Abstract
Cbl was originally discovered in 1989 as the cellular homologue of the v-Cbl oncogene, the transforming gene of the Cas NS-1 murine retrovirus which causes myeloid leukemia and lymphomas in mice. Cbl is a member of a family of RING finger ubiquitin ligases which negatively regulate signaling by tyrosine kinases and which function as adaptor proteins to regulate signaling positively. Until the past two years, there was little evidence that Cbl proteins were involved in human malignancies. Recent publications have demonstrated homozygous mutations in Cbl in human myeloid neoplasms. While in vitro and animal transformation models suggested that mutant forms of Cbl acted as an oncogene, the cellular role suggested that the protein could serve as a tumor suppressor gene. The recent data begin to reconcile this paradox as the loss of ubiquitin ligase function (the tumor suppressor function) is coupled to the maintenance of the positive signaling function (the oncogene function). These data also provide insight into potential therapeutic approaches to myeloid disorders harboring Cbl mutations.
Keywords: Cbl, ubiquitin ligase, myeloid neoplasms
The Cbl family
Cbl proteins are a highly conserved family of RING finger (RF) ubiquitin ligases (E3s) which regulate signaling by tyrosine kinases (TKs) in many pathways (for comprehensive review of the structure and function of Cbl proteins, see (1) and the chapters therein). There are three mammalian Cbl proteins encoded by separate genes: Cbl (a.k.a. c-Cbl, CBL2, RNF55), Cbl-b (a.k.a. RNF56), and Cbl-c (a.k.a. Cbl-3, Cbl-SL, RNF57) (Fig. 1A). Cbl and Cbl-b are widely expressed in mammalian cells, and data from Cbl and Cbl-b null mice indicate a prominent role in T and B cell function (1). The expression of Cbl-c is restricted to epithelial cells (2). Cbl-c null mice have no overt phenotype, and thus the physiological function of Cbl-c is not known (2).
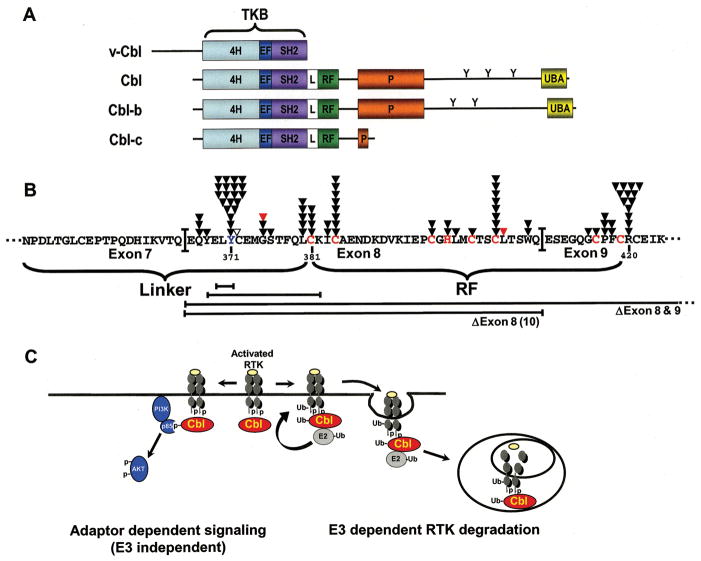
Figure 1.
A, The domain organization of the mammalian Cbl proteins. v-Cbl: the Gag-Cbl fusion protein of the Cas NS-1 murine retrovirus; Cbl: the human proto-oncogene of v-Cbl; Cbl-b: the second human Cbl protein; Cbl-c: the third human Cbl protein The tyrosine kinase binding domain (TKB) is comprised of a 4-helix bundle (4H), an EF hand (EF), and a variant SH2 domain (SH2). The linker (L), RING finger (RF), proline-rich (P), and the ubiquitin-associated (UBA) domains are indicated on the diagram. The tyrosines in the C-terminus of Cbl and Cbl-b that are phosphorylated are indicated. B, Compilation of Cbl mutations in the linker and RF domains from human myeloid neoplasms. Sequence shown includes the linker region and RF. The critical linker tyrosine (Y371) is shown in blue. The zinc coordinating amino acids are shown in red. Missense mutations in the linker and RF of Cbl are indicated above the sequence by black arrows, frame shift mutations are indicated by red arrows, and an insertion is indicated by an open arrow. Deletion mutations are indicated by the brackets below the sequence. Exon 8 deletions were found in 5 samples indicated in the parentheses. C, Cbl pathways. Cbl functions as a tumor suppressor by E3 dependent ubiquitination and downregulation of activated RTKs as indicated to the right of the figure. Cbl can function also as an oncogene by mediating adaptor function dependant activation of downstream signaling to the PI3K/AKT pathway.
Cbl proteins are tyrosine phosphorylated upon activation of a variety of growth factor and immune receptors and they associate with many proteins containing SH2 and SH3 domains (1). They have been implicated as regulators of signaling by receptor and non-receptor tyrosine kinases (RTKs and non-RTKs respectively), immune receptors such as the T-cell receptor, B-cell receptors, and Fc receptors. Mechanistic studies using transfected proteins and mammalian cell lines have demonstrated that Cbl proteins regulate negatively RTKs and non-RTKs by functioning as E3s which mediate the ubiquitination and degradation of activated TKs (Fig. 1C) (1). In addition, Cbl proteins can serve as adaptor proteins and through this function serve as positive regulators of signal transduction (1). For example, phosphorylation of Cbl on Y731 in the C-terminus has been shown to recruit the p85 subunit of phosphoinositide 3 kinase (PI3K) leading to activation of PI3K and downstream signaling pathways (Fig. 1C) (1).
Ubiquitination of proteins is a multi-enzyme process which regulates the degradation, trafficking, and activity of many proteins (reviewed in (3–5)). Ubiquitination is initiated by the activation and conjugation of ubiquitin to a cysteine on one of two ubiquitin activating enzymes (E1). Ubiquitin is then transferred from the E1 to a cysteine on one of more than 30 ubiquitin conjugating enzymes (E2). There are several hundred E3s and they are primarily responsible for determining the substrate specificity of the ubiquitination process. There are two classes of E3s, HECT and RF E3s. The HECT E3s form a thioester covalent intermediate with ubiquitin before attaching the ubiquitin to a lysine on the substrate protein. The RF E3s catalyze the direct transfer of the ubiquitin from the E2 to a lysine on the substrate without forming a covalent intermediate. Cbl proteins belong to the RF class of E3s. The catalytic domain of the Cbl proteins is a C3HC4 RF which serves to bind to the ubiquitin conjugating enzyme (E2) and to catalyze the transfer of ubiquitin from the E2 to the substrate (Fig. 1A) (1, 6). The RF is surrounded by protein interaction motifs. All Cbl proteins contain a highly conserved N-terminal tyrosine kinase binding (TKB) domain composed of a 4-helix bundle, a calcium binding EF hand, and an atypical SH2 domain. The TKB domain mediates interactions between Cbl proteins and phosphorylated tyrosines on other proteins. The RF is separated from the TKB by an alpha helical structure known as the linker region. This region is critical for the regulation of Cbl E3 function and is implicated in the transforming mutations which will be discussed below. The crystal structure of the linker region has demonstrated that it forms contacts with the TKB, the RF, and the E2. All Cbl proteins have a conserved tyrosine within the linker region (Y371 in Cbl) that becomes phosphorylated upon interaction of the Cbl protein with a TK, leading to a conformational change in the Cbl protein and activation of the E3 activity (Fig. 1B) (1, 6–8). C-terminal to the RF, Cbl proteins have proline rich domains that mediate interactions with SH3 containing proteins. Cbl and Cbl-b share additional areas of homology in the C-terminal half of the proteins. This includes more extensive proline rich regions and tyrosines which can be phosphorylated and mediate interactions with SH2 proteins (such as the p85 subunit of PI3K). Another region of homology shared by Cbl and Cbl-b is an ubiquitin-associated (UBA) domain.
Cbl and transformation
The first Cbl gene, v-Cbl, was discovered as the transforming gene of the Cas NS-1 virus which causes B-cell lymphomas and myeloid leukemias in mice and which transforms fibroblasts in culture (reviewed in (1)). v-Cbl is a gag-Cbl fusion protein containing the N-terminal one-third of the Cbl protein (Fig. 1A). Two other transforming forms of Cbl, 70Z Cbl, and p95Cbl, have been identified in murine cell lines derived from carcinogen-induced murine lymphomas (9, 10). These forms of Cbl have deletions within the linker region and RF of Cbl. Additional transforming mutations of Cbl have been identified from mutation analysis studies (11). These transforming mutations all have deletions or mutations within the linker region and RING finger and have lost E3 activity. The transforming forms of Cbl are believed to work, at least in part, as dominant negative proteins which inhibit the negative regulatory function of the endogenous Cbl proteins. However, mutation analysis has revealed that not all Cbl mutants that have lost the E3 activity are transforming and that alterations of the linker region are critical for transformation by Cbl (11). Consistent with this, there is no evidence for increased tumors or leukemia in mice that are null for any of the three Cbl genes (1). Thus, the loss of E3 activity appears to be a prerequisite for transformation, but it is not sufficient. Interestingly, the transforming 70Z form of Cbl which has a deletion that includes the linker region and the beginning of the RF (and has lost E3 activity), activates the EGFR in the absence of ligand and enhances activity of the EGFR and downstream signaling upon ligand stimulation (12). These results suggest that, in addition to the E3 dependent negative role that Cbl plays in signaling, Cbl mediates positive pro-oncogenic effects that are unmasked when the E3 activity is lost (this concept will be revisited below) (Fig. 1C).
Cbl mutations in myeloid neoplasms
As described above, v-Cbl causes myeloid leukemia in mice. Additional evidence that Cbl mutations may contribute to myeloid leukemogenesis comes from analysis of secondary mutations in an animal model of myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML) (13). Transgenic mice expressing the fusion gene NUP98-HOXD13 develop MDS at an early age which progresses to AML with a latency of 4–14 months. Analysis of mutations that arise as the MDS progresses to AML has identified a Cbl message lacking exon 8 in the AML cells from one mouse. In these cells, one allele of the Cbl gene contains a deletion within the splice acceptor site for exon 8 and this deletion results in the loss of Cbl exon 8 via missplicing. Exon 8 contains the C-terminal portion of the linker and the beginning of the RF (Fig. 1B).
The first example that Cbl proteins may contribute to myeloid leukemogenesis in humans was described before the function of Cbl proteins was appreciated. Mutations of a tyrosine in the cytoplasmic domain of the human Colony Stimulating Factor-1 (CSF-1) receptor were found in children with secondary myelodysplasia and secondary AML (sAML) (14, 15). Cbl binds to this tyrosine and ubiquitinates the CSF-1 receptor upon ligand activation (16). Mutations in the Cbl binding site of the CSF-1 receptor enhances transforming activity of this receptor in vitro (17).
More recently, mutations of Cbl were described in the leukemic cells of patients with AML (18, 19). Sargin et al. demonstrated that Cbl can down regulate the Flt3 protein, an RTK that is activated frequently in AML, and that the oncogenic 70Z Cbl can enhance transformation by activated Flt3 in vitro (19). This prompted the authors to look for evidence of Cbl mutations in AML. They found a novel mutation (R420Q) in the amino acid immediately downstream of the RF in one of 150 patients with AML (Fig. 1B)(19). A second group identified mutations of Cbl in the AML cell line MOLM-13 and in 4 of 12 primary AML samples (18). Similar to the results described in the NUP98-HOXD13 transgenic mice, the MOLM-13 cell line and one patient sample contained a deletion of exon 8 in one allele (Fig. 1B). Two other groups have subsequently identified the same deletion within the MOLM-13 cell line and in a second line, MOLM-14, derived from the same patient (20, 21). Two patient samples contained missense mutations within the linker domain, and one sample had a frame shift mutation within the linker region that would result in a truncated Cbl protein (Fig. 1B).
Using SNP based analysis, Dunbar et al. identified loss of heterozygosity based on uniparental disomy (UPD) at chromosome 11q in 12 of 301 patients with myeloid neoplasms (22). Cbl is located within the region of UPD, and sequence analysis revealed homozygous mutations affecting the RF of Cbl in 7 of 12 samples with UPD. These mutations included ones in the zinc coordinating complex (which would be predicted to abrogate E3 activity) as well as mutations of R420 (R420Q and R420P) (Fig. 1B). The R420Q mutation was the same as the one previously characterized by Sargin et al. (19). These and additional sequencing studies of more than 2000 samples from patients with myeloid neoplasms have identified Cbl mutations in approximately 5% of these samples (18–27). The frequency of homozygous Cbl mutations was approximately 85–90% in samples with UPD of chromosome 11q (22–27). Sequence analysis of the ~250 samples without UPD of chromosome 11q found only four Cbl mutations (26, 27). Cbl mutations have been found in cells from a wide variety of myeloid neoplasms including MDS, myelofibrosis, refactory anemia with excess blasts, AML, sAML, atypical chronic myelogenous leukemia (aCML), CML in blast crisis, chronic myelomonocytic leukemia (CMML), and juvenile myelomonocytic leukemia (JMML) (18, 19, 21–27). However, the frequency of Cbl mutations appears to be highest in JMML (~15%), CMML (~13%), sAML (~10%), and aCML (8%). By contrast, while v-Cbl also caused B-cell lymphomas in mice, UPD at 11q is rare in lymphoid malignancies. Sequencing of Cbl in more than 200 lymphoid malignancies found no mutations in the linker or RF of Cbl (21, 28).
A compilation of the mutations found in human myeloid neoplasms is shown in Fig. 1B (18–27, 29). The vast majority of the mutations described occur within the linker region or the RF. A few mutations have been described outside of this region (representing ~2–3% of the reported mutations), but most of the studies have evaluated only the linker and RF regions for mutations. Thus, the true frequency of mutations outside of the linker and RF regions in myeloid neoplasms is unknown. Most mutations are missense mutations that cluster within the linker region and at or near the zinc coordinating amino acids of the RF. Also, deletions of all or portions of exon 8 have been described (Fig. 1B) (18, 20, 21, 23, 24, 26, 27). The deletions of the entire exon 8 result from missplicing due to mutation, insertions, or deletions in the splice donor and acceptor sites surrounding exon 8. Frame shift mutations and insertions within the linker and RF regions have been found as well (Fig. 1B). The amino acids of the linker and RF regions have multiple contacts with the TKB of the Cbl protein and with the E2 (1, 6). Mutations within these regions are likely to disrupt these interactions. Two particular hot spots for mutation include Y371 in the linker and R420 just C-terminal to the RF (Fig. 1B). Y371 forms hydrogen bonds with amino acids in the TKB region (1, 6). As described above, Y371 serves an important regulatory role in Cbl function and phosphorylation of Y371 which is required for E3 activity disrupts these hydrogen bonds (6–8). The mutations at Y371 occur mostly in patients with JMML and CMML (18, 21, 23, 24, 26, 27). R420 is conserved across all orthologues and homologues of Cbl (1, 6). This amino acid interacts with the E2 and mutations of this residue have been shown to disrupt E3 function of Cbl (19). There is not a clear subtype of myeloid neoplasm with R420 mutations.
Interestingly, the missense mutations are usually homozygous while the deletions that arise from splicing mutations are usually heterozygous (18–27, 29). Of the published missense mutations approximately 83% are homozygous. By contrast, 80% of the deletion mutations are heterozygous. The deletion mutations found in the NUP98-HOXD13 transgenic mouse and the MOLM-13 and MOLM-14 cell lines were also heterozygous (13, 18, 20, 21). This raises an issue of whether the proteins containing deletions are more transforming than those with point mutations. In vitro transformation assays in NIH 3T3 cells found that deletions of the linker domain were transforming while point mutations in the linker or RF were not (11). In addition, one group found that 70Z Cbl induces greater ligand independent proliferation and survival than the R420Q mutation (30). However, Sanada et al. found no difference in transformation efficiency between 70Z Cbl and a variety of point mutants found in patients (27). Thus it is unclear why most missense mutations are homozygous and the deletion mutations are heterozygous.
The majority of the Cbl mutations in myeloid neoplasms occur in the absence of other mutations known to be involved in the pathogenesis of these diseases (e.g., Flt3 internal tandem duplications, V617F Jak2 mutations, NF1 mutations, PTPN11 mutations, and Ras mutations) although some cases of overlap have been described (23, 24, 26, 27, 29). The mutual exclusivity of Cbl and Ras or PTPN11 mutations appears to be the most consistent finding in these studies. Several of the studies have found mutations of Cbl in myeloid neoplasms containing core binding factor (CBF) mutations (20, 21, 27). One study found that two of three AML samples containing heterozygous Cbl exon 8 or 8/9 deletion mutations had CBF mutations based on the presence of either t(8;21) or inv(16) (21). A second report identified two Cbl exon 8 deletion mutants from a randomly selected sample of 183 AML cases (20). Both of these cases carried an inv(16) indicative of a CBF mutation. Subsequently, they screened 79 CBF AML samples and found three additional cases with Cbl exon 8 deletion mutations. A third study found four of 15 samples with homozygous Cbl point mutations also had mutations in RUNX1 (27). Together these studies indicate that Cbl mutations and CBF mutations may cooperate in the pathogenesis of myeloid neoplasms. Whether Cbl mutations confer unique clinical characteristics on the myeloid neoplasms is not yet clear. Some of the studies have found that myeloid neoplasms containing Cbl mutations have a poor prognosis while others have not (23, 25–27). However, each report contained a limited number of cases with Cbl mutations and, thus, correlation with clinical outcomes may be difficult to discern until larger numbers are evaluated.
The frequent occurrence of Cbl mutations in patients with sAML with an antecedent MDS/MPN suggests that Cbl mutations contribute to the progression of the disease (22, 25). Consistent with this model, one patient described by Grand et al. had essential thrombocythemia (ET) that progressed to myelofibrosis (MF) over a 15 year period (23). The Cbl mutation (R420Q) was absent in the ET sample and present upon progression to MF, reminiscent of the animal model of Slape et al. (13, 23).
While mutations of Cbl frequently have been found in myeloid neoplasms, mutations of the other Cbl genes, Cbl-b and Cbl-c, are much less common. Two studies have described a total of five mutations within Cbl-b, all of which are either frame shift or missense mutations within the RF domain (18, 25). Two of the Cbl-b mutations identified by Makishima et al. were heterozygous and one was hemizygous (25). These Cbl-b mutants have not been characterized further but the locations would be predicted to disrupt E3 function (18, 25). Other studies have not found mutations of Cbl-b suggesting that the frequency of Cbl-b mutations is low in myeloid neoplasms relative to Cbl (19, 23, 26). One report describes four samples with a frame shift polymorphism in the RF domain of Cbl-c (25). The expression of Cbl-c appears restricted to epithelial cells, so the significance of these abnormalities remains to be determined (1, 6).
Functional studies of Cbl mutants: the two personalities of Cbl
All transforming variants of Cbl (e.g., 70Z Cbl) that have been characterized in vitro have lost their E3 activity (11). Similarly, all of the novel Cbl missense mutations found in myeloid neoplasms that have been studied abrogate the ability of Cbl to ubiquitinate RTKs such as Flt3, Kit, and EGFR (19, 23, 27). This includes missense mutations in the linker region (i.e., Q367P, Y371S, and S376F) and missense mutations in the RF (i.e., H398Y, P417A, and R420Q) (19, 23, 27). Co-expression of Cbl missense mutants (i.e., S376F, H398Y, p417A, and R420QA) and Flt3 in the IL-3 dependent 32D myeloid cell line stimulates ligand independent growth and enhanced ligand stimulated growth – consistent with a transformed phenotype in these cells (19, 23). Similarly, Reindl et al. demonstrated that Cbl 70Z, exon 8, and 8/9 deletion mutants caused ligand independent growth and enhanced ligand dependent growth of the IL-3 dependent Ba/F myeloid cell line (21). Coexpression of the deletion mutants with Flt3 resulted in activation of Flt3 in the absence of ligand and enhanced activation of Flt3 upon the addition of ligand (21). This result is similar to the activation of EGFR in the absence of ligand caused by the 70Z Cbl mutant (31). Sanada et al. demonstrated that a number of Cbl missense mutations (i.e., Q367P, Q367K, Y368C, Y371C, Y371S, I383L, C384G, and L399P) transformed NIH3T3 cells as assessed by in vitro and in vivo assays (27). Animal studies demonstrated that reconstitution of lethally irradiated mice with bone marrow cells that have been retrovirally transduced with the R420Q or 70Z mutants of Cbl develop mastocytosis, MPN, or AML (30).
The loss of E3 activity in transforming mutants suggests a tumor suppressor function for the E3 activity of Cbl (Fig. 1C). Consistent with this, the loss of Cbl accelerated the evolution of blast crisis in mice expressing a BCR-ABL transgene (27). The activation of Flt3 or EGFR in the absence of ligand suggested that, in addition, the Cbl mutants may function as oncogenes. However, an alternative explanation could be that Cbl functions as a tumor suppressor and that the mutants act by a dominant negative mechanism to inhibit the function of endogenous Cbl proteins. Sanada et al. addressed these possibilities by expressing wild type or mutant Cbl proteins in hematopoietic stem/progenitor cells (HSPCs) from either Cbl+/+ or Cbl−/− mice (27). HSPCs from Cbl−/− mice showed an increased proliferative response to a variety of cytokines. This response was enhanced significantly by the introduction of Cbl mutants, consistent with a gain of function role for the Cbl mutants. Importantly, this enhanced response to cytokines induced by Cbl mutants was not observed in the HSPCs from Cbl+/+ mice – suggesting that the loss of normal Cbl alleles is required to unmask the gain of function or oncogene function of Cbl.
Cbl null mice containing a RF mutant knock-in have increased activation of the PI3K/AKT pathway upon T-cell receptor activation which is due to increased phosphorylation of Cbl on Y731, the recruitment site for the p85 subunit of PI3K (32). Investigation of the perturbations in signaling of cells expressing mutant Cbl proteins from myeloid neoplasms found enhanced activation of of the PI3K/AKT and STAT5 pathways in the absence and presence of ligand (19, 21, 27). Importantly, the ligand independent growth was inhibited by PI3K pathway inhibitors (21).
Together, the data suggest that Cbl has both tumor suppressor and oncogene activities and that the transforming effects of Cbl mutations are due to the loss of the tumor suppressor E3 function and the maintenance of the oncogenic adaptor function. This is consistent with the homozygous nature of the mutations found in myeloid neoplasms.
Beyond Myeloid Neoplasms
There is some evidence that disruption of Cbl function may contribute to the pathogenesis of solid tumors. A recent publication identified somatic mutations of Cbl in 8 non-small cell lung tumors out of 119 samples. All but one of the mutations were outside the linker and RF regions, and all were heterozygous. Further analysis of three mutants that were outside of the linker and RF domains revealed that E3 activity was maintained, but overexpression of these mutants in lung cancer cells resulted in increased viability and motility (33). The mechanism by which these mutants affected viability or motility is not known.
A number of studies suggest that disruption of Cbl function in epithelial malignancies may occur by alterations in target proteins that are downregulated by Cbl proteins. For example, activating mutations of the EGFR found in non-small cell lung cancer tumors may interfere with Cbl mediated downregulation of the EGFR (34). Overexpression of ErbB-2, can inhibit Cbl-mediated downregulation of EGFR by forming heterodimers with the activated EGFR and preventing Cbl binding to the activated EGFR (35). Also, evidence exists that proteins which negatively regulate Cbl-mediated downregulation of RTKs (e.g., Src, Cdc42, cortactin, and DUB-2) are aberrantly active or overexpressed in human epithelial malignancies and thus may contribute to transformation by inhibiting downregulation RTKs (reviewed in (36)). The importance of these abnormalities to the pathogenesis of solid tumors is an open question.
Conclusions
The recent identification of recurrent transforming Cbl mutations in patients with myeloid neoplasms has clearly established that Cbl, first described in 1989, plays a role in the pathogenesis of human malignancies. The studies of Cbl mutations in myeloid neoplasms, along with older in vitro and animal studies, have led to the understanding that Cbl has both tumor suppressor and oncogene functions. The challenge now will be to elucidate further the relative contributions that the E3 dependent and E3 independent functions play in the pathogenesis of myeloid neoplasms. Importantly the activation of PI3K/AKT and STAT5 signaling by the Cbl mutants suggests that inhibition of these signaling pathways may provide a therapeutic opportunity in myeloid neoplasms with Cbl mutations.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research
We thank Peter Aplan for critical review of this manuscript.
Footnotes
Potential conflicts of interest: NONE
References
- 1.Tsygankov A, editor. Cbl Proteins. New York: Nova Science Publishers, Inc; 2008. [Google Scholar]
- 2.Tezuka T, Kim M, Yamamoto T. Cbl-c/Cbl-3: The third member of teh mammalian Cbl family. In: Tsygankov AY, editor. Cbl Proteins. New York: Nova Science Publishers; 2008. pp. 195–9. [Google Scholar]
- 3.Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–61. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–5. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 6.Nau MM, Lipkowitz S. Welcome to the family: Cbl-family gene organization, overview of stucture and functions of cbl-related proteins in various taxonomical groups. In: Tsygankov A, editor. Cbl proteins. New York: Nova Science Publishers, Inc; 2008. pp. 3–25. [Google Scholar]
- 7.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem. 2004;279:28017–27. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 8.Levkowitz G, Waterman H, Ettenberg SA, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–40. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 9.Andoniou CE, Thien CBF, Langdon WY. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. EMBO. 1994;13:4515–23. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisson SA, Ujack EE, Robbins SM. Isolation and characterization of a novel, transforming allele of the c-Cbl proto-oncogene from a murine macrophage cell line. Oncogene. 2002;21:3677–87. doi: 10.1038/sj.onc.1205510. [DOI] [PubMed] [Google Scholar]
- 11.Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol Cell. 2001;7:355–65. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 12.Thien CB, Langdon WY. Tyrosine kinase activity of the EGF receptor is enhanced by the expression of oncogenic 70Z-Cbl. Oncogene. 1997;15:2909–19. doi: 10.1038/sj.onc.1201468. [DOI] [PubMed] [Google Scholar]
- 13.Slape C, Liu LY, Beachy S, Aplan PD. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood. 2008;112:2017–9. doi: 10.1182/blood-2008-01-135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridge SA, Worwood M, Oscier D, Jacobs A, Padua RA. FMS mutations in myelodysplastic, leukemic, and normal subjects. Proc Natl Acad Sci U S A. 1990;87:1377–80. doi: 10.1073/pnas.87.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker A, Cachia P, Ridge S, et al. FMS mutations in patients following cytotoxic therapy for lymphoma. Leuk Res. 1995;19:309–18. doi: 10.1016/0145-2126(94)00128-w. [DOI] [PubMed] [Google Scholar]
- 16.Mancini A, Koch A, Wilms R, Tamura T. c-Cbl associates directly with the C-terminal tail of the receptor for the macrophage colony-stimulating factor, c-Fms, and down-modulates this receptor but not the viral oncogene v-Fms. J Biol Chem. 2002;277:14635–40. doi: 10.1074/jbc.M109214200. [DOI] [PubMed] [Google Scholar]
- 17.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–23. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 18.Caligiuri MA, Briesewitz R, Yu J, et al. Novel c-CBL and CBL-B ubiquitin ligase mutations in human acute myeloid leukemia. Blood. 2007 doi: 10.1182/blood-2006-12-061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sargin B, Choudhary C, Crosetto N, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007 doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 20.Abbas S, Rotmans G, Lowenberg B, Valk PJ. Exon 8 splice site mutations in the gene encoding the E3-ligase CBL are associated with core binding factor acute myeloid leukemias. Haematologica. 2008;93:1595–7. doi: 10.3324/haematol.13187. [DOI] [PubMed] [Google Scholar]
- 21.Reindl C, Quentmeier H, Petropoulos K, et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res. 2009;15:2238–47. doi: 10.1158/1078-0432.CCR-08-1325. [DOI] [PubMed] [Google Scholar]
- 22.Dunbar AJ, Gondek LP, O’Keefe CL, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68:10349–57. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grand FH, Hidalgo-Curtis CE, Ernst T, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113:6182–92. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 24.Loh ML, Sakai DS, Flotho C, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114:1859–63. doi: 10.1182/blood-2009-01-198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makishima H, Cazzolli H, Szpurka H, et al. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol. 2009;27:6109–16. doi: 10.1200/JCO.2009.23.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muramatsu H, Makishima H, Jankowska AM, et al. Mutations of E3 ubiquitin ligase Cbl family members but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia. Blood. 2009 doi: 10.1182/blood-2009-06-226340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanada M, Suzuki T, Shih LY, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–8. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 28.McKeller MR, Robetorye RS, Dahia PL, Aguiar RC. Integrity of the CBL gene in mature B-cell malignancies. Blood. 2009;114:4321–2. doi: 10.1182/blood-2009-08-239988. [DOI] [PubMed] [Google Scholar]
- 29.Beer PA, Delhommeau F, Lecouedic JP, et al. Two routes to leukemic transformation following a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2009 doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 30.Bandi SR, Brandts C, Rensinghoff M, et al. E3 ligase-defective Cbl mutants lead to a generalized mastocytosis and myeloproliferative disease. Blood. 2009;114:4197–208. doi: 10.1182/blood-2008-12-190934. [DOI] [PubMed] [Google Scholar]
- 31.Thien CB, Langdon WY. EGF receptor binding and transformation by v-cbl is ablated by the introduction of a loss-of-function mutation from the Caenorhabditis elegans sli-1 gene. Oncogene. 1997;14:2239–49. doi: 10.1038/sj.onc.1201193. [DOI] [PubMed] [Google Scholar]
- 32.Thien CB, Blystad FD, Zhan Y, et al. Loss of c-Cbl RING finger function results in high-intensity TCR signaling and thymic deletion. Embo J. 2005;24:3807–19. doi: 10.1038/sj.emboj.7600841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan YH, Krishnaswamy S, Nandi S, et al. CBL Is Frequently Altered in Lung Cancers: Its Relationship to Mutations in MET and EGFR Tyrosine Kinases. PLoS One. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–50. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 35.Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–57. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan PE, Davies GC, Nau MM, Lipkowitz S. Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem Sci. 2006;31:79–88. doi: 10.1016/j.tibs.2005.12.004. [DOI] [PubMed] [Google Scholar]