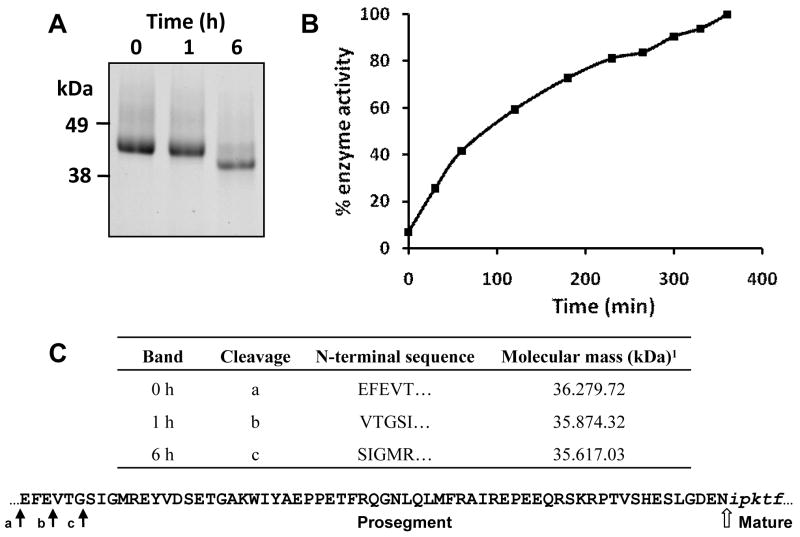
The Ov-CB-1 zymogen was expressed in the yeast P. pastoris and purified from culture supernatants as a single major band migrating at ∼ 44 kDa which was confirmed by N-terminal sequencing as the un-processed zymogen (with the addition of Glu-Phe at the N-terminal that was introduced by the EcoRI cloning site used in the pPIC ZαA expression vector). Since the theoretical molecular mass of the zymogen is 36 kDa, the additional 8 kDa likely resulted from addition of N-linked glycans on the recombinant enzyme, a phenomenon consistent with the smearing observed by SDS-PAGE. Attempts to produce recombinant Ov-CB-2 in P. pastoris were unsuccessful so detailed biochemical analysis was performed using Ov-CB-1 only. In order to determine if Ov-CB-1 undergoes autocatalytic activation, the recombinant protease was incubated at pH 4.5 for 6 h and the reaction monitored by SDS-PAGE. At pH 4.5, the 44 kDa Ov-CB-1 zymogen was clipped to bands migrating at ∼ 43 kDa and ∼ 40 kDa after 1 h and 6 h incubation respectively (Figure 3). However, N-terminal sequencing showed that only the first six residues at the N-terminal of the zymogen were removed by cleavage at Phe-64-Glu-63 ↓ Val-62 and then at Thr-61-Gly-60 ↓ Ser-59. The prosegment of Ov-CB-1 was not removed even after extended incubation periods in a range of buffers (pH 4.0 – 8.0) for up to 6 h (not shown).
Figure 3.
Ov-CB-1 is expressed as an active zymogen. (A) Purified recombinant Ov-CB-1 (50 μg) was incubated in 0.1 M sodium acetate, pH 4.5 for 6 h. Aliquots of the reaction mixtures were removed at time 0 h, 1 h and 6 h, halted with E-64 on ice, and analyzed on 4-12 % Bis-Tris NuPage gels (Invitrogen). The recombinant enzyme showed a progressive decrease in molecular mass during this incubation period suggesting that partial auto-processing had occurred. (B) The auto-activation of Ov-CB-1 shown in (A) was analysed by following the initial rates of hydrolysis of the fluorogenic dipeptide substrate Z-Leu-Arg-NHMec, measured by monitoring the release of the fluorogenic leaving group (-NHMec) over 360 min (6 h) at 37°C. (C) N-terminal sequences obtained for the 0 h, 1 h and 6 h Ov-CB-1 samples shown in (A). The cleavage sites (arrows) identified by N-terminal sequencing are also mapped onto the primary amino acid sequence of the Ov-CB-1 prosegment (bottom). The EF found at the N-terminal was introduced by the EcoR I cloning site used in the pPIC ZαA expression vector and the open arrow represents the predicted juncture of the prosegment and mature enzyme domain. 1Theoretical molecular mass of the Ov-CB-1 polypeptides calculated by Compute pI/MW.
The potential activation of the Ov-CB-1 zymogen was examined in a continuous fluorescence assay by mixing the enzyme with substrate Z-Leu-Arg-NHMec at pH 4.5 and monitoring the release of -NHMec over time. As shown in Figure 3B, the enzyme activity increases rapidly in the first 60 min and although the rate slows slightly after this, the activity continues to increase until the 6 h time point. The data show that, unlike most other papain-like cysteine proteases, the Ov-CB-1 zymogen is highly active even when the N-terminal prosegment is still attached to the mature enzyme domain. The recombinant Ov-CB-1 cleaved fluorogenic -NHMec peptide substrates over a wide pH range (pH 3.5 - 8.5 with an optimum of pH 5.0; Figure 4B) whether the enzyme had been pre-incubated at pH 4.5 or not and showed the preference: Gly-Pro-Arg > Gly-Pro-Lys ≥ Arg-Arg > Ala-Gly-Pro-Arg > Leu-Arg > Phe-Arg > Val-Pro-Arg. There was little or no detectable activity against Pro-Arg, Val-Leu-Lys or Val-Val-Arg (Figure 4A).
Ov-CF-1 is trans-processed and activated by Ov-CB-1
Ov-CF-1 does not undergo autocatalytic processing and activation at pH values greater than 4.5 (Figure 1). However, it is possible that the Ov-CF-1 zymogen is trans-processed and activated by other O. viverrini enzymes such as Ov-CB-1. To investigate this, Ov-CF-1 was incubated in the absence or presence of Ov-CB-1 at pH 5.5 (the Ov-CF-1 zymogen is inactive at pH 5.5) for 1 h at 37°C. The reaction mixtures were then assayed for specific proteolytic activity against the fluorogenic substrate Z-Leu-Arg-NHMec as described above (Figure 5). In these studies, Ov-CF-1 which had been incubated in the absence of Ov-CB-1 at pH 5.5 displayed only minimal activity against Z-Leu-Arg-NHMec which is consistent with the low activity of the enzyme at pH 5.5 shown in Figure 1. At the low concentrations used, Ov-CB-1 alone also displayed very low activity against the Z-Leu-Arg-NHMec substrate at pH 5.5. However, pre-incubation of Ov-CF-1 with Ov-CB-1 led to a marked increase in the specific activity of the cathepsin F against Z-Leu-Arg-NHMec, which increased over the course of the experiment (Figure 5B).
Figure 5.
Exogenous activation of Ov-CF-1 by Ov-CB-1. Purified recombinant Ov-CF-1 was pre-incubated with Ov-CB-1 in 0.1 M sodium acetate containing 1 mM EDTA and 1 mM DTT (pH 5.5) for 1 h at 37°C. Each recombinant was also incubated alone as control reactions. (A) Reactions were analysed on 4-12 % Bis-Tris gels. A band with a molecular mass of ∼ 30 kDa (arrowhead) consistent with the mass of mature cathepsin F, that appeared when Ov-CF-1 and Ov-CB-1 were co-incubated was confirmed as a processed variant of Ov-CF-1 by nanoLC-ESI-MS/MS. (B) Following the 1 h pre-incubation step, the dipeptide substrate Z-Leu-Arg-NHMec was added and the reaction was assayed for proteolytic activity by monitoring the release of the fluorogenic leaving group (-NHMec) over 90 min at 37°C. The specific activity of Ov-CF-1 against Z-Leu-Arg-NHMec increased over the course of the assay when the enzyme was pre-incubated with Ov-CB-1. (C) Prolonged incubation (up to 24 h) of Ov-CF-1 with Ov-CB-1 resulted in a SDS-PAGE profile consistent with the molecular sizes of the Ov-CF-1 mature enzyme and liberated prosegment that was identical to that observed when Ov-CF-1 was trans-activated by exogenous FhCL1 (Pinlaor et al., 2009). (D) The ∼ 30 kDa processed variant of Ov-CF-1 (A, arrowhead) was digested with trypsin and analysed by nano-LC-ESI-MS/MS. A high-scoring putative N-terminal peptide MDNSNFDWR (m/z 592.773) was detected (shaded in grey) and indicates that Ov-CB-1 removes the prosegment of Ov-CF-1 via cleavage at Val-1-Asp-1 ↓ Met1 (arrow) which is also the site used during Ov-CF-1 autocatalysis and exogenous cleavage of the Ov-CF-1 zymogen by FhCL1 (Pinlaor et al., 2009). An annotated MS/MS spectra for the 592.773 precursor ion is shown in Supplementary Figure 1.
Aliquots of the Ov-CF-1/Ov-CB-1 pre-incubation reaction were removed at various time points and analysed by SDS-PAGE. During the reaction, Ov-CF-1 was progressively clipped to faster migrating bands and by 24 h a profile consistent with the molecular sizes of the Ov-CF-1 mature enzyme, 30 kDa, and liberated prosegment was observed (Figure 5C). This SDS-PAGE profile was identical to that observed when Ov-CF-1 was trans-activated by FhCL1 via cleavage of the prosegment at Val-2-Thr-1↓Met1 (Pinlaor et al., 2009). However, for undetermined reasons N-terminal sequencing attempts to identify the cleavage site were not successful.
An alternative approach was therefore undertaken. The 30 kDa band was excised, digested with trypsin and analysed by nanoLC-ESI-MS/MS. A high-scoring doubly charged ion matching with a peptide corresponding to the putative N-terminal of Ov-CF-1 following trans-processing by Ov-CB-1 was identified (Figure 5C; Supplementary file 1). Matched peptide MDNSNFDWR (m/z 592.773), corresponding to residues 1-9 of the mature domain could not be generated by tryptic digest alone since the amino acid preceding this sequence in Ov-CF-1 is Thr (trypsin can only cleave after Lys or Arg). Thus, this peptide is likely to form the N-terminus of the ∼ 30 kDa species that appeared when Ov-CF-1 was trans-processed by Ov-CB-1 making the cleavage site Val-2-Thr-1↓Met1: this site is used by Ov-CF-1 during auto-processing at pH 4.5 (Figure 1) and is the site of exogenous cleavage of the Ov-CF-1 prosegment by FhCL1 (Pinlaor et al., 2009).
Ov-CF-1 and Ov-CB-1 can work in concert to hydrolyse haemoglobin at low pH
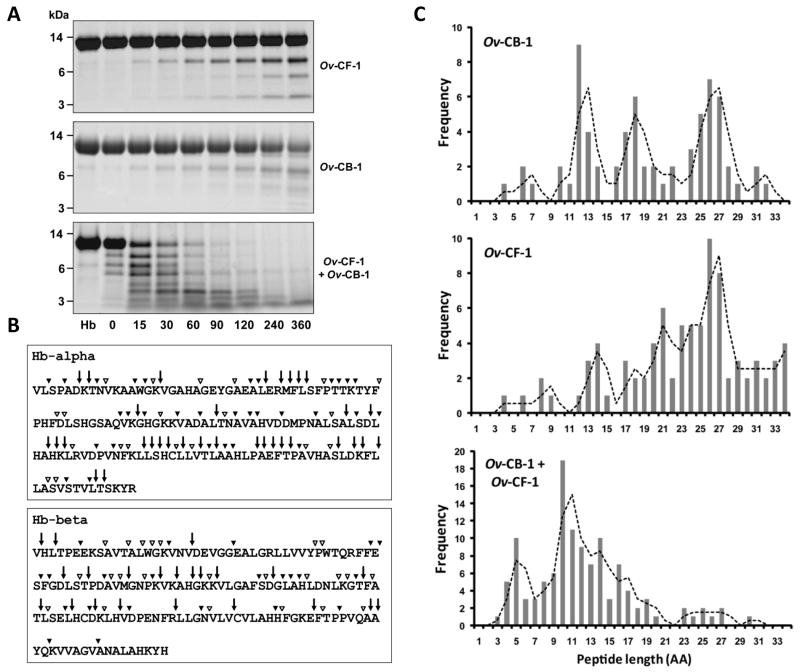
Since blood is a major source of nutrition for O. viverrini the ability of Ov-CF-1 and Ov-CB-1 to hydrolyse Hb was investigated. To examine the process of Hb degradation by Ov-CF-1 and Ov-CB-1, Hb was mixed with each protease individually as well as together at pH 4.0 for up to 360 minutes at 37°C. Reactions were stopped at several time points by addition of E-64 (an irreversible inhibitor of cysteine proteases) and the degradation products analysed by SDS-PAGE. When incubated at pH 4.0 the Hb molecule migrates as a major band at ∼15 kDa representing the Hb-alpha and Hb-beta monomers (Lowther et al., 2009). However, this band was gradually degraded to smaller peptides in the molecular size region of 3 – 10 kDa following incubation with Ov-CF-1 or Ov-CB-1 (Figure 6A). Strikingly, when both proteases were incubated together, Hb was rapidly digested to smaller protein bands (3 – 10 kDa) within the first 15 minutes of the reaction and almost completely degraded between 240 and 360 min (Figure 6A).
Figure 6.
Hydrolysis of human haemoglobin by Ov-CB-1 and Ov-CF-1 and analysis of digests by nanoLC-ESI-MS/MS. (A) Purified human haemoglobin (Hb) was digested by Ov-CB-1 and Ov-CF-1 in 0.1 M sodium acetate buffer (pH 4.0), containing 1 mM DTT and 1mM EDTA at 37°C. Reactions were stopped at time 0 and at various time-points (10, 15, 30, 60, 90, 120, 240 and 360 min) by the addition of the cysteine protease inhibitor E-64 and aliquots analysed on 4-12 % Bis-Tris NuPage gels. (B) Map of Hb α- and β-chains indicating sites of Ov-CB-1 and Ov-CF-1 cleavage. Cleavage sites within Hb present in 15 min reactions as determined by nanoLC-ESI-MS/MS are shown. Arrows, cleavage sites shared by Ov-CB-1 and Ov-CF-1; open arrowheads, Ov-CB-1-specific cleavage sites; filled arrowheads, Ov-CF-1-specific cleavage sites. (C) Frequency of peptides of varying length released following hydrolysis of Hb alpha and beta chains by Ov-CB-1 and Ov-CF-1.
To identify the cleavage sites for Ov-CF-1 and Ov-CB-1 within Hb, the 15 min reaction aliquots were analysed by nanoLC-ESI-MS/MS to determine the masses and sequence identities of the resulting hydrolytic products. Liberated peptides were mapped onto the primary amino acid sequences of human Hb-alpha and Hb-beta to identify the cleavage sites of the O. viverrini proteases (Figure 6B). By 15 min, Ov-CF-1 cleaved Hb-alpha at 64 sites and Hb-beta at 44 sites while Ov-CB-1 cleaved Hb-alpha at 53 sites and Hb-beta at 45 sites. When Ov-CF-1 and Ov-CB-1 were added together, Hb-alpha was cleaved at 70 sites and Hb-beta at 48 sites.
Within a 15 min time-frame Ov-CF-1 and Ov-CB-1 could both generate small peptides (ranging from 4 - 34 amino acids) from Hb but not dipeptides or free amino acids. The average length of the released peptides (from both the Hb alpha and beta chains) was 23 amino acids for Ov-CF-1 with 26-residue peptides occurring most frequently and 19 amino acids for Ov-CB-1 with 12-residue peptides occurring most frequently. When both enzymes digested Hb together, the average length of the released peptides was 12 amino acids with 10-residue peptides occurring most often (Figure 6C). It is unlikely that Ov-CB-1 and Ov-CF-1 cleave all Hb molecules in the same manner and, thus, the cleavage map shown in Figure 6B represents a composite of cleavage sites.
Ov-CB-1 and Ov-CF-1 cleavage sites within Hb indicate different substrate specificities
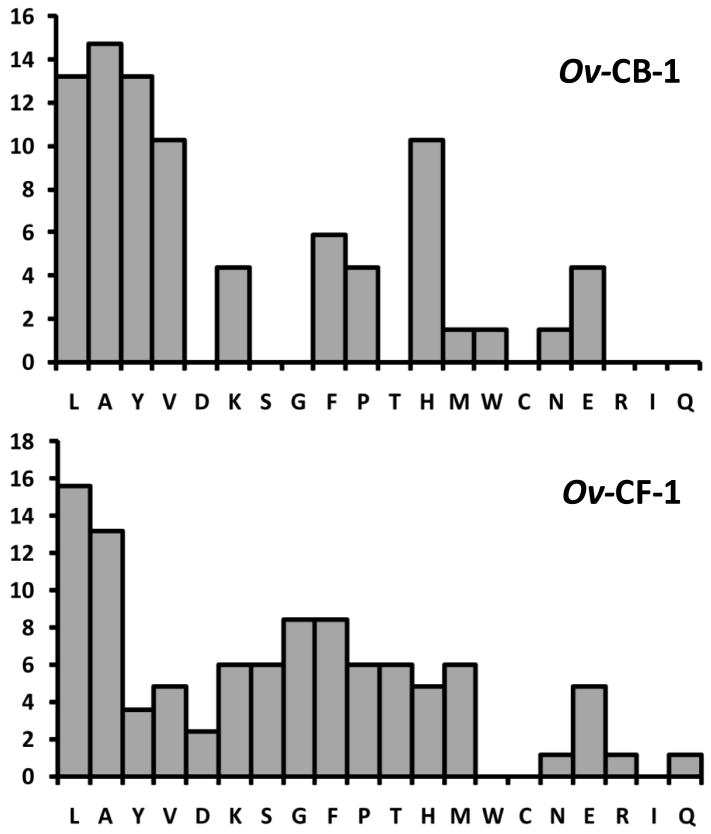
Substrate residues present at the P2 position from the scissile bond interact with the S2 subsite of the active site of papain-like cysteine proteases and determine the efficiency by which the bond is cleaved (Schechter and Berger, 1968). Therefore, we examined the frequency of each amino acid in the P2 site of the proteolytic cleavage site identified in aliquots of the 15 min Hb digest described above. Consistent with our previous findings using fluorogenic peptide substrates (Pinlaor et al., 2009) Ov-CF-1 preferentially cleaved Hb at peptide bonds where the P2 position was occupied with hydrophobic residues such as Leu, Ala and Phe. However, Ov-CF-1 could also accommodate a range of other amino acids at the P2 position, notably Gly and to a lesser extent Pro, Met, Lys, Ser and Thr (Figure 7B). In contrast, Ov-CB-1 showed a more specific P2 preference within Hb. Ov-CB-1 also digested at bonds where Leu and Ala occupied the P2 position but also showed a marked preference for other residues including Tyr, Val and His that was not evident in the Ov-CF-1 digests (Figure 7B). Finally, in similarity to our recent findings that used mass spectrometry to map Hb cleavage sites for F. hepatica cathepsin L1 (Lowther et al., 2009), the P1 position in human Hb could be occupied by many amino acids but most preferentially Leu or Ala for both O. viverrini proteases (not shown).
Figure 7.
P2 residues in peptides released from Hb following digestion by Ov-CB-1 and Ov-CF-1 were determined by nanoLC-ESI-MS/MS analysis of digest samples. The frequency by which amino acids occur at the P2 positions of Hb α- and β-chains (converted to a percentage of the total) are plotted for the 15 min reactions shown in (Figure 6A).
Differential degradation of extracellular matrix proteins by Ov-CB-1 and Ov-CF-1
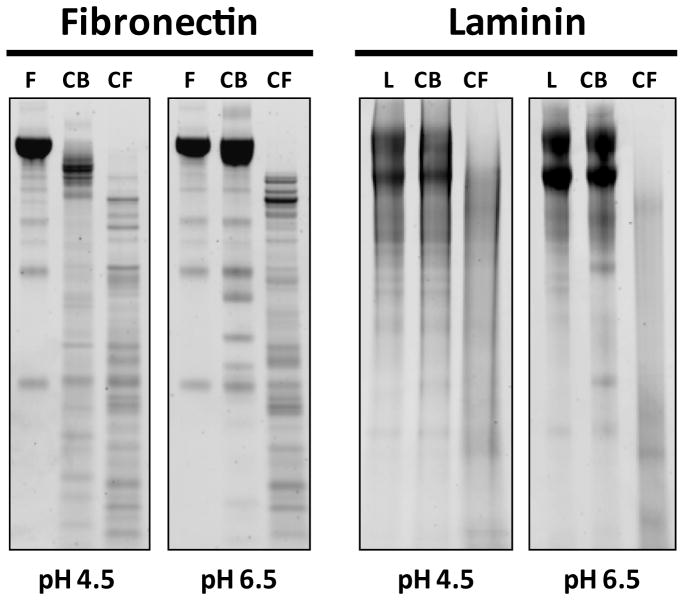
The ability of Ov-CB-1 and Ov-CF-1 to hydrolyse two major components of the extracellular matrix (ECM) was investigated as an indication of their potential roles in tissue invasion and nutrition (Figure 8). Fibronectin was digested by both O. viverrini proteases. At pH 4.5, the ∼ 200 kDa band corresponding to co-migrating fibronectin α- and β-chains was partially digested by Ov-CB-1 to a major band of approximately 150 kDa and a number of smaller degradation products. In contrast, Ov-CF-1 completely digested the fibronectin α- and β-chains at pH 4.5 into a large number of well-defined fragments. This pattern of fibronectin digestion was repeated by Ov-CF-1 at pH 6.5 whereas at this pH Ov-CB-1 displayed negligible activity against fibronectin. Ov-CB-1 was not capable of degrading laminin at either pH 4.5 or 6.5. In contrast, both high molecular mass bands (∼ 200 kDa) representing the laminin B1- and B2-chains were readily hydrolysed by Ov-CF-1 at both pH 4.5 and pH 6.5 to a range of breakdown products indicated by the smearing shown by SDS-PAGE.
Figure 8.
Digestion of extracellular matrix proteins by Ov-CB-1 and Ov-CF-1. Fibronectin and laminin were incubated with Ov-CB-1 and Ov-CF-1 at pH 4.5 and pH 6.5 at 37 °C for 3 hours. F, fibronectin alone; L, laminin alone; CB, Ov-CB-1 digest; CF, Ov-CF-1 digest.
Discussion
Papain-like cathepsin cysteine proteases are predominant molecules secreted by trematode pathogens and perform a number of functions in parasite-host interactions including facilitating tissue penetration (Curwen et al., 2006; McGonigle et al., 2008; Robinson et al., 2008a), obtaining nutrients (Dalton et al., 2004; Na et al., 2008; Lowther et al., 2009) and disarming the soluble and cellular arms of the host immune system (Dalton et al,, 2003). However, there are clear differences in the array of cathepsins expressed by each trematode species; for example, parasites of the genus Fasciola, which burrow through host liver tissue and reside in the bile ducts, express a large family of cathepsin L proteases and several cathepsin B proteases (reviewed in Robinson et al., 2008b), while those of the genus Schistosoma, that live in the blood vessels, secrete a mixture of cathepsin L, cathepsin F, cathepsin B and cathepsin C cysteine proteases (Dvorak et al., 2008). As presented in this study, the major proteases expressed by O. viverrini are cathepsin F and cathepsin B, which is in agreement with reports for the related fish-borne trematodes including Clonorchis sinensis and Paragonimus westermani (Park et al., 2001; Na et al., 2008; Pinlaor et al., 2009). These differences likely have a biological significance that relate to the organ or tissue site in which the parasites reside, and hence the protein macromolecules they consume. Understanding the complexity and specificity of the proteases secreted by these flukes should expose critical features in host-parasite relationships and stimulate novel means by which we can devise future control mechanisms.
Papain-like cathepsin proteases are synthesised as inactive zymogens consisting of a mature enzyme domain with an N-terminal extension, or prosegment that lies within the active site cleft of the enzyme and prevents unwanted proteolysis during folding, trafficking and storage. The proteases become active following removal of the prosegment to produce a mature protease with a substrate-accessible active site (Coulombe et al. 1996; Stack et al. 2008). Much of our knowledge of the synthesis, secretion and activation of cysteine protease zymogens in trematodes is inferred from studies of the cathepsin L protease family from the liver fluke, Fasciola hepatica (Robinson et al., 2008a). Our present data shows that Opisthorchis cathepsin F and Fasciola cathepsin L proteases share similar mechanisms of secretion and auto-catalytic activation. In F. hepatica cathepsin Ls are synthesised within specialised gastrodermal epithelial cells and are stored in secretory vesicles as inactive zymogens (Collins et al., 2004). Upon secretion, the prosegment is removed by specific auto-catalytic processing events, facilitated by the low pH environment of the parasite gut lumen, to release a mature active enzyme in the digestive milieu (Robinson et al., 2008a). Ov-CF-1 is also secreted by gastrodermal cells surrounding the gut of O. viverrini (Pinlaor et al., 2009), and here we have shown that recombinant Ov-CF-1 undergoes auto-catalytic activation in the range pH 4.5 – 6.5, with the rate of auto-activation increasing with decreasing pH. It is likely that auto-catalysis rather than trans-processing is the major mechanism of Ov-CF-1 activation within O. viverrini eggs given the absence of cathepsin B transcripts (Figure 2).
When incubated at pH 5.5 for 6 hours Ov-CF-1 underwent an autocatalytic process that produced a 41 kDa intermediate via cleavage at Phe-26-Lys-25↓Thr-24. However, at pH 4.5 the prosegment was completely removed via cleavage at Val-2-Thr-1↓Met1 to release a 30 kDa fully mature enzyme that efficiently cleaves synthetic peptides and macromolecular substrates. This process of auto-activation of O. viverrini cathepsin F is similar to that reported for F. hepatica cathepsin L1 (FhCL1) (Collins et al., 2004; Stack et al., 2007; Lowther et al., 2009). Notably, the prosegment of Ov-CF-1 contains a conserved GXTXFXD motif similar to the GXNXFXD motif found FhCL1 and in other cathepsin L-like proteases. This motif is implicated in triggering the pH-dependent intra-molecular cleavage of the prosegment whereby the interaction of conserved charged Asp with residues on the mature portion of the enzyme is perturbed by reduced pH (Vernet et al., 1995). This triggering event may occur in Ov-CF-1 at pH 5.5 allowing the initial cleavage at Phe-26-Lys-25↓Thr-24 which is C-terminal to the GXNXFXD motif. Primary sequence alignments and comparative analysis with the atomic structure of FhCL1 (Stack et al., 2008; not shown) show that the 24 residue C-terminal region of the Ov-CF-1 prosegment that remains attached to the mature domain of the enzyme lies within the active site cleft of the enzyme. This would explain why retention of this 41 kDa intermediate cleavage form of Ov-CF-1 exhibited a reduced (∼ 20 %) activity.
A number of studies have shown that as well as being capable of autocatalytic activation helminth cathepsin proteases can be trans-activated by other proteases that cleave at residues that lie at the junction between the prosegment and mature enzyme domain (Dalton and Brindley, 1996; Sajid et al., 2003; Beckham et al., 2006; Delcroix et al., 2006). Here, pre-incubation of proOv-CF-1 with exogenously added active Ov-CB-1 zymogen at pH 5.5 for 60 minutes was sufficient to trans-activate the cathepsin F and release a fully mature and active enzyme. However, the activity of the cathepsin B-trans-activated Ov-CF-1 against the fluorogenic substrate Z-Leu-Arg-NHMec was very low when measured immediately after pre-incubation with Ov-CB-1 (time zero). This suggests that only a small population of Ov-CF-1 zymogens are initially trans-activated by Ov-CB-1. Nevertheless, this exogenous proteolysis appears to have generated sufficient amounts of mature Ov-CF-1 to trans-activate other cathepsin F zymogens since a rapid increase in Z-Leu-Arg-NHMec hydrolysis occurs as the reaction proceeds. Indeed, when the Ov-CF-1/Ov-CB-1 reactions were analysed by SDS-PAGE, Ov-CF-1 did not show a significant shift in molecular mass; a discreet band of ∼ 30 kDa appeared following 60 minutes at pH 5.5 that was absent when Ov-CF-1 was incubated alone. The presence of peptide MDNSNFDWR (m/z 592.773) in tryptic digests of the ∼ 30 kDa product shows that this represents a fully matured protein formed by exogenous cleavage of the cathepsin F prosegment at Val-2-Thr-1↓Met1. This was the same cleavage site observed when Ov-CF-1 was trans-activated by exogenously-added FhCL1 (Pinlaor et al., 2009) and when the enzyme was auto-catalytically activated at pH 4.5 (see above). Therefore, our data shows that the Val-2-Thr-1↓ Met1 cleavage site represents a critical protease-susceptible region between the prosegment and mature domain, which serves to regulate cathepsin F activation.
In contrast to cathepsin F, the major secreted cathepsin B of O. viverrini did not undergo typical auto-processing events that lead to removal of the prosegment. This contrasts with a number of cathepsin Bs that are secreted from related trematode species including F. hepatica, S. mansoni and T. regenti which release the prosegment following auto-activation at low pH (Gotz et al., 1992; Dvorak et al., 2005; Beckham et al., 2006). However, although the prosegment was still covalently attached to the mature domain of Ov-CB-1, the zymogen exhibited activity against a range of fluorogenic –NHMec substrates in the pH range 3.5 - 8.5, and, at pH 4.5, efficiently hydrolysed haemoglobin to small peptides. These data demonstrate that the Ov-CB-1 zymogen can cleave physiologically relevant substrate molecules and, in addition, its ability to trans-activate cathepsin F suggests that one of its functions is to regulate the protease network in O. viverrini.
Atypical active human procathepsin B has been reported previously (Pungercar et al., 2009) and crystallographic studies indicate that cathepsin B prosegments (which are considerably shorter than their cathepsin L and cathepsin F counterparts) are susceptible to acid-induced conformational changes making them less efficient inhibitors of the mature domain (Cygler et al., 1996; Coulombe et al., 1996; Groves et al., 1998). Thus the low-pH environment of the Opisthorchis gut may loosen the tertiary fold of Ov-CB-1 increasing the mobility of the prosegment which then dissociates from the mature domain, albeit still attached. This would allow entry of the protease-susceptible region of the Ov-CF-1 zymogen into the Ov-CB-1 active site cleft for trans-activation. Thus, bringing all of our observations together we can propose a model of cathepsin F activation which involves initial processing of a small population of cathepsin F zymogens by an active cathepsin B zymogen followed by rapid activation of additional cathepsin F zymogens by these active mature molecules (Figure 9).
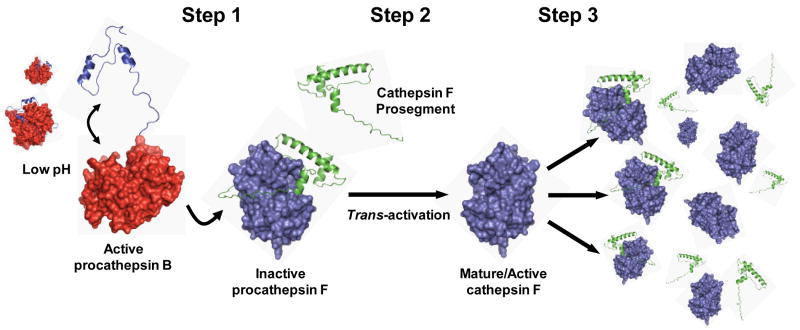
Figure 9.
Proposal for a three-step mechanism of cathepsin F trans-activation in O. viverrini. Step 1: under the acidic microenvironment of the O. viverrini gut, the short pH-sensitive prosegment of cathepsin B undergoes conformational relaxation and dissociates from the mature domain sufficient to allow the zymogen to become active. Step 2: subsequently, in a bi-molecular process, a small number of cathepsin F zymogens are trans-activated by the active cathepsin B zymogens which remove the cathepsin F prosegment via cleavage at Val-1-Asp-1↓Met1 at the junction of the prosegment and mature domain. Step 3: the rapid cleavage of prosegments from other cathepsin F zymogens by the trans-activated cathepsin F molecules again through cleavage at Val-1-Asp-1↓Met1. Molecular models of Ov-CB-1 and Ov-CF-1 were established (for illustrative purposes only) by the SWISS-MODEL homology modelling pipeline (http://swissmodel.expasy.org) using the atomic structures of human procathepsin B (PBD ID: 3PBH) and F. hepatica cathepsin L1 (PBD ID: 2O6X) as templates, respectively. Figures were produced with Pymol (http://www.pymol.org).
Trematodes produce prodigious numbers of eggs, which requires a reliable source of amino acids for anabolism of eggshell proteins. Within the bile ducts, adult F. hepatica is an obligate blood feeder and liberates 30,000 eggs per hour. To obtain nutrient, the liver fluke secretes the FhCL1 protease that has evolved an active site with a strong preference for hydrophobic amino acids, Leu, Ala, Phe and Val, which comprise 42 % of haemoglobin (Robinson et al., 2008b; Lowther et al., 2009). Adult O. viverrini also use blood as a food source and likely employ the Ov-CF-1 and Ov-CB-1 to digest human haemoglobin. Although both proteases can degrade this substrate into short peptides the digestion process is much more rapid and complete when both enzymes act together. It is noteworthy that the SDS-PAGE profile of haemoglobin digestion by Ov-CF-1 and Ov-CB-1 together was similar to the pattern of digestion when haemoglobin was digested with FhCL1 alone (Lowther et al., 2009). Thus, adult O. viverrini may require both Ov-CF-1, Ov-CB-1, and possibly a cathepsin D-like aspartic protease (Ov-APR-1) (Suttiprapa et al. 2009), to accomplish complete digestion of haemoglobin into small peptides that can be used for protein anabolism. Such multi-enzyme networks have been reported in other blood-feeding parasites including the human blood fluke S. mansoni (Delcroix et al., 2006) and the hookworm Ancylostoma caninum (Williamson et al., 2004) representing a mechanism common to evolutionarily distant haemotophagous parasites.
In addition to blood, O. viverrini flukes also graze on bile duct epithelial cells and mucus (Rim, 2005; Sripa et al., 2007). These food preferences would require the ability to cleave a variety of macromolecular substrates and may explain why Ov-CF-1 and/or Ov-CB-1 exhibit different substrate specificity to FhCL1. We observed that a broader range of haemoglobin residues can be accommodated at the P2 site by the two O. viverrini cathepsins compared with the more specific usage of hydrophobic P2 amino acids by FhCL1 (Stack et al., 2008; Lowther et al., 2009). The broader specificity of Ov-CF-1 and Ov-CB-1, and overlapping pH optima for activity (pH 6.5 and 4.5, respectively), may allow them to work in concert to digest a number of physiologically relevant extracellular matrix (ECM) proteins. Ov-CF-1 effectively digested fibronectin and laminin close to physiological pH (pH 6.5) and under acidic conditions (pH 4.5). In contrast, Ov-CB-1 partially degraded fibronectin at low pH but could not digest either of the ECM proteins at pH 6.5. The combined action of the secreted O. viverrini cathepsins would result in the degradation of interstitial laminin and fibronectin within the bile duct, and lead to disruption of the cellular integrity of the cholangiocytes thus allowing the parasite to access underlying liver cells. A cathepsin F (CsCF-6) secreted by adult Clonorchis sinensis displayed similar biochemical properties to Ov-CF-1 and also degraded a range of host macromolecules including collagen, fibronectin and haemoglobin (Na et al., 2008). The evolution of ECM-degrading activity in trematodes is significant and likely to be pivotal to the ability of these pathogens to infect and survive within their mammalian hosts.
Adult O. viverrini express both cathepsin F and B endoproteases at similar levels; an analysis of ∼ 5000 O. viverrini ESTs using Ov-CF-1 and Ov-CB-1 primary sequences as queries gave 60 and 50 significant (e < 1) matches, respectively. The Ov-CF-1 and Ov-CB-1 combination clearly constitutes effective molecular machinery for tissue degradation. One of the most intriguing aspects of O. viverrini biology is the link between liver fluke infection and the development of CCA. Given the potential of Ov-CF-1 and Ov-CB-1 to cause tissue destruction it is likely that these proteases contribute to CCA progression. Since local changes in ECM microenvironment contribute to the induction of intra-hepatic CCA (Farazi et al., 2006) it is likely that degradation of ECM proteins by the battery of proteases secreted by O. viverrini has a similar effect during the aetiology of liver fluke-associated CCA. As degradation of the ECM is a prerequisite step for the invasion and metastasis of cancer cells, O. viverrini infection may even promote the spread of invasive carcinoma as a result of ECM and basement membrane instability within the bile ducts (Mon et al., 2009; Yasoshima et al., 2009).
Supplementary Material
Acknowledgments
This research was supported by a grant from the Strategic Scholarships for Frontier Research Network (Thai Doctoral degree) from the Office of the Higher Education Commission, Thailand and by grants from the Sandler Family Foundation and from the National Institute of Allergy and Infectious Diseases (award number UO1AI065871): the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the NIH. JPD is supported by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada. MWR is supported by a UTS Chancellor's Postdoctoral Fellowship.
Footnotes
References
- Beckham SA, Law RH, Smooker PM, Quinsey NS, Caffrey CR, McKerrow JH, et al. Production and processing of a recombinant Fasciola hepatica cathepsin B-like enzyme (FhcatB1) reveals potential processing mechanisms in the parasite. Biol Chem. 2006;387:1053–1061. doi: 10.1515/BC.2006.130. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 30. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens - Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- Brady MT, O'Neill SM, Dalton JP, Mills KHG. Fasciola hepatica suppresses a protective Th1 response against Bordetella pertussis. Infect Immun. 1999;67:5372–5378. doi: 10.1128/iai.67.10.5372-5378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JY, Murrell KD, Lymbery AJ. Fish-borne parasitic zoonoses: status and issues. Int J Parasitol. 2005;35:1233–1254. doi: 10.1016/j.ijpara.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Collins PR, Stack CM, O'Neill SM, Doyle S, Ryan T, Brennan GP, et al. Cathepsin L1, the major protease involved in liver fluke (Fasciola hepatica) virulence: propetide cleavage sites and autoactivation of the zymogen secreted from gastrodermal cells. J Biol Chem. 2004;279:17038–17046. doi: 10.1074/jbc.M308831200. [DOI] [PubMed] [Google Scholar]
- Coulombe R, Grochulski P, Sivaraman J, Menard R, Mort JS, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- Curwen RS, Ashton PD, Sundaralingam S, Wilson RA. Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Mol Cell Proteomics. 2006;5:835–844. doi: 10.1074/mcp.M500313-MCP200. [DOI] [PubMed] [Google Scholar]
- Cygler M, Sivaraman J, Grochulski P, Coulombe R, Storer AC, Mort JS. Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion. Structure. 1996;4(4):405–416. doi: 10.1016/s0969-2126(96)00046-9. [DOI] [PubMed] [Google Scholar]
- Dalton JP, Brindley PJ. Schistosome asparaginyl endopeptidase Sm 32 in hemoglobin digestion. Parasitol Today. 1996;12:125. doi: 10.1016/0169-4758(96)80676-4. [DOI] [PubMed] [Google Scholar]
- Dalton JP, Neill SO, Stack C, Collins P, Walshe A, Sekiya M, et al. Fasciola hepatica cathepsin L-like proteases: biology, function, and potential in the development of first generation liver fluke vaccines. Int J Parasitol. 2003;33:1173–1181. doi: 10.1016/s0020-7519(03)00171-1. [DOI] [PubMed] [Google Scholar]
- Dalton JP, Skelly P, Halton DW. Role of the tegument and gut in nutrient uptake by parasitic platyhelminths. Can J Zool. 2004;82:211–232. [Google Scholar]
- Delcroix M, Sajid M, Caffrey CR, Lim KC, Dvorák J, Hsieh I, et al. A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. J Biol Chem. 2006;281:39316–39329. doi: 10.1074/jbc.M607128200. [DOI] [PubMed] [Google Scholar]
- Dowd AJ, Tort J, Roche L, Ryan T, Dalton JP. Isolation of a cDNA encoding Fasciola hepatica cathepsin L2 and functional expression in Saccharomyces cerevisiae. Mol Biochem Parasitol. 1997;88:163–174. doi: 10.1016/s0166-6851(97)00090-x. [DOI] [PubMed] [Google Scholar]
- Dvorák J, Delcroix M, Rossi A, Vopálenský V, Pospísek M, Sedinová M, et al. Multiple cathepsin B isoforms in schistosomula of Trichobilharzia regenti: identification, characterisation and putative role in migration and nutrition. Int J Parasitol. 2005;35(8):895–910. doi: 10.1016/j.ijpara.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Dvorák J, Mashiyama ST, Braschi S, Sajid M, Knudsen GM, Hansell E, Lim KC, et al. Differential use of protease families for invasion by schistosome cercariae. Biochemie. 2008;90:345–358. doi: 10.1016/j.biochi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Farazi PA, Zeisberg M, Glickman J, Zhang Y, Kalluri R, DePinho RA. Chronic bile duct injury associated with fibrotic matrix microenvironment provokes cholangiocarcinoma in p53-deficient mice. Cancer Res. 2006;66(13):6622–6627. doi: 10.1158/0008-5472.CAN-05-4609. [DOI] [PubMed] [Google Scholar]
- Gabay T, Ginsburg H. Hemoglobin denaturation and iron release in acidified red blood cell lysate – a possible source of iron for intraerythrocytic malaria parasites. Exp Parasitol. 1993;77:261–272. doi: 10.1006/expr.1993.1084. [DOI] [PubMed] [Google Scholar]
- Götz B, Felleisen R, Shaw E, Klinkert MQ. Expression of an active cathepsin B-like protein Sm31 from Schistosoma mansoni in insect cells. Trop Med Parasitol. 1992;43(4):282–284. [PubMed] [Google Scholar]
- Groves MR, Coulombe R, Jenkins J, Cygler M. Structural basis for specificity of papain-like cysteine protease proregions toward their cognate enzymes. Proteins. 1998;32(4):504–514. [PubMed] [Google Scholar]
- Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J. Helminth infections: The great neglected tropical diseases. J Clin Invest. 2008;118(4):1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsuksuntigul P, Imsomboon T. Opisthorchiasis control in Thailand. Acta Trop. 2003;88(3):229–232. doi: 10.1016/j.actatropica.2003.01.002. [DOI] [PubMed] [Google Scholar]
- Kaewpitoon N, Kaewpitoon SJ, Pengsaa P, Sripa B. Opisthorchis viverrini: the carcinogenic human liver fluke. World J Gastroenterol. 2008;14(5):666–674. doi: 10.3748/wjg.14.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser J, Utzinger J. Emerging foodborne trematodiasis. Emerg Infect Dis. 2005;11:1507–1514. doi: 10.3201/eid1110.050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha T, Pinlaor P, Mulvenna J, Sripa B, Sripa M, Smout MJ, et al. Gene discovery for the carcinogenic human liver fluke, Opisthorchis viverrini. BMC Genomics. 2007;8:189. doi: 10.1186/1471-2164-8-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lowther J, Robinson MW, Donnelly SM, Xu W, Stack CM, Matthews JM, Dalton JP. The importance of pH in regulating the function of Fasciola hepatica cathepsin L1 cysteine protease. PLoS Negl Trop Dis. 2009;3(1):e369. doi: 10.1371/journal.pntd.0000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle L, Mousley A, Marks NJ, Brennan GP, Dalton JP, Spithill TW, et al. The silencing of cysteine proteases in Fasciola hepatica newly excysted juveniles using RNA interference reduces gut penetration. Int J Parasitol. 2008;38:149–155. doi: 10.1016/j.ijpara.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Mon NN, Kokuryo T, Hamaguchi M. Inflammation and tumor progression: a lesson from TNF-alpha-dependent FAK signaling in cholangiocarcinoma. Methods Mol Biol. 2009;512:279–293. doi: 10.1007/978-1-60327-530-9_15. [DOI] [PubMed] [Google Scholar]
- Na BK, Kang JM, Sohn WM. CsCF-6, a novel cathepsin F-like cysteine protease for nutrient uptake of Clonorchis sinensis. Int J Parasitol. 2008;38(5):493–502. doi: 10.1016/j.ijpara.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Park H, Hong KM, Sakanari JA, Choi JH, Park SK, Kim KY, Hwang HA, et al. Paragonimus westermani: cloning of a cathepsin F-like cysteine proteinase from the adult worm. Exp Parasitol. 2001;98:223–227. doi: 10.1006/expr.2001.4634. [DOI] [PubMed] [Google Scholar]
- Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- Pinlaor S, Sripa B, Sithithaworn P, Yongvanit P. Hepatobiliary changes, antibody response, and alteration of liver enzymes in hamsters re-infected with Opisthorchis viverrini. Exp Parasitol. 2004;108(1-2):32–39. doi: 10.1016/j.exppara.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Pinlaor P, Kaewpitoon N, Laha T, Sripa B, Kaewkes S, Morales ME, Mann VH, et al. Cathepsin F cysteine protease of the human liver fluke, Opisthorchis viverrini. PLoS Negl Tropl Dis. 2009;3(3):e398. doi: 10.1371/journal.pntd.0000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungercar JR, Caglic D, Sajid M, Dolinar M, Vasiljeva O, Pozgan U, et al. Autocatalytic processing of procathepsin B is triggered by proenzyme activity. FEBS J. 2009;276(3):660–668. doi: 10.1111/j.1742-4658.2008.06815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rim HJ. Clonorchiasis: An update. J Helminthol. 2005;79:269–281. doi: 10.1079/joh2005300. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Connolly B. Proteomic analysis of the excretory-secretory proteins of the Trichinella spiralis L1 larva, a nematode parasite of skeletal muscle. Proteomics. 2005;5:4525–4532. doi: 10.1002/pmic.200402057. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Greig R, Beattie K, Lamont D, Connolly B. Comparative analysis of the excretory-secretory proteome of the muscle larva of Trichinella pseudospiralis and Trichinella spiralis. Int J Parasitol. 2007;37:139–148. doi: 10.1016/j.ijpara.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Dalton JP, Donnelly S. Helminth pathogen cathepsin proteases: it's a family affair. Trends Biochem Sci. 2008a;33(12):601–608. doi: 10.1016/j.tibs.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Tort JF, Wong E, Donnelly SM, Lowther J, Xu W, Stack CM, et al. Proteomic and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen, Fasciola hepatica: expansion of a repertoire of virulence-associated factors. Mol Cell Proteomics. 2008b;7:1111–1123. doi: 10.1074/mcp.M700560-MCP200. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S. An integrated transcriptomic and proteomic analysis of the secretome of the helminth pathogen, Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics. 2009;8(8):1891–1907. doi: 10.1074/mcp.M900045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid M, McKerrow JH, Hansell E, Mathieu MA, Lucas KD, Hsieh I, et al. Functional expression and characterization of Schistosoma mansoni cathepsin B and its trans-activation by an endogenous asparaginyl endopeptidase. Mol Biochem Parasitol. 2003;131:65–75. doi: 10.1016/s0166-6851(03)00194-4. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1968;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Smout MJ, Laha T, Mulvenna J, Sripa B, Suttiprapa S, Jones A, et al. A granulin-like growth factor secreted by the carcinogenic liver fluke, Opisthorchis viverrini, promotes proliferation of host cells. PLoS Pathog. 2009;5(10):e1000611. doi: 10.1371/journal.ppat.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripa B, Pairojkul C. Cholangiocarcinoma: lessons from Thailand. Curr Opin Gastroenterol. 2008;24:349–356. doi: 10.1097/MOG.0b013e3282fbf9b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, Smout M, et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007;4(7):e201. doi: 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripa B. Concerted action is needed to tackle liver fluke infections in Asia. PLoS Negl Trop Dis. 2008;2(5):e232. doi: 10.1371/journal.pntd.0000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack CM, Donnelly S, Lowther J, Xu W, Collins PR, Brinen LS, Dalton JP. The major secreted cathepsin L1 protease of the liver fluke, Fasciola hepatica: a Leu-12 to Pro-12 replacement in the nonconserved C-terminal region of the prosegment prevents complete enzyme autoactivation and allows definition of the molecular events in prosegment removal. J Biol Chem. 2007;282:16532–16543. doi: 10.1074/jbc.M611501200. [DOI] [PubMed] [Google Scholar]
- Stack CM, Caffrey CR, Donnelly SM, Seshaadri A, Lowther J, Tort JF, et al. Structural and functional relationships in the virulence-associated cathepsin L proteases of the parasitic liver fluke, Fasciola hepatica. J Biol Chem. 2008;283:9896–9908. doi: 10.1074/jbc.M708521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttiprapa S, Loukas A, Laha T, Wongkham S, Kaewkes S, Gaze S, et al. Characterization of the antioxidant enzyme, thioredoxin peroxidase, from the carcinogenic human liver fluke, Opisthorchis viverrini. Mol Biochem Parasitol. 2008;160(2):116–122. doi: 10.1016/j.molbiopara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttiprapa S, Mulvenna J, Huong NT, Pearson MS, Brindley PJ, Laha T, et al. Ov-APR-1, an aspartic protease from the carcinogenic liver fluke, Opisthorchis viverrini: functional expression, immunolocalization and subsite specificity. Int J Biochem Cell Biol. 2009;41:1148–1156. doi: 10.1016/j.biocel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuwajit C, Thuwajit P, Kaewkes S, Sripa B, Uchida K, Miwa M, Wongkham S. Increased cell proliferation of mouse fibroblast NIH-3T3 in vitro induced by excretory/secretory product(s) from Opisthorchis viverrini. Parasitology. 2004;129:455–464. doi: 10.1017/s0031182004005815. [DOI] [PubMed] [Google Scholar]
- Vernet T, Berti PJ, de Montigny C, Musil R, Tessier DC, Menard R, et al. Processing of the papain precursor. The ionization state of a conserved amino acid motif within the Pro region participates in the regulation of intramolecular processing. J Biol Chem. 1995;270:10838–10846. doi: 10.1074/jbc.270.18.10838. [DOI] [PubMed] [Google Scholar]
- Williamson AL, Lecchi P, Turk BE, Choe Y, Hotez PJ, McKerrow JH, et al. A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J Biol Chem. 2004;279(34):35950–35957. doi: 10.1074/jbc.M405842200. [DOI] [PubMed] [Google Scholar]
- Yasoshima M, Sato Y, Furubo S, Kizawa K, Sanzen T, Ozaki S, et al. Matrix proteins of basement membrane of intrahepatic bile ducts are degraded in congenital hepatic fibrosis and Caroli's disease. J Pathol. 2009;217(3):442–451. doi: 10.1002/path.2472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.