Abstract
Insulin-induced gene 2 (INSIG2) plays an important role in the regulation of cholesterol and fatty acids synthesis. A polymorphism, rs7566605, located 10 kb upstream of the INSIG2 gene was identified in a genome-wide association study of obesity. We conducted an association study of 12 INSIG2 tagSNPs with longitudinal measures of body size (BMI and waist circumference) and lipid metabolism (plasma HDL-, LDL-cholesterol and triglycerides levels). We investigated their interaction with age in 4,304 CARDIA participants (49.5% Blacks; 50.5% Whites) followed prospectively for 20 years. rs7566605 was not associated with variation in body size or lipid metabolism at any age, in either racial group. However, rs1352083 and rs10185316 were associated with age-related decline in HDL-cholesterol in Whites (P=0.0005 and 0.04, respectively). A similar trend was observed in Blacks who consistently maintained a BMI<25 kg/m2 over the study period. These data support a role of INSIG2 sequence variation in the regulation of cholesterol metabolism.
1. Introduction
Insulin-induced gene 2 (INSIG2) encodes a protein of the endoplasmic reticulum that prevents the proteolytic processing of sterol regulatory element-binding proteins (SREBPs) into active transcription factors, which regulate cholesterol and fatty acid synthesis [1]. A study integrating QTL mapping using a comprehensive single nucleotide polymorphism (SNP) map and gene expression profiling analyses identified INSIG2 as a susceptibility gene influencing plasma cholesterol levels in mice [2]. However, few studies have examined the association between sequence variation in the INSIG2 gene and measures of cholesterol metabolism in humans [3].
In a genome-wide association study, a SNP (rs7566605) upstream of the INSIG2 gene was recently identified, which was associated with increased body mass index (BMI) in 923 individuals from the Framingham Heart study [4]. This association was replicated in several but not all cohorts of varying ethnicity and age [5]. Many additional studies have failed to reproduce this finding [6–11]. Because the functional relevance of the rs7566605 polymorphism is uncertain and linkage disequilibrium (LD) patterns may vary across populations, investigation of additional polymorphisms in the INSIG2 gene may provide further information about the relationship of this gene with metabolic risk factors, including obesity-related phenotypes and plasma lipids. Moreover, it has been suggested that age may play an important role in modifying the association between INSIG2 gene variation and BMI [5]. To address these issues, we conducted an association study of 12 tagSNPs of the INSIG2 gene, selected based on publicly available LD information, with longitudinal measures of body size (waist circumference and BMI) and plasma lipid levels (HDL- and LDL-cholesterol and triglycerides), and we investigated their interaction with age in black and white CARDIA study participants followed prospectively for 20 years.
2. Material and Methods
Study sample and data collection
Participants were from the Coronary Artery Risk Development in Young Adults (CARDIA) study. Details about the study design have been previously published [12]. Briefly, 5,115 black and white men and women, 18–30 years of age, were initially recruited from the total community in Birmingham, AL; from selected census tracts in Chicago, IL and Minneapolis, MN; and from the Kaiser-Permanente health plan membership in Oakland, CA. The initial study population was approximately balanced with respect to race, age, gender, and education groups. From the time of initiation of the study in 1985–1986, participants have completed seven sequential examinations in years 0 (baseline), 2, 5, 7, 10, 15 and 20. Retention rates for the follow-up examinations were 90%, 86%, 81%, 79%, 74%, and 72%, respectively. Written informed consent was obtained from the participants at each examination and all study protocols were approved by the institutional review boards of the participating institutions.
Each participant’s age, race, and sex were self-reported during the recruitment phase and verified during the baseline clinic visit. Body weight, height, and waist circumference were assessed at each examination. Body weight was measured to the nearest 0.1 kg, using a calibrated scale, with the participant in light clothing without shoes. Height was measured to the nearest 0.5 cm with a vertical ruler. Body mass index (BMI) was computed as body weight/height2 (kg/m2). Blood samples were drawn after an overnight fast at each examination. Total plasma cholesterol, HDL-, LDL-cholesterol and triglyceride levels were measured according to standardized methods [13]. Participants eligible for the current study included 4,304 individuals, including 2,129 Blacks and 2,175 Whites, who consented to isolation of genomic DNA from a blood sample obtained at the year 10, 15, or 20 examination. Individuals without genotype data were more likely to be Black, male, and were slightly younger and less educated, but were otherwise similar to those with genotype data (not shown).
Polymorphism selection and genotyping
Single nucleotide polymorphisms (SNPs) were chosen from publicly available genotype data on individuals of African and European ancestry from the HapMap project [14, 15]. Within each race, a minimal set of tagSNPs was selected based on pairwise linkage disequilibrium relationships (r2), as described by Carlson et al. [16] and implemented in the Tagger algorithm [17]. Briefly, bins of SNPs are created based on a specified r2 threshold, and then one SNP is selected to represent the remainder of SNPs in that bin. In this study, we used an r2 threshold of 0.8 and minimum allele frequency of 0.05. A total of 12 SNPs spanning a 28 kb region around the INSIG2 gene were genotyped in the sample of CARDIA black and white participants using the TaqMan assay (Applied Biosystems, Foster City, CA) as previously described [18]. Primer and probes are available from the authors upon request. Polymorphism genotyping in the CARDIA study adheres to a rigorous quality control (QC) program, which includes barcode identification of samples, robotic sample handling, and blind replicate genotype assessment on 5% of the total sample (n=219). The overall genotyping rate was 97% and the concordance rate for blind duplicates was greater than 99%.
Statistical analyses
Genotype frequencies were estimated in each racial group by direct counting. Agreement of the genotype frequencies with Hardy Weinberg equilibrium expectations was tested using a Χ2 goodness-of-fit test.
To examine the association of INSIG2 polymorphisms with 20-year measures of body size (BMI) and lipid metabolism (plasma HDL-cholesterol, LDL cholesterol, and triglycerides levels), we first performed serial cross-sectional analyses on the subset of the CARDIA participants who completed all 7 examinations from year 0 to year 20. In each racial group, associations between each phenotype and individual tagSNPs were assessed using multiple linear regression models adjusting for baseline age, sex, and field center. Additional models adjusting for baseline BMI and current use of lipid lowering medication were estimated in the analyses of plasma lipid levels. Triglyceride levels were log-transformed to reduce skewness. For each polymorphism, genotypes were coded as the number of copies (0, 1, or 2) of the reference allele. Additive models have been shown to perform well even when the underlying inheritance model is recessive or dominant.[19, 20] Significance of allelic effects on each trait was determined empirically by permutation-derived P values. Both point-wise and family-wise (i.e., corrected for multiple tests) permutation-derived P values were estimated.
To examine the age-dependent effects of INSIG2 polymorphisms on measures of body size and lipid metabolism independently from secular trends or other age-unrelated changes in the phenotypes, we performed longitudinal analyses using Generalized Estimating Equations (GEE). In each racial group, models were estimated with INSIG2 genotype (coded as previously), time-dependent age, and genotype x time-dependent age interaction as independent variables and adjusting for sex, field center, and examination. Additional models adjusting for time-dependent BMI and lipid-lowering medication use were estimated for the analyses of plasma lipids.
3. Results
Baseline characteristics of the CARDIA participants included in this study are shown in Table 1. Of the 4,304 CARDIA participants with available DNA, there were 2,705 individuals who participated in all 7 exams. Compared to the total cohort, individuals in the constant cohort were more likely to be female, white, and slightly better educated, and less likely to smoke. Allele frequencies of the 12 INSIG2 polymorphisms are shown in Table 2 by race. Genotype frequency distributions of all polymorphisms were in agreement with Hardy-Weinberg equilibrium expectations in each racial group. Pairwise linkage disequilibrium (LD) between the INSIG2 SNPs is shown in Figure 1. As expected based on our tagSNP selection strategy, there was little LD between the selected SNPs, with the exception of rs10185316 and rs1352083 in Blacks (r2=0.83).
Table 1.
Baseline characteristics of the CARDIA participants genotyped for INSIG2 tagSNPs
| Characteristic | Total cohort (N=4,304) | Constant cohort (N=2,705) |
|---|---|---|
| Age (years) | 25.0 (3.6) | 25.3 (3.5) |
| Male (%) | 44.3 | 43.4 |
| Black (%) | 49.5 | 42.5 |
| Education (years) | 13.9 (2.2) | 14.2 (2.2) |
| Current smoking (%) | 29.1 | 24.1 |
| Body mass index (kg/m2) | 24.5 (5.1) | 24.4 (4.8) |
| LDL-cholesterol (mg/dL) | 109.5 (31.1) | 109.8 (30.6) |
| HDL-cholesterol (mg/dL) | 53.1 (13.1) | 53.3 (12.7) |
| Log-triglycerides (mg/dL) | 4.15 (0.48) | 4.14 (0.47) |
| Diabetes (%) | 0.6% | 0.6% |
Table 2.
Allele frequencies and P-value for test of significance of departure from Hardy-Weinberg equilibrium expectations for 12 INSIG2 tagSNPs in each racial group.
| Blacks (N=2, 129) | Whites (N=2,175) | |||||||
|---|---|---|---|---|---|---|---|---|
| Polymorphism | Chromosomal position | Gene Location | Allele 1 | Allele 2 | Freq (allele 1) | HWE P | Freq (allele 1) | HWE P |
| rs7566605 | 118552495 | 5′ flanking | C | G | 0.26 | 0.15 | 0.33 | 0.32 |
| rs10185316 | 118560948 | 5′ flanking | G | C | 0.30 | 0.40 | 0.32 | 0.66 |
| rs4848492 | 118561741 | 5′ flanking | C | T | 0.20 | 0.59 | 0.10 | 0.61 |
| rs13428113 | 118563355 | Intron 1 | C | T | 0.41 | 0.24 | 0.58 | 0.65 |
| rs1352083 | 118564311 | Intron 1 | T | C | 0.27 | 0.70 | 0.25 | 0.82 |
| rs12464355 | 118566320 | Intron 1 | G | A | 0.01 | 1.00 | 0.09 | 0.11 |
| rs17528324 | 118572626 | Intron 2 | A | G | 0.01 | 1.00 | 0.05 | 0.67 |
| rs889904 | 118576941 | Intron 2 | G | A | 0.40 | 0.96 | 0.52 | 0.28 |
| rs10490624 | 118578962 | Intron 3 | G | A | 0.11 | 0.14 | 0.09 | 0.59 |
| rs13409050 | 118580240 | Intron 3 | A | G | 0.25 | 0.48 | 0.07 | 0.63 |
| rs9308762 | 118580344 | Intron 3 | C | T | 0.14 | 0.06 | 0.17 | 0.75 |
| rs17047764 | 118585051 | 3′ flanking | C | G | 0.39 | 0.37 | 0.16 | 0.81 |
Figure 1.
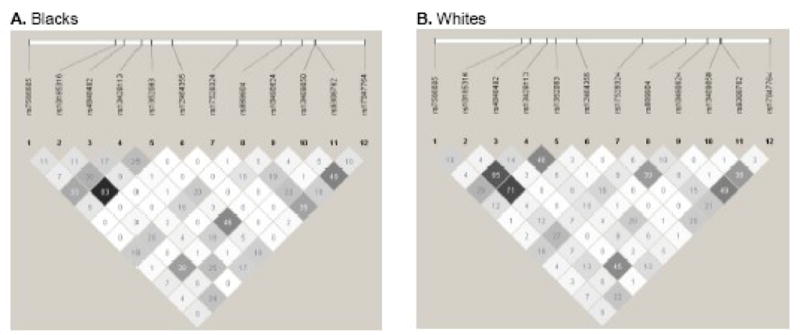
Pairwise inkage disequilibrium (r2) among 12 INSIG2 tagSNPs by racial group
We first performed serial cross-sectional analyses of association between each INSIG2 SNP and lipid levels and measures of body size in the constant cohort by race. In Blacks, rs13409050 was associated with variation in BMI at all exams and both significance of association and magnitude of the effect tended to increase over time. At baseline, individuals carrying the common rs13409050_G allele had a 0.60 kg/m2 higher BMI per copy (P=0.03). At Year 20, these individuals had a 1.1 kg/m2 higher BMI per copy (P=0.01) (Table 3a). Similar results were obtained in the association of the rs13409050 polymorphism with waist circumference. At baseline, individuals carrying the common rs13409050_G allele had a 1.1 cm higher waist circumference per copy (P=0.05). At Year 20, these individuals had a 2.1 cm higher waist circumference per copy (P=0.01). In Blacks from the CARDIA study, there was no significant association of INSIG2 polymorphisms with plasma HDL-cholesterol levels (Table 3b) at any of the 7 examinations; similarly for plasma LDL-cholesterol and triglyceride levels (not shown).
Table 3.
| Table 3a. Serial cross-sectional association of INSIG2 tagSNPs with BMI in the constant cohort by race. | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year 0 | Year 2 | Year 5 | Year 7 | Year 10 | Year 15 | Year 2 | |||||||||
| SNP | Ref. allele | Difference a (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb |
| African-Americans (N=1,149) | |||||||||||||||
| rs7566605 | C | 0.05 (0.27) | 0.86 | 0.02 (0.30) | 0.96 | 0.04 (0.31) | 0.89 | −0.05 (0.32) | 0.87 | 0.04 (0.33) | 0.90 | 0.38 (0.35) | 0.28 | 0.20 (0.36) | 0.56 |
| rs10185316 | G | −0.46 (0.25) | 0.07 | −0.50 (0.27) | 0.07 | −0.6 (0.28) | 0.03 | −0.60 (0.30) | 0.04 | −0.54 (0.31) | 0.08 | −0.57 (0.33) | 0.08 | −0.58 (0.33) | 0.09 |
| rs4848492 | C | 0.30 (0.29) | 0.32 | 0.32 (0.32) | 0.32 | 0.34 (0.34) | 0.33 | 0.30 (0.35) | 0.39 | 0.36 (0.36) | 0.32 | 0.09 (0.38) | 0.80 | 0.15 (0.39) | 0.69 |
| rs13428113 | C | −0.07 (0.23) | 0.75 | 0.002 (0.25) | 0.98 | 0.01 (0.26) | 0.96 | 0.004 (0.28) | 0.99 | −0.03 (0.28) | 0.90 | 0.21 (0.30) | 0.48 | 0.20 (0.31) | 0.49 |
| rs1352083 | T | −0.45 (0.26) | 0.07 | −0.56 (0.29) | 0.05 | −0.64 (0.30) | 0.04 | −0.70 (0.31) | 0.02 | −0.62 (0.32) | 0.06 | −0.66 (0.34) | 0.05 | −0.73 (0.35) | 0.05 |
| rs12464355 | G | 0.05 (0.97) | 0.95 | 0.19 (1.06) | 0.85 | 0.21 (1.13) | 0.85 | 0.12 (1.16) | 0.91 | −0.70 (1.21) | 0.59 | −0.63 (1.28) | 0.63 | −0.58 (1.30) | 0.66 |
| rs17528324 | A | −1.71 (0.96) | 0.08 | −1.20 (1.07) | 0.23 | −1.81 (1.11) | 0.08 | −1.17 (1.15) | 0.33 | −0.96 (1.19) | 0.40 | −1.38 (1.27) | 0.27 | −0.75 (1.31) | 0.57 |
| rs889904 | G | 0.12 (0.24) | 0.61 | 0.13 (0.26) | 0.63 | 0.07 (0.27) | 0.81 | −0.07 (0.28) | 0.83 | 0.03 (0.30) | 0.89 | 0.01 (0.31) | 0.95 | 0.18 (0.32) | 0.58 |
| rs10490624 | G | 0.02 (0.37) | 0.95 | 0.12 (0.41) | 0.78 | 0.43 (0.43) | 0.31 | 0.55 (0.45) | 0.22 | 0.66 (0.46) | 0.15 | 0.22 (0.49) | 0.65 | 0.66 (0.50) | 0.21 |
| rs13409050 | G | 0.60 (0.27) | 0.03 | 0.68 (0.30) | 0.02 | 0.80 (0.31) | 0.02 | 0.83 (0.33) | 0.01* | 0.81 (0.34) | 0.02 | 1.0 (0.36) | 0.005** | 0.95 (0.34) | 0.01* |
| rs9308762 | C | 0.19 (0.34) | 0.58 | 0.24 (0.37). | 0.51 | 0.07 (0.39) | 0.87 | 0.04 (0.41) | 0.93 | −0.04 (0.42) | 0.93 | −0.08 (0.45) | 0.87 | −0.17 (0.46) | 0.70 |
| Whites (N=1,556) | |||||||||||||||
| rs7566605 | C | −0.12 (0.15) | 0.43 | −0.10 (0.16) | 0.46 | −0.03 (0.17) | 0.86 | 0.02 (0.19) | 0.90 | 0.002 (0.20) | 0.99 | −0.08 (0.22) | 0.72 | −0.21 (0.26) | 0.43 |
| rs10185316 | G | 0.14 (0.15) | 0.33 | 0.11 (0.16) | 0.48 | 0.17 (0.17) | 0.33 | 0.09 (0.19) | 0.66 | 0.17 (0.20) | 0.37 | 0.14 (0.22) | 0.54 | −0.06 (0.25) | 0.82 |
| rs4848492 | C | 0.17 (0.23) | 0.47 | 0.09 (0.25) | 0.74 | 0.01 (0.27) | 0.97 | −0.01 (0.30) | 0.96 | −0.05 (0.32) | 0.83 | −0.26 (0.36) | 0.50 | −0.13 (0.40) | 0.73 |
| rs13428113 | T | 0.19 (0.14) | 0.15 | 0.15 (0.15) | 0.32 | 0.17 (0.17) | 0.30 | 0.08 (0.18) | 0.64 | 0.14 (0.19) | 0.46 | 0.05 (0.22) | 0.81 | −0.09 (0.25) | 0.73 |
| rs1352083 | T | −0.001 (0.16) | 0.99 | −0.04 (0.17) | 0.81 | 0.11 (0.19) | 0.53 | 0.07 (0.21) | 0.72 | 0.09 (0.22) | 0.65 | 0.08 (0.24) | 0.75 | −0.19 (0.28) | 0.50 |
| rs12464355 | G | −0.03 (0.25) | 0.90 | −0.01 (0.27) | 0.96 | −0.03 (0.29) | 0.90 | 0.13 (0.32) | 0.66 | 0.07 (0.34) | 0.84 | 0.33 (0.38) | 0.38 | 0.38 (0.44) | 0.36 |
| rs17528324 | A | 0.32 (0.30) | 0.28 | 0.33 (0.32) | 0.32 | −0.001 (0.35) | 0.99 | −0.11 (0.39) | 0.79 | 0.16 (0.41) | 0.67 | 0.25 (0.46) | 0.60 | 0.26 (0.53) | 0.61 |
| rs889904 | A | 0.22 (0.14) | 0.12 | 0.22 (0.15) | 0.12 | 0.26 (0.16) | 0.10 | 0.24 (0.18) | 0.19 | 0.30 (0.19) | 0.10 | 0.41 (0.21) | 0.06 | 0.35 (0.24) | 0.15 |
| rs104906624 | G | 0.07 (0.24) | 0.77 | 0.06 (0.26) | 0.80 | −0.02 (0.26) | 0.94 | −0.11 (0.31) | 0.75 | 0.04 (0.33) | 0.89 | 0.11 (0.37) | 0.76 | 0.06 (0.42) | 0.87 |
| rs13409050 | G | −0.46 (0.27) | 0.09 | −0.30 (0.30) | 0.31 | −0.74 (0.32) | 0.02 | −0.70 (0.35) | 0.05 | −0.81 (0.38) | 0.04 | −0.91 (0.41) | 0.03 | −0.74 (0.48) | 0.14 |
| rs9308762 | C | 0.17 (0.19) | 0.36 | 0.07 (0.20) | 0.72 | 0.02 (0.22) | 0.92 | −0.02 (0.25) | 0.91 | −0.08 (0.26) | 0.73 | −0.07 (0.29) | 0.83 | 0.21 (0.33) | 0.54 |
| Table 3b. Serial cross-sectional association of INSIG2 tagSNPs with HDL-cholesterol levels in the constant cohort by race. | |||||||||||||||
| Year 0 | Year 2 | Year 5 | Year 7 | Year 10 | Year 15 | Year 20 | |||||||||
| SNP | Ref. allele | Differencea (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb | Difference (SE) In trait value per copy of ref. allele | Pb |
| African-Americans (N=1,149) | |||||||||||||||
| rs7566605 | C | 0.41 (0.61) | 0.53 | 0.52 (0.69) | 0.44 | 0.01 (0.68) | 0.98 | −0.08 (0.68) | 0.90 | 0.27 (0.66) | 0.68 | 0.11 (0.66) | 0.87 | 0.84 (0.76) | 0.29 |
| rs10185316 | G | 0.65 (0.57) | 0.26 | 0.77 (0.64) | 0.24 | 0.56 (0.64) | 0.39 | 0.93 (0.63) | 0.14 | 0.42 (0.62) | 0.49 | 0.18 (0.62) | 0.77 | 0.05 (0.72) | 0.95 |
| rs4848492 | C | 1.12 (0.66) | 0.10 | −0.76 (0.76) | 0.31 | 0.93 (0.76) | 0.21 | 0.57 (0.74) | 0.44 | 0.76 (0.72) | 0.31 | 0.67 (0.73) | 0.38 | 0.47 (0.84) | 0.56 |
| rs13428113 | C | −0.98 (0.52) | 0.06 | −0.21 (0.59) | 0.71 | −1.22 (0.59) | 0.03 | −0.95 (0.58) | 0.10 | −0.70 (0.57) | 0.22 | −0.40 (0.57) | 0.47 | −0.24 (0.66) | 0.73 |
| rs1352083 | T | 0.50 (0.60) | 0.41 | 0.58 (0.67) | 0.38 | 0.54 (0.67) | 0.39 | 1.15 (0.66) | 0.08 | 0.44 (0.64) | 0.47 | 0.36 (0.65) | 0.58 | −0.15 (0.75) | 0.81 |
| rs12464355 | G | 1.91 (2.21) | 0.38 | 1.75 (2.45) | 0.45 | −2.97 (2.52) | 0.22 | −1.25 (2.45) | 0.61 | −1.19 (2.39) | 0.63 | 1.50 (2.40) | 0.51 | −0.46 (2.79) | 0.87 |
| rs17528324 | A | 0.57 (2.23) | 0.80 | −0.38 (2.52) | 0.87 | 0.52 (2.49) | 0.84 | −0.78 (2.46) | 0.75 | −0.23 (2.44) | 0.93 | −0.70 (2.46) | 0.77 | 1.17 (2.81) | 0.66 |
| rs889904* | G | −0.57 (0.54) | 0.29 | −0.57 (0.61) | 0.34 | −0.73 (0.62) | 0.21 | −0.54 (0.61) | 0.37 | 0.18 (0.59) | 0.76 | −0.15 (0.59) | 0.81 | 0.46 (0.69) | 0.51 |
| rs10490624 | G | 0.75 (0.85) | 0.36 | 0.21 (0.96) | 0.83 | 0.52 (0.96) | 0.57 | −0.19 (0.93) | 0.83 | 0.11 (0.92) | 0.91 | 0.25 (0.93) | 0.79 | 0.02 (1.0) | 0.98 |
| rs13409050 | G | −0.84 (0.62) | 0.19 | −0.90 (0.70) | 0.20 | −1.12 (0.71) | 0.11 | −1.81 (0.69) | 0.01* | −1.07 (0.68) | 0.11 | −1.20 (0.68) | 0.08 | −0.91 (0.79) | 0.24 |
| rs9308762 | C | 0.56 (0.77) | 0.45 | −1.27 (0.89) | 0.15 | 0.16 (0.88) | 0.85 | 0.28 (0.86) | 0.74 | 0.49 (0.85) | 0.55 | 0.28 (0.84) | 0.75 | 0.60 (0.99) | 0.53 |
| Whites (N=1,556) | |||||||||||||||
| rs7566605 | C | 0.06 (0.45) | 0.90 | 0.21 (0.49) | 0.66 | 0.12 (0.49) | 0.81 | −0.03 (0.49) | 0.96 | 0.25 (0.48) | 0.62 | 0.15 (0.50) | 0.77 | 0.28 (0.58) | 0.60 |
| rs10185316 | G | −0.84 (0.45) −0.71 (0.43) |
0.05 0.09 |
−1.1 (0.40) 0.80 (0.47) |
0.04 0.10 |
−1.1 (0.49) −0.72 (0.46) |
0.030 0.10 |
−1.2 (0.4.9) −0.91 (0.47) |
0.02 0.05 |
−1.6 (0.48) −1.3 (0.46) |
0.004** 0.005* |
−1.5 (0.51) −1.3 (0.49) |
0.005** 0.005** |
−1.8 (0.58) −1.6 (0.56) |
0.003** 0.004** |
| rs4848492 | C | 0.04 (0.71) | 0.96 | −0.21 (0.78) | 0.79 | 0.33 (0.77) | 0.64 | 0.89 (0.78) | 0.27 | 0.74 (0.77) | 0.33 | 1.33 (0.81) | 0.10 | 1.3 (0.92) | 0.17 |
| rs13428113 | T | −0.71 (0.43) | 0.10 | −0.94 (0.47) | 0.05 | −0.60 (0.46) | 0.19 | −0.91 (0.47) | 0.19 | −1.0 (0.46) | 0.04 | −0.85 (0.48) | 0.07 | −1.07 (0.55) | 0.06 |
| rs1352083 | T | −0.3 (0.48) −0.15 (0.46) |
0.74 0.74 |
−0.3 (0.53) −0.16 (0.51) |
0.74 0.75 |
−0.6 (0.52) −0.34 (0.50) |
0.27 0.49 |
−0.7 (0.53) −0.54 (0.51) |
0.27 0.29 |
−1.2 (0.53) −1.0 (0.50) |
0.03 0.04 |
−1.3 (0.55) −1.1 (0.53) |
0.02 0.03 |
−1.8 (0.62) −1.6 (0.60) |
0.007** 0.01* |
| rs12464356 | G | −1.0 (0.76) | 0.19 | 0.85 (0.83) | 0.32 | −0.15 (0.81) | 0.85 | 0.34 (0.82) | 0.66 | 0.05 (0.82) | 0.93 | −0.57 (0.85) | 0.52 | −1.0 (0.97) | 0.30 |
| rs17528324 | A | −1.9 (0.92) | 0.05 | −2.44 (1.0) | 0.01 | −1.71 (0.99) | 0.10 | −1.6 (1.0) | 0.10 | −1.7 (0.99) | 0.09 | −1.67 (1.04) | 0.12 | −0.99 (1.19) | 0.41 |
| rs889904 | A | −0.71 (0.43) | 0.10 | −0.51 (0.47) | 0.27 | −0.52 (0.46) | 0.24 | −0.36 (0.47) | 0.45 | −0.58 (0.46) | 0.22 | −1.10 (0.48) | 0.02 | −1.21 (0.55) | 0.03 |
| rs10490624 | G | −0.82 (0.73) | 0.27 | −0.93 (0.81) | 0.25 | −0.67 (0.79) | 0.38i | −0.23 (0.81) | 0.76 | −0.87 (0.79) | 0.28 | −1.08 (0.83) | 0.20 | −0.23 (0.95) | 0.79 |
| rs13409050 | G | −0.71 (0.83) | 0.40 | 0.64 (0.91) | 0.49 | 0.06 (0.89) | 0.95 | 1.04 (0.91) | 0.25 | 0.85 (0.89) | 0.34 | 0.95 (0.93) | 0.32 | 1.18 (1.06) | 0.24 |
| rs9308762 | C | 0.21 (0.58) | 0.71 | 0.16 (0.63) | 0.79 | 0.28 (0.62) | 0.66 | 0.65 (0.63) | 0.32 | 0.53 (0.63) | 0.41 | 1.33 (0.65) | 0.05 | 1.10 (0.74) | 0.15 |
expressed in kg/m2;
point-wise empirical P-value derived from permutation;
family-wise empirical P value < 0.10;
family-wise empirical P value < 0.05; Models were adjusted for age, sex, and field center; Ref.: Reference; SE: standard error
expressed in mg/dL;
point-wise empirical P-value derived from permutation;
family-wise empirical P value < 0.10;
family-wise empirical P value < 0.05; Models were adjusted for age, sex, and field center;
model further adjusted for baseline BMI and current use of lipid lowering medication; Ref.: Reference; SE: standard error
In Whites, none of the INSIG2 polymorphisms were significantly associated with variation in body size at any of the 7 examinations (Table 3a). However, two polymorphisms in moderate LD (r2=0.7), rs10185316 and rs1352083, showed similar associations with variation in HDL cholesterol (Table 3b). At baseline, individuals carrying the variant allele rs10185316_G had a 0.84 mg/dL lower plasma HDL-cholesterol level per copy (P=0.05) compared to those who did not carry this allele. The magnitude and statistical significance of this difference increased over time. At year 20, these individuals had a 1.8 mg/dL lower plasma HDL-cholesterol per copy (P=0.003). A similar trend was observed for the rs1352083_T variant, although both the magnitude and statistical significance of the difference in plasma HDL-cholesterol between variant and wild-type allele were slightly less than for rs10185316. Adjusting for BMI and current use of lipid-lowering medication did not significantly affect these associations (Table 3b). There was no significant association of INSIG2 polymorphisms with plasma LDL-cholesterol or triglycerides levels at any of the 7 examinations (not shown).
We next examined the association of INSIG2 polymorphisms with age-related changes in measures of body size and plasma lipid levels using GEE models in the total cohort. Consistent with results of the serial cross-sectional analyses, we observed a significant association of rs13409050 with variation in BMI in Blacks and this association was age-dependent. At age 18, individuals carrying the common allele of rs13409050 (G) had a slightly higher BMI per copy (0.4 kg/m2; P=0.07) (Table 4a) and slightly larger waist circumference (0.7 cm; P=0.09), compared to individuals who did not carry this allele. However, with increasing age, these individuals had a significant increase in BMI (0.02 kg/m2 per year of age and per copy of the allele; P=0.02) (Table 4a) and waist circumference (0.05 cm per year of age and per copy of the allele). In Whites, we also observed an age-dependent association between INSIG2 SNPs and HDL-cholesterol levels, consistent with results from the serial cross-sectional analyses (Table 4b). At age 18, individuals carrying the variant allele rs10185316_G had a slightly lower plasma HDL-cholesterol level per copy (0.7 mg/dL; P=0.05) compared to those who did not carry this allele. With increasing age, these individuals tended to have increasingly lower plasma HDL-cholesterol levels (0.03 mg/dl per year of age and per copy of the allele; P=0.08). Similarly for rs1352083, individuals aged 18 years carrying the variant rs1352083_T allele had slightly, although not significantly, lower HDL-cholesterol levels than those who did not carry this allele. However, as they got older, carriers of the rs1352083_T allele had a greater decline in HDL-cholesterol levels (0.07 mg/dl per year of age and per copy of the allele; P=0.002). Further adjusting for BMI and use of lipid-lowering medication did not significantly modify these age-dependent associations (Table 4b).
Table 4.
| Table 4a. Age-dependent association of selected INSIG2 tagSNPs with BMI in the total cohort by race. | |||||
|---|---|---|---|---|---|
| African-Americans (N=2,129) | |||||
| SNP | Reference | Differencea in trait value (95% CI) at age 18 per copy of reference allele | P | Changea in trait value (95% CI) par year of age per copy of reference allele | P |
| rs7566605 | C | −0.03 (−0.46; 0.39) | 0.88 | 0.004 (−0.018; 0.023) | 0.65 |
| rs10185316 | G | 0.02 (−0.37; 0.41) | 0.92 | −0.006 (−0.024; 0.011) | 0.49 |
| rs4848492 | C | 0.01 (−0.48; 0.51) | 0.95 | −0.008 (−0.029; 0.013) | 0.44 |
| rs13428113 | C | −0.003 (−0.36; 0.36) | 0.99 | 0.009 (−0.008; 0.025) | 0.32 |
| rs1352083 | T | −0.06 (−0.45; 0.34) | 0.77 | −0.011 (−0.020; 0.007) | 0.23 |
| rs12464355 | G | 0.47 (−1.44; 2.39) | 0.62 | −0.03 (−0.11; 0.05) | 0.51 |
| rs17528324 | A | −0.16 (−1.83; 1.50) | 0.85 | 0.003 (−0.063; 0.069) | 0.92 |
| rs889904 | G | 0.10 (−0.27; 0.47) | 0.6 | 0.001 (−0.016; 0.018) | 0.93 |
| rs10490624 | G | −0.04 (−0.62; 0.54) | 0.89 | 0.011 (−0.017; 0.039) | 0.44 |
| rs13409050 | G | 0.4 (0.03; 0.80) | 0.07 | 0.02 (0.01; 0.03) | 0.02 |
| rs9308762 | C | 0.12 (−0.43; 0.68) | 0.66 | −0.006 (−0.031; 0.018) | 0.61 |
| Whites (N=2,175) | |||||
| rs7566605 | C | 0.19 (−0.09; 0.47) | 0.2 | −0.003 (−0.019; 0.012) | 0.69 |
| rs10185316 | G | 0.10 (−0.17; 0.37) | 0.48 | −0.005 (−0.019; 0.010) | 0.51 |
| rs4848492 | C | 0.20 (−0.30; 0.69) | 0.44 | −0.014 (−0.043; 0.015) | 0.35 |
| rs13428113 | C | −0.18 (−0.45; 0.08) | 0.17 | 0.009 (−0.005; 0.023) | 0.18 |
| rs1352083 | T | 0.07 (−0.22; 0.37) | 0.62 | −0.003 (−0.018; 0.013) | 0.75 |
| rs12464355 | G | −0.26 (−0.68; 0.16) | 0.23 | 0.017 (−0.010; 0.044) | 0.22 |
| rs17528324 | A | −0.07 (−0.56; 0.42) | 0.77 | −0.005 (−0.032; 0.023) | 0.74 |
| rs889904 | G | −0.004 (−0.26; 0.26) | 0.98 | −0.008 (−0.021; 0.006) | 0.26 |
| rs10490624 | G | −0.15 (−0.54; 0.23) | 0.44 | 0.004 (−0.018; 0.027) | 0.7 |
| rs13409050 | G | 0.002 (−0.47; 0.48) | 0.99 | 0.023 (−0.002; 0.048) | 0.08 |
| rs9308762 | C | 0.08 (−0.31; 0.46) | 0.69 | −0.006 (−0.027; 0.014) | 0.55 |
| Table 4b. Age-dependent association of selected INSIG2 tagSNPs with HDL-cholesterol levels in the total cohort by race. | |||||
| African-Americans (N=2,129) | |||||
| SNP | Reference allele | Differencea in trait value (95% CI) at age 18 per copy of reference allele | P | Changea in trait value (95% CI) par year of age per copy of reference allele | P |
| rs7566605 | C | −0.07 (−0.93; 0.80) | 0.88 | 0.006 (−0.043; 0.055) | 0.81 |
| rs10185316 | G | 0.27 (−0.53; 1.08) | 0.51 | −0.024 (−0.068; 0.020) | 0.29 |
| rs4848492 | C | 0.21 (−0.76; 1.18) | 0.67 | 0.02 (−0.03; 0.07) | 0.43 |
| rs13428113 | C | −0.47 (−1.22; 0.28) | 0.22 | 0.008 (−0.030; 0.047) | 0.66 |
| rs1352083 | T | −0.05 (−0.79; 0.88) | 0.91 | −0.015 (−0.061; 0.030) | 0.5 |
| rs12464355 | G | −0.48 (−3.84; 2.87) | 0.78 | 0.02 (−0.12; 0.16) | 0.78 |
| rs17528324 | A | 1.01 (−1.63; 3.65) | 0.45 | 0.03 (−0.14; 0.20) | 0.76 |
| rs889904 | G | −0.65 (−1.43; 0.13) | 0.1 | 0.025 (−0.017; 0.066) | 0.24 |
| rs10490624 | G | 0.50 (−0.76; 1.77) | 0.44 | −0.012 (−0.076; 0.051) | 0.7 |
| rs13409050 | G | 0.72 (−0.16; 1.60) | 0.11 | 0.014 (−0.036; 0.063) | 0.59 |
| rs9308762 | C | −0.41 (−1.48; 0.67) | 0.46 | −0.012 (−0.068; 0.043) | 0.66 |
| Whites (N=2,175) | |||||
| rs7566605 | C | 0.20 (−0.93; 0.53) | 0.59 | −0.002 (−0.042; 0.037) | 0.9 |
| rs10185316 | G | −0.7 (−1.4; 0.01) −0.6 (−1.3; 0.06)* |
0.05 0.07 |
−0.03 (−0.07; 0.01) −0.04 (−0.07; 0.00)* |
0.09 0.05 |
| rs4848492 | C | 0.22 (−1.03; 1.47) | 0.73 | 0.06 (−0.005; 0.118) | 0.07 |
| rs13428113 | C | 0.56 (−0.14; 1.28) | 0.11 | 0.013 (−0.025;0.051) | 0.49 |
| rs1352083 | T | −0.19 (−0.98; 0.59) −0.14 (−0.88; 0.60)* |
0.62 0.71 |
−0.07 (−0.11; −0.02) −0.07 (−0.11; −0.03)* |
0.002 0.0007 |
| rs12464355 | G | −0.99 (−0.46; 2.43) | 0.18 | −0.046 (−0.115; 0.023) | 0.19 |
| rs17528324 | A | −1.48 (−3.06; 0.10) | 0.06 | 0.07 (−0.01; 0.15) | 0.09 |
| rs889904 | G | 0.11 (−0.56; 0.80) | 0.74 | 0.02 (−0.01; 0.06) | 0.27 |
| rs10490624 | G | −0.74 (−1.91; 0.44) | 0.22 | 0.012 (−0.051; 0.076) | 0.7 |
| rs13409050 | G | 0.13 (−1.28; 1.54) | 0.85 | −0.09 (−0.16; −0.02) | 0.01 |
| rs9308762 | C | 0.09 (−0.83; 1.02) | 0.84 | 0.053 (0.004; 0.103) | 0.03 |
Models were adjusted for examination, sex, and field center.
expressed in kg/m2
Models were adjusted for examination, sex, end field center.
also adjusted for time-dependent BMI and use of lipid-lowering medication;
expressed in mg/dL
We also examined the association of INSIG2 sequence variants with plasma lipid levels in a subset of the CARDIA participants who consistently maintained their BMI below 25 kg/m2 over the 20 year study period (N=246 Blacks; 615 Whites). In Whites who were consistently lean, rs1352083_T and rs10185316_G were significantly associated with a greater age-related decrease in HDL-cholesterol (0.13 and 0.11 mg/dl per year of age and per copy of the allele, P=0.0004 and 0.003, respectively). In Blacks, a similar trend was observed although statistical significance was not reached (0.08 and 0.10 mg/dl per year of age and per copy of the allele, P=0.10 and 0.12, respectively).
In an effort to determine whether the identified SNPs had potential functional consequences, we screened the 2 tagSNPs against a set of previously reported expression-associated SNPs (eSNPs) in lymphoblastoid cell lines [21]. Neither polymorphism (or their proxy) was associated with variation in INSIG2 gene expression levels in cell lines.
4. Discussion
In the CARDIA cohort of young adults of African and European ancestry, sequence variants of the INSIG2 gene were significantly associated with BMI and HDL-cholesterol and these associations were age-dependent.
We failed to reproduce an association of rs7566605 with measures of body size, including BMI at any age, in either Blacks or Whites, consistent with our previous report [11] and similar to many studies [7–10]. Although it was previously suggested this polymorphism was associated with an increased BMI in overweight individuals [10], additional analyses of a subgroup of CARDIA participants who were consistently overweight over the 20-year study period (N=537 Black, 475 Whites), did not reveal a significant association of rs7566605 with body size (not shown). Our results also did not support an earlier suggestion of an interaction of this polymorphism with age, namely a stronger effect on BMI in younger individuals and a decreasing effect size with increasing age [5]. A recent report also showed suggestive evidence of an association of this polymorphism with BMI in children [22]. Nonetheless, we report a significant association of the common allele of rs13409050 – a tagSNP located in intron 3 of the INSIG2 gene – with a greater BMI and waist circumference in Blacks. We also provide evidence of an interaction of this polymorphism with age on body size, specifically an increasing effect size with increasing age. These associations were not observed in Whites, possibly due to differences in allele frequency, in linkage disequilibrium patterns, or in genetic or environmental contexts. The mechanisms that underlie the association of INSIG2 variants and increased BMI remain unclear. INSIG2 is ubiquitously expressed and adipocyte differentiation is characterized by an enhanced INSIG2 expression [23]. It has been recently suggested that INSIG2 may regulate adipogenesis via the sterol regulatory element-binding protein 1 (SREBP1) [23, 24]. A recent study in Hispanic individuals also reported an association of INSIG2 gene variants with computed tomography (CT)-measured adiposity [25]. Future characterization of functional variation in the INSIG2 gene may shed light on the relationship of this gene with obesity-related traits.
We provide evidence of an association of two INSIG2 variants, rs1352083 and rs10185316, with age-dependent decline in HDL cholesterol in the 20-year period from young adulthood to middle age. This association was observed in both black and white individuals who maintained a normal weight (BMI<25 kg/m2) over the study period, although statistical significance was reached only for Whites. rs10185316 is located in the promoter region of the gene and rs1352083 is located in intron 1. These variants have similar allele frequencies in the 2 racial groups and are in linkage disequilibrium. The relationship between INSIG2 and cholesterol metabolism has been well documented in experimental studies. In the presence of sterols, INSIG2 prevents the proteolytic activation of SREBPs by Golgi enzymes and, hence, blocks cholesterol metabolism [1]. Moreover, INSIG2 has been shown to regulate the degradation of HMG-CoA reductase, a rate-limiting enzyme in cholesterol synthesis [26, 27]. However, few studies have investigated the association of INSIG2 polymorphism with plasma lipid levels. No significant association of rs7566605 was reported with plasma levels of either total, HDL-, or LDL-cholesterol in middle-aged individuals from European ancestry [3] or Japanese ancestry [28]. However, none of these studies examined the relationship of other INSIG2 gene variants with longitudinal profiles of plasma lipid levels.
Despite the clear strengths of our study, including a well-characterized bi-racial sample of young adults followed prospectively for 20 years, and a more comprehensive tagSNP strategy in the INSIG2 gene, some limitations must be acknowledged. First, we have not examined possible interactions of INSIG2 polymorphisms with environmental or other genetic factors known to influence body size and plasma lipid levels. Therefore, we cannot exclude that the associations (or lack thereof) reported here may vary in different environmental contexts, such as physical activity or dietary fat intake, or in the presence of other susceptibility genes [29]. Second, association studies cannot distinguish the causal SNP (or combination of SNPs), among those in linkage disequilibrium (LD). Therefore, as the functional relevance of the identified SNPs is uncertain, we cannot exclude that a polymorphism(s) unrelated to INSIG2 function may be responsible for the effects observed in this study. Finally, the novel SNP associations reported here require validation through analysis in other cohorts. In particular, replication in other African-American cohorts taking into account genetic measures of ancestry is warranted to rule out any spurious findings due to population stratification.
In summary, common variants of the INSIG2 gene are associated with age-dependent increase in body size and decrease in plasma HDL cholesterol in the period of young adulthood to early middle-age. The molecular mechanisms that mediate these relationships remain to be investigated. In particular, characterization of functional variants in the INSIG2 gene will contribute toward a better understanding of the role of this gene in susceptibility to metabolic and cardiovascular dysfunction.
Acknowledgments
This study is supported by grant R01-HL69126 to MF and by contracts N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050 and N01-HC-95095 from the National Heart, Lung, and Blood Institute to the CARDIA investigators. The authors thank the staff and participants of the CARDIA study for their important contributions.
Footnotes
Conflict of Interest
None
Institutional Approval
The University of Texas Health Science Center at Houston HSC-MS-03-221 and HSC-IMM-05-0253.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci U S A. 2002;99(20):12753–8. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cervino AC, Li G, Edwards S, Zhu J, Laurie C, Tokiwa G, et al. Integrating QTL and high-density SNP analyses in mice to identify Insig2 as a susceptibility gene for plasma cholesterol levels. Genomics. 2005;86(5):505–17. doi: 10.1016/j.ygeno.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Boes E, Kollerits B, Heid IM, Hunt SC, Pichler M, Paulweber B, et al. INSIG2 Polymorphism Is Neither Associated With BMI Nor With Phenotypes of Lipoprotein Metabolism. Obesity (Silver Spring) 2008 doi: 10.1038/oby.2007.132. [DOI] [PubMed] [Google Scholar]
- 4.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312(5771):279–83. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 5.Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, Zhu X, et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007;3(4):e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreasen CH, Mogensen MS, Borch-Johnsen K, Sandbaek A, Lauritzen T, Sorensen TI, et al. Non-replication of genome-wide based associations between common variants in INSIG2 and PFKP and obesity in studies of 18,014 Danes. PLoS ONE. 2008;3(8):e2872. doi: 10.1371/journal.pone.0002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, et al. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315(5809):187. doi: 10.1126/science.1129402. author reply. [DOI] [PubMed] [Google Scholar]
- 8.Hall DH, Rahman T, Avery PJ, Keavney B. INSIG-2 promoter polymorphism and obesity related phenotypes: association study in 1428 members of 248 families. BMC Med Genet. 2006;7:83. doi: 10.1186/1471-2350-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loos RJ, Barroso I, O’Rahilly S, Wareham NJ. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315(5809):187. doi: 10.1126/science.1130012. author reply. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosskopf D, Bornhorst A, Rimmbach C, Schwahn C, Kayser A, Kruger A, et al. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science (New York, NY) 2007;315(5809):187. doi: 10.1126/science.1130571. author reply. [DOI] [PubMed] [Google Scholar]
- 11.Bressler J, Fornage M, Hanis CL, Kao WH, Lewis CE, McPherson R, et al. The INSIG2 rs7566605 genetic variant does not play a major role in obesity in a sample of 24,722 individuals from four cohorts. BMC Med Genet. 2009;10(1):56. doi: 10.1186/1471-2350-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR, Jr, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41(11):1105–16. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 13.Warnick GR. Enzymatic methods for quantification of lipoprotein lipids. Methods Enzymol. 1986;129:101–23. doi: 10.1016/0076-6879(86)29064-3. [DOI] [PubMed] [Google Scholar]
- 14.A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. American journal of human genetics. 2004;74(1):106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 18.Fornage M, Doris PA. Single-Nucleotide Polymorphism Genotyping for Association Studies. In: Fennell JP, Baker AH, editors. Hypertension Methods and Protocols. Totowa, NJ: Humana Press; 2004. pp. 159–72. [Google Scholar]
- 19.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7(10):781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 20.Horvath S, Wei E, Xu X, Palmer LJ, Baur M. Family-based association test method: age of onset traits and covariates. Genet Epidemiol. 2001;21 (Suppl 1):S403–8. [PubMed] [Google Scholar]
- 21.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39(10):1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Bradfield JP, Li M, Wang K, Zhang H, Kim CE, et al. The Role of Obesity-associated Loci Identified in Genome-wide Association Studies in the Determination of Pediatric BMI. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krapivner S, Popov S, Chernogubova E, Hellenius ML, Fisher RM, Hamsten A, et al. Insulin-induced gene 2 involvement in human adipocyte metabolism and body weight regulation. J Clin Endocrinol Metab. 2008;93(5):1995–2001. doi: 10.1210/jc.2007-1850. [DOI] [PubMed] [Google Scholar]
- 24.Kolehmainen M, Vidal H, Alhava E, Uusitupa MI. Sterol regulatory element binding protein 1c (SREBP-1c) expression in human obesity. Obes Res. 2001;9(11):706–12. doi: 10.1038/oby.2001.95. [DOI] [PubMed] [Google Scholar]
- 25.Talbert ME, Langefeld CD, Ziegler JT, Haffner SM, Norris JM, Bowden DW. INSIG2 SNPs Associated With Obesity and Glucose Homeostasis Traits in Hispanics: The IRAS Family Study. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sever N, Song BL, Yabe D, Goldstein JL, Brown MS, DeBose-Boyd RA. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. The Journal of biological chemistry. 2003;278(52):52479–90. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- 27.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Molecular cell. 2003;11(1):25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 28.Hotta K, Nakamura M, Nakata Y, Matsuo T, Kamohara S, Kotani K, et al. INSIG2 gene rs7566605 polymorphism is associated with severe obesity in Japanese. J Hum Genet. 2008;53(9):857–62. doi: 10.1007/s10038-008-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franks PW, Jablonski KA, Delahanty LM, McAteer JB, Kahn SE, Knowler WC, et al. Assessing gene-treatment interactions at the FTO and INSIG2 loci on obesity-related traits in the Diabetes Prevention Program. Diabetologia. 2008;51(12):2214–23. doi: 10.1007/s00125-008-1158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]


