Abstract
All cells count on precise mechanisms that regulate protein homeostasis to maintain a stable and functional proteome. A progressive deterioration in the ability of cells to preserve the stability of their proteome occurs with age and contributes to the functional loss characteristic of old organisms. Molecular chaperones and the proteolytic systems are responsible for this cellular quality control by assuring continuous renewal of intracellular proteins. When protein damage occurs, such as during cellular stress, the coordinated action of these cellular surveillance systems allows detection and repair of the damaged structures or, in many instances, leads to the complete elimination of the altered proteins from inside cells. Dysfunction of the quality control mechanisms and intracellular accumulation of abnormal proteins in the form of protein inclusions and aggregates occur in almost all tissues of an aged organism. Preservation or enhancement of the activity of these surveillance systems until late in life improves their resistance to stress and is sufficient to slow down aging. In this work, we review recent advances on our understanding of the contribution of chaperones and proteolytic systems to the maintenance of cellular homeostasis, the cellular response to stress and ultimately to longevity.
Keywords: Autophagy, chaperones, proteases, proteasome, proteolysis, ubiquitin
1. Introduction: Importance of protein quality control
The cellular proteome – the entire pool of proteins of all types located inside cells and in their plasma membrane – is subjected to a very tight regulation that assures that each given protein is properly synthesized, folded and subcompartmentalized (Balch et al., 2008; Hutt et al., 2009). Several cellular systems are actively involved in protein quality control and their failure has severe negative consequences for cellular homeostasis and cellular functioning. In fact, alterations in different components of the protein quality control systems have been shown to underlie the basis of devastating human diseases, generically known as protein conformational disorders, and which include pathologies such as neurodegenerative diseases, metabolic disorders, myopathies, liver diseases and systemic disorders type amyloidosis (Esser et al., 2004; Morimoto, 2008). The toxic effect of altered proteins in cells has even gained its own name and it is currently known as “proteotoxicity”, and the complex cellular systems that contribute to preserve protein homeostasis are referred to as the “proteostasis network” (Balch et al., 2008).
Surveillance systems inside cells detect the altered proteins and coordinate their folding, repair or elimination from the cells, often through their breakdown by different cellular proteases. The consequences of poor quality control depend on the type of protein alteration, the location of the altered protein and on the defective step in quality control. As reviewed in more detail in the following sections, different modifications may transform proteins into cytotoxic products. Protein unfolding, abnormal cleavage or undesirable posttranslational modifications can all promote protein self-assembling into toxic oligomeric structures or aggregation into cytosolic inclusions, often bringing along other proteins (Kopito, 2000; Kourie and Henry, 2001; Ravikumar et al., 2002). The cellular response to protein aggregation and the consequences of protein deposition are different depending on the location of these structures (cytosol, inside organelles or in the nucleus). Terms as endoplasmic reticulum stress or mitochondrial stress, have become common nowadays to describe functional failures in these compartments as consequence of the accumulation of unfolded proteins in their lumen. Dedicated quality control mechanisms in each cellular compartment assist in the handling of these misbehaving proteins. Enhanced formation of toxic protein products, defective functioning of the cellular surveillance mechanisms that normally detect altered proteins or impairment in the proteolytic systems responsible for their elimination from inside cells, lead to intracellular accumulation of altered proteins. This feature is in fact a common characteristic of almost all terminally differentiated tissues in old organisms. However, for a long time, the abnormally high levels of damaged protein products in these tissues were solely attributed to increase in “damaging” agents with age. Leader among the damaging products are the reactive oxygen species, proposed as central components to the aging process by the free radical theory of aging (Harman, 1956). The harmful effect of the accumulation of the oxidized proteins target of the oxidizing agents and their contribution to increased frailty and morbidity has been recognized since the early days of the formulation of this and similar theories. However, historically the main emphasis has been placed on the prevention of protein damage (i.e. interest in antioxidants as an effort to prevent oxidative damage) rather than in the mechanisms that normally handle these damaged products. In recent years, the better understanding of the cellular mechanisms that contribute to protein quality control, the fact that some of the genes coding for the components of these systems have been shown necessary for lifespan extension, and the growing number of evidence supporting that failure of the proteostasis network represents an early event in aging, have all motivated a growing interest in the systems responsible for protein homeostasis in the field of biogerontology. In addition, lifespan extension is almost invariably associated with resistance to environmental stress (thermal stress, osmotic stress, oxidative stress) (Lithgow, 2000; Munoz, 2003; Olsen et al., 2006) and the mechanisms responsible for protein homeostasis are critical for the cellular adaptation to stress. In this review, we describe the main systems involved in protein quality control and comment on recent findings supporting the contribution of their failure with age to the functional loss and health deterioration characteristic of the aging process.
2. Protein homeostasis and the major gate-keepers
Intracellular proteins are highly dynamic molecules that undergo frequent conformational changes and partial or complete unfolding/refolding to translocate across membranes, assemble into functional structures or regulate their own function and that of other interacting proteins (Fig. 1). All these structural changes often expose hydrophobic regions of the proteins normally hidden in their core. Chaperones are essential during these processes to cover these regions and prevent undesired non-specific binding of other proteins with them. In addition, a subset of cellular chaperones, known as chaperonins, play a more active role by creating a small chamber or microenvironment that facilitates protein folding or refolding away from the pro-aggregating intracellular milieu (Sakahira et al., 2002; Willis et al., 2009). A complex chaperone network is dedicated to assure that proteins acquire a stable folded conformational state, and in the event that they fail to reach this state, the same chaperones often act as a surveillance mechanism that target the faulty proteins for degradation.
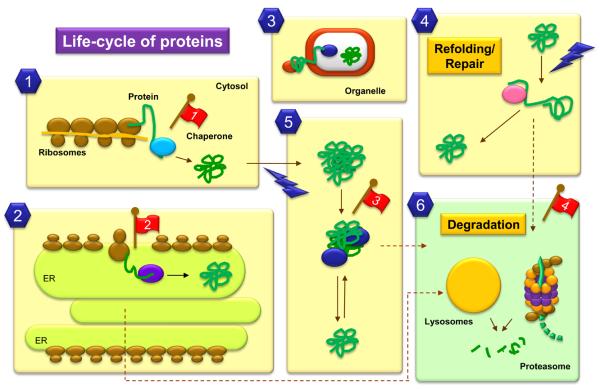
Figure 1. Cellular events that involve protein folding, refolding and degradation.
Most cytosolic proteins fold spontaneously after synthesis, but those that fail to acquire a proper folded conformation are aided by chaperones and chaperonins which provide a favorable folding environment (1). Chaperones also assist folding of proteins synthesized in the endoplasmic reticulum (ER) (2). Further failure to fold will destine both cytosolic and ER luminal proteins for degradation (6). Unfolding of previously folded protein is required for their trafficking across membranes (3) and for their assembly into protein complexes. Chaperones also assist these proteins in their refolding after such events. Lastly, proteins can be targets for damaging agents (4 and 5) which lead to their unfolding and/or their aggregation. Chaperones will assist in refolding (4) or disaggregation (5). However, when the damage is irreversible molecular chaperones mediate the degradation of the altered proteins by the different proteolytic systems (6). Red flags indicate some steps altered with aging: 1. Reduced content of cytosolic chaperones; 2. Impaired ER response to stress; 3. Increased content of aggregated proteins; 4. Deficient targeting of damaged proteins toward degradation.
This control of quality is exerted over all cellular proteins and along the full life of a protein inside the cell (Broadley and Hartl, 2009; Liberek et al., 2008; Sloan et al., 2009). Abnormally synthesized proteins are directly sent for degradation, whereas de novo synthesized proteins that fail to spontaneously fold are recognized by chaperones and folding chaperonins that attempt to drive them into a stably folded conformation (Fig. 1) (Liberek et al., 2008). Only if these folding attempts are futile, they are delivered to the proteolytic machinery. A similar “two-step” control (folding/degradation) regulates the fate of previously folded proteins that unfold, often as result of undesired posttranslational modifications or as consequence of the effect of damaging agents. Both chaperones and the intracellular proteolytic systems are thus the “gate keepers” or main effectors in protein quality control.
3. Molecular Chaperones in protein quality control
Molecular chaperones or heat shock proteins (HSP) are stress factors rapidly induced in response to elevated temperatures and other stress stimuli. In addition, cells count on a subset of chaperones constitutively expressed that participate in the continuous control of quality for proteins located either in the cytosol or in intracellular compartments (Fig. 1) (Bukau et al., 2006; True, 2006). Chaperones are highly conserved molecules from bacteria to mammals and can be classified according to their molecular weight in five major classes: HSP100, HSP90, HSP70, HSP60 and the small heat shock proteins (sHSP) with molecular weights between 12 and 43 kDa (Kappe et al., 2003; Liberek et al., 2008). Members of each family can be located in different cellular subcompartments and contribute to the protection/control of a different subset of proteins. Some HSPs are promiscuous and can act over a large number of proteins in the cells (i.e. hsp70 or hsp60) (Bukau et al., 2006; True, 2006). This group of chaperones recognizes very common motifs in proteins, such as hydrophobic patches or frequent posttranslational modifications. There are also chaperones very specific and solely dedicated to the surveillance of a very limited subset of proteins in which they recognize a defined binding region (i.e. those acting over collagen or modulating the dynamics of actin or intermediate filaments) (Mounier and Arrigo, 2002).
3.1. Cytosolic chaperones
This group of chaperones modulates folding and unfolding events that take place in the cytosol and that involve mainly proteins synthesized in polysomes or proteins from other compartments that are translocated into the cytosol to avoid luminal clogging (i.e. ER proteins) (Fig. 1) (Bukau et al., 2006; Frydman, 2001; True, 2006). Most of these chaperones belong to the hsp70, hsp60 and hsp90 family of chaperones that often act cooperatively in their surveillance function. Thus for example, if a de novo synthesized protein fails to fold spontaneously, hsc70/hsp40 may attempt to fold it and if they fail to do it they may send the unfolded protein to the hsp60 chaperonin folding chamber or to the hsp90/HOP stabilizing chaperone complex (Spiess et al., 2004). However the succession of events is not always necessarily in this order.
sHSP and members of the hsp70 family are perhaps the most prominent subset of cytosolic proteins for which tight connections with the cellular response to stress have been already established. Both groups of proteins are highly conserved (up to 50% amino acid identity among species) and they all bear a chaperone-like function. Induction of these chaperones is closely related to tolerance to high temperature and their overexpression confers cells resistance to heat shock (Nollen et al., 1999) and makes whole organisms, such as flies, stress tolerant (Welte et al., 1993).
The function of cytosolic chaperones in quality control is closely linked to the two major proteolytic systems in this compartment, the ubiquitin/proteasome system and the lysosomes (Fig. 1). The mechanisms that determine delivery to one system or another are currently object of intensive investigation, although growing evidence supports that the same chaperone can mediate re-routing of proteins to both systems and that, probably, particular posttranslational modifications in the substrate protein determine its ultimate fate.
Cytosolic chaperones are key for the maintenance of cellular homeostasis and, as described in more detail in later sections, failure of this surveillance system has been linked to the pathogenesis of severe human disorders and to the loss of proteostasis characteristic of old organisms.
3.2. Organelle-specific chaperones
Proper folding of proteins located in the membrane and lumen of the different organelles inside cells is essential for normal functioning of these organelles. A subset of cellular chaperones localizes in these compartments and is fully dedicated to maintenance of their homeostasis (Fig. 1) (Hetz and Glimcher, 2009; Scheper and Hoozemans, 2009; Todd et al., 2008). As in the case of the cytosol, folding may be required for de novo synthesized proteins (i.e. secretory proteins that gain access to the ER lumen as they are being synthesized in the ER-associated ribosomes). Folding may be also needed for mature already folded proteins that reach the organelle lumen after crossing its membrane (i.e. luminal mitochondria proteins synthesized in the cytosol that reach the lumen through the translocation complexes at the mitochondria membrane).
The best characterized organelle chaperones for their role in quality control are those responsible for maintenance of protein homeostasis in the ER. The direct involvement of the ER in protein synthesis demands a high content of chaperones in its lumen, which sense unfolded protein products and facilitate their folding. As in the case of cytosol, some of the ER chaperones are rather promiscuous and act on a large subset of proteins in which they recognize hydrophobic patches (i.e. BiP) or oligosaccharide chains (i.e. the calnexin/calreticulin complex) (Hetz and Glimcher, 2009; Scheper and Hoozemans, 2009; Todd et al., 2008). There are also chaperones specialized in the folding of specific substrates (i.e. hsc47 is key for collagen folding). The importance of the ER chaperones is such that a very complex network of proteins and factors is in place in this organelle to upregulate ER chaperone synthesis when the amount of unfolded products reaches levels that could compromise ER homeostasis. This response, known as the unfolding protein response (UPR), increases the content of ER chaperones and reduces protein translation to alleviate ER clogging (Ron and Walter, 2007).
Similar to the cytosolic proteins, when organelle proteins cannot be folded they are sent for degradation. In most cases this degradation takes place in the cytosol after retrotranslocation of the unfolded products from the organelle lumen. A tight connection has been described between the UPR and proteolysis through the proteasome (known as ERAD or ER-associated degradation) (Meusser et al., 2005; Vembar and Brodsky, 2008). Proteins that fail to fold in the ER are actively retrotranslocated into the cytosol and, after tagging with ubiquitin, are degraded by the 26S proteasome. Recent studies support that lysosomes may also play an important role in the clearance of misfolded ER proteins although in this case instead of retrotranslocation, these proteins are degraded along with other ER integral components upon engulfment of whole ER regions by lysosomes (Yorimitsu et al., 2006).
The characterization of the mitochondria unfolding response is still at its very beginning but it already points towards having many similarities with the UPR and ERAD (Broadley and Hartl, 2008).
3.3. Molecular chaperones in longevity and aging
There is a long list of pathologies, many of them classified as age-related diseases, in which primary or secondary deficits in chaperone function have been reported. In other pathologies, changes in chaperone content represent the cellular attempt to accommodate to the pathogenic condition and preserve cellular homeostasis, at least during the first stages of the disease. For example hsp27, a heat inducible sHSP ubiquitously expressed in human brain (Renkawek et al., 1993) has been detected at high levels in degenerating areas of Alzheimer's disease brains (Wilhelmus et al., 2006). A vast literature also supports that stress-induced synthesis of heat shock proteins is impaired in aging. In all these cases, the extent of changes likely depends on the chaperone, the tissue and even the organism.
Decreased transcriptional upregulation of hsp70 in response to different stressors has been reported in senescent fibroblasts in culture and tissues from old organisms in different species up to monkeys (Fargnoli et al., 1990; Hall et al., 2000; Pahlavani et al., 1995) (Fig. 1). Caloric restriction, the most successful intervention to slow down aging, restores normal chaperone response in many cell types (Ehrenfried et al., 1996; Heydari et al., 1993; Moore et al., 1998 ). Furthermore, cells from human centenarians display preserved chaperone upregulation during stress (Ambra et al., 2004; Marini et al., 2004).
Numerous studies support now that increased chaperone induction leads to increased longevity both in uni- and multicellular organisms (Lithgow et al., 1995; Shama et al., 1998; Tatar et al., 1997). Flies and worms carrying extra copies of an hsp-70 family member or sHSPs have been shown to be long-lived (Morrow et al., 2004a; Morrow et al., 2004b; Walker and Lithgow, 2003; Wang et al., 2004). Likewise, over-expression of the gene encoding the hsp gene transcriptional activator, heat shock factor (HSF), also increases lifespan in nematodes (Hsu et al., 2003; Morley and Morimoto, 2004). In addition, many long-lived mutants (such as those with lower insulin signaling or with mutations that mimic caloric restriction) have up-regulated hsp-16 genes (Wadhwa et al., 2005) and higher levels of the protein product have also been detected in long-lived mutant worms during and after heat stress.
The reason behind the failure to upregulate chaperone transcription with age, at least in the case of hsp70, has been narrowed to the inability of HSF to bind the heat shock element on the chaperone gene promoter (Ambra et al., 2004; Heydari et al., 2000; Locke and Tanguay, 1996; Singh et al., 2006). In fact, certain polymorphisms in the promoter region of Hsp70 gene affect the probability to attain longevity (Altomare et al., 2003). Different factors also connected to longevity have been shown to modulate HSF, such as the histone deacetylase 6 (HDAC6) that is required for activation of HSF1 during proteasome inhibition (Haggarty et al., 2003), or Sirt1, another tubulin deacetylase, recently shown to activate HSF1 in mammals (Westerheide et al., 2009). Interestingly, HSF activation and binding to the promoter is preserved in certain tissues in old organisms (Locke, 2000), but whether these differences could contribute to organ-specific rates of aging, particularly noticeable in mammals (Swindell et al., 2009), requires further investigation. The contribution to the altered chaperone response with age of changes in other mechanisms that regulate cytosolic levels of chaperones, such as their own degradation, is currently under study (Marques et al., 2006).
The carboxyl-terminus of hsp70-interacting protein (CHIP) has been a chaperone recently in the spotlight because CHIP-defective mice have reduced longevity and display an accelerated aging phenotype (Marques et al., 2006). It is not surprising that these animals also present higher levels of damaged proteins and declined proteasome activity, since this chaperone is known to deliver cytosolic proteins for degradation through this proteolytic complex (Min et al., 2008).
Interest is also growing in recent years on the contribution of organelle-specific chaperones to longevity. Levels and activity of BiP, calnexin and PDI, key ER chaperones, decrease with age in many tissues of old rodents (Hussain and Ramaiah, 2007; Naidoo et al., 2008; Nuss et al., 2008; Paz Gavilan et al., 2006; Rabek et al., 2003). In addition, despite the higher levels of essential components of the UPR in older organisms, several arms of this stress response are not properly activated in aging (Naidoo et al., 2008; Paz Gavilan et al., 2006) (Fig. 1). Unbalanced expression with age of pro-apoptotic versus pro-survival markers of the UPR has also been proposed responsible for the lower resistance to stress of old organisms (Gavilan et al., 2009). Regarding the mitochondrial chaperones, an increase in lifespan in human fibroblasts in culture and in worms has been obtained through the overexpression of mortalin, a mitochondrial hsp70 (Kaul et al., 2003; Kaula et al., 2000; Yokoyama et al., 2002). Expression of another mitochondrial chaperone, the sHSP hsp22, also increases flies life- and health-span (Morrow et al., 2004a). A better characterization of the key players in the organelles' responses to stress and the contributions of alterations in these systems to altered proteostasis with age require further investigation.
An exciting spin has recently been added by Morimoto's group to the contribution of chaperone failure to proteotoxicity in aging. They have demonstrated the existence of a pool of metastable proteins in all cells, highly dependent on chaperones for their normal folding and function, and how these proteins can become unstable and aggregate in the presence of additional pathogenic proteins, such as those altered in protein conformational disorders (Gidalevitz et al., 2006). This and subsequent studies have revealed that compromised folding of these proteins occurs early in adulthood and leads to the collapse of proteostasis characteristic of old organisms (Ben-Zvi et al., 2009).
All these examples support the hypothesis that a better capacity to adapt to various stresses, in which chaperones play a leading role, makes a major contribution to lifespan extension.
4. Proteolytic systems
4.1. The ubiquitin/proteasome system
The ubiquitin/proteasome (UPS) is one of the main proteolytic systems that participate in protein quality control. The two main components of this pathway, the ubiquitinization machinery – responsible for substrate targeting – and the proteasome core – where degradation takes place, undergo age-dependent changes that contribute to the lower efficiency of this system in old organisms and the consequent alterations in proteostasis.
4.1.1. Coordinated action of a catalytic complex and a tagging system
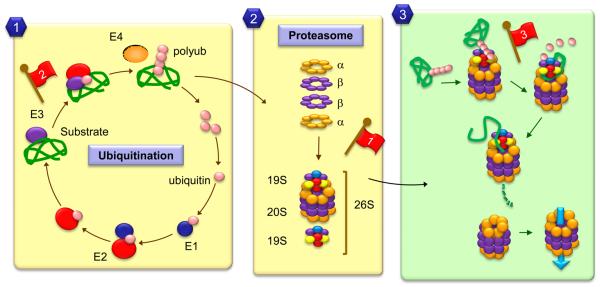
The proteasome is a multicatalytic complex with a proteolytic core, known as the 20S proteasome, which in high eukaryotes results from the association of 28 subunits in 4 rings stacked as a cylinder-like structure (Goldberg, 2003; Pickart and Cohen, 2004) (Fig. 2). Three major types of proteolytic activities have been described in 20S proteasomes, but because some of the subunits of this core are exchangeable, proteasomes with different catalytic activities coexist in all cells. The activity of the catalytic core is modulated through the assembly of different regulatory subunits (19S or 11S) that dock on one or both sides of the 20S proteasome to form several proteasome species (Murata et al., 2009; Navon and Ciechanover, 2009). The 26S proteasome, formed by the association of 20S proteasome to the 19S regulatory complex, has a prominent role in quality control. The components of the regulatory subunits are primarily chaperones, ATPases and enzymes that remove degradation tags from the substrate proteins. Degradation of most substrates requires the regulatory complexes, which mediate substrate recognition, unfolding and often act as the driving force that opens the proteasome barrel and “pushes” substrates into the catalytic core (Fig. 2).
Figure 2. Schematic model of the ubiquitin/proteasome system.
The main events that mediate targeting and degradation of soluble proteins by the proteasome are depicted here. 1. Most proteins are targeted for degradation through the covalent attachment of 4-5 ubiquitins through a lysine residue in their sequence. Ubiquitinization requires the coordinated action of catalytic enzymes (E1, E2 and E3) that act sequentially to activate the ubiquitin and ligate it to the substrate presented by the E3. 2. The proteolytic component, the proteasome or 26S, has a catalytic core (the 20S) formed by 4 rings containing two types of catalytic subunits (α and β), and a regulatory complex (the 19S). 3. Polyubiquitin chains are recognized by components of the regulatory subunit, where deubiquitinases reverse the covalent conjugation releasing free ubiquitin for recycling. The substrate is unfolded by unfoldases in the regulatory lid and ATPases in this complex provide the energy required for the injection of the substrate protein into the catalytic barrel or 20S proteasome. Red flags indicate described age-related changes in the different steps of this process: 1. Lower levels of catalytic and/or regulatory subunits and inefficient assembling of the 26S proteasome; 2. Reduced content/activity of free ubiquitin and conjugating enzymes; 3. Posttranslational modifications and crosslinking of the substrates can interfere with proteasome activity.
Most proteasome substrates are targeted for degradation through the covalent linkage of ubiquitin, a small (8 kDa) heat-stable protein that also undergoes self-conjugation, resulting in the formation of polyubiquitin chains. Linkage of ubiquitin to the cargo proteins is mediated by a group of enzymes – generically known as E ligases – that act sequentially to activate ubiquitin, presenting it to the substrate and catalyzing the conjugation (Fig. 2) (Finley, 2009; Hochstrasser, 2009). Repeated cycles of ubiquitinization generate the polyubiquitin chain that is then recognized by the chaperones and the ubiquitin binding subunits of the regulatory complex of the proteasome.
The UPS plays a critical role in cellular homeostasis and protein quality control, and also contributes to the regulatory degradation of essential intracellular proteins involved in cell cycle progression, cell division, transcription and cell signaling (Goldberg, 2007; Murata et al., 2009; Navon and Ciechanover, 2009).
4.1.2. How does the UPS change with age?
Many reports have shown different degrees of decrease in the activity of the proteasome with age in several tissues, although, in contrast to the lysosomal system, the decline in activity with age does not seem to be universal (Carrard et al., 2003; Ferrington et al., 2005; Keller et al., 2000 b; Shibatani et al., 1996). The decreased activity may obey to different reasons depending on the tissue. For example, down-regulated expression of proteasomal subunits (Keller et al., 2000 b), unbalanced levels of α and β catalytic subunits (Chondrogianni et al., 2003), defective expression of regulatory subunits (Ferrington et al., 2005) and damaging posttranslational modifications in critical proteasome subunits (Bulteau et al., 2000; Carrard et al., 2003), have all been proposed to contribute to the functional decline of this protease (Fig. 2). In addition, factors extrinsic to the proteasome could also interfere with their function in aging cells. For example, the reduced ATP content in aging flies may be behind the reduced assembly of the 26S proteasome (Vernace et al., 2007), and massive oxidation and crosslinking of proteasome substrates has been shown to have a direct inhibitory effect on this protease (Bulteau et al., 2001b; Friguet and Szweda, 1997; Okada et al., 1999; Sitte et al., 2000).
Age-related changes in specific components of the ubiquitin system have also been explored. A decrease in levels of free ubiquitin (Jahngen et al., 1990) and transcriptional downregulation of two ubiquitin-conjugating enzymes (Ruotolo et al., 2003) and an E3 ligase (Hawse et al., 2004) have been reported in some aged tissues. In other tissues, constitutive levels of these enzymes are higher in the old animals that instead fail to upregulate their activity in response to stressors (Scrofano et al., 1998a) (Fig. 2). The increased content with age of mutant forms of ubiquitin due to molecular misreading could also be behind the inefficient clearance of polyubiquitinated proteins in aging (Tsirigotis et al., 2001).
Connections between longevity and the UPS have been established in different model systems. Cultured fibroblasts from healthy centenarians have a more active proteasome (Chondrogianni et al., 2000), and part of the anti-aging effect attained in human skin fibroblasts by hormesis through repeated exposure to mild stress could also be attributed to an increase in proteasome activities in these cells (Rattan and Ali, 2007). In C. elegans, expression of RLE-1, an E3 ubiquitin ligase, regulates aging by determining degradation of DAF-16 by the UPS (Li et al., 2007). Also in worms, defects in the regulatory components of the proteasome have an impact on lifespan. Thus, worms lacking AIP-1, a homologue of the regulatory mammalian subunit AIRAP, exhibit shortened lifespan and hypersensitivity to misfolding proteins (Yun et al., 2008). Proper proteasome function is required in the extension of lifespan observed in some of the long-lived worm mutants. Thus, mutations in the genes of the CUL-1 E3 ligase complex shorten the extended lifespan of different insulin signaling C. elegans mutants (Ghazi et al., 2007) and, as described in the previous section, studies in CHIP defective mice also support an important role for the UPS in longevity and health-span (Min et al., 2008).
In summary, functional decline of the UPS occurs with age and maintained UPS activity is required to attain maximal lifespan extension in long-lived mutants, underlying an important role for this proteolytic system in lifespan.
4.2. The autophagic/lysosomal system
Lysosomes are intracellular organelles fully devoted to the degradation of intra- and extracellular components. Because the main emphasis of this review is on protein quality control, we will primarily focus on the removal of intracellular components through lysosomal degradation or autophagy, and will emphasize their role on protein turnover. Readers are encouraged to consult recent reviews on degradation of extracellular components and organelles by lysosomes (Farre et al., 2009; Fortini and Bilder, 2009; Kirkin et al., 2009; Kraft et al., 2009; Nichols, 2009; Pandey, 2009; Sorkin and von Zastrow, 2009; Tolkovsky, 2009).
4.2.1. Major autophagic pathways and their physiological relevance
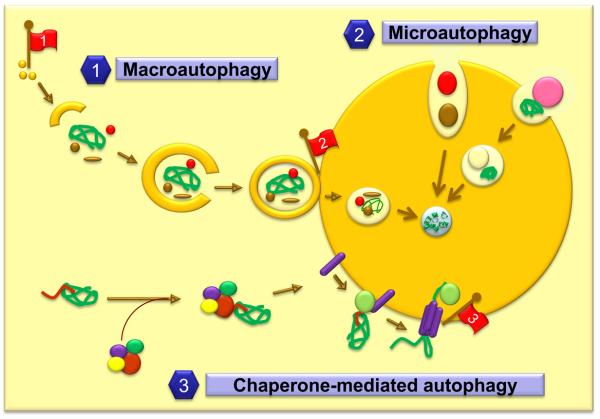
Three different forms of autophagy have been described in mammalian cells: macroautophagy, microautophagy and chaperone-mediated autophagy (Fig. 3) (Cuervo, 2009; He and Klionsky, 2009; Mizushima et al., 2008).
Figure 3. Intracellular autophagic pathways and changes with age.
Three different mechanisms contribute to the delivery of cytosolic cargo to lysosomes: 1. macroautophagy, 2. microautophagy and 3. chaperone-mediated autophagy (CMA). Red flags indicate changes in the autophagic system with age: 1. Defects in the induction of macroautophagy; 2. Inefficient lysosomal clearance of the double membrane vesicles that sequester the cargo; 3. Decrease in the levels of the lysosomal receptor that mediates docking of CMA substrates at the lysosomal membrane and their translocation into the lumen.
In Microautophagy, the lysosomal membrane invaginates or tubulates to engulf whole regions of the cytosol (Fig.3). Microautophagy is well characterized in yeast (Dubouloz et al., 2005), but it is poorly understood in mammals (Marzella et al., 1981; Mortimore et al., 1988) and consequently there is no available information on its changes with age.
In Macroautophagy, a whole region of cytosol (including both organelles and soluble proteins) is sequestered inside double membrane vesicles (autophagosomes), which then fuse with lysosomes to degrade their cargo (Fig. 3) (He and Klionsky, 2009; Mizushima et al., 2008). When macroautophagy is activated, the combined action of two different conjugation cascades - a protein to protein and a protein to lipid conjugation - and a kinase nucleation complex induces formation of a limiting membrane or phagophore, which elongates and fuses to form the autophagosome (Ohsumi and Mizushima, 2004). All these autophagy-related genes and proteins are known as ATG and Atg, respectively (Klionsky et al., 2003). The main components of the kinase complex are vps34, a class III PI3Kinase, and beclin-1 (Itakura et al., 2008; Liang et al., 1999). Both the conjugation cascades and the class III PI3K complex are essential for autophagy. Formation of autophagosomes can be blocked by knocking down Atg5, 7 or 12 (the Atgs involved in conjugation) or with type III PI3Kinase inhibitors (i.e. 3-methyladenine). Induction of autophagy is negatively regulated by a second kinase complex, mTOR (Kamada et al., 2000; Noda and Ohsumi, 1998). mTOR inhibitors, such as rapamycin, are widely used as macroautophagy activators. Notably, mTOR is also a modulator of lifespan in a number of invertebrate species (see below). Basal and inducible macroautophagy are essential for cellular homeostasis in many different tissues. Although basal macroautophagic activity occurs in different tissues (Hara et al., 2006; Komatsu et al., 2006; Nakai et al., 2007), macroautophagy is often a stress-induced pathway essential for the maintenance of cellular homeostasis and energetic balance (Kuma et al., 2004; Mizushima et al., 2004; Singh et al., 2009), for the defense against exogenous and endogenous aggressors (Deretic, 2009), and in circumstances requiring extensive cellular remodeling (Tsukamoto et al., 2008). Macroautophagy malfunctioning has been involved in the pathogenesis of detrimental human pathologies, such as cancer, neurodegenerative and metabolic diseases, muscular diseases, etc. (He and Klionsky, 2009; Mizushima et al., 2008).
In Chaperone-mediated autophagy (CMA), substrate proteins are directly translocated across the lysosomal membrane (Cuervo, 2009; Dice, 2007). All CMA substrates contain a pentapeptide motif (Dice, 1990), which is selectively recognized by the cytosolic heat shock cognate chaperone of 70KDa (hsc70) (Chiang et al., 1989) (Fig. 3). The substrate/chaperone complex is targeted to the lysosomal surface where it interacts with a receptor protein, the lysosome-associated membrane protein type 2A (LAMP-2A) (Cuervo and Dice, 1996). Once unfolded, the substrate translocates across the membrane, assisted by a lumenal chaperone (lys-hsc70) (Agarraberes et al., 1997) and then is degraded rapidly. Although basal CMA can be detected in almost all cell types, CMA is maximally activated in response to stress (starvation, oxidative stress and conditions that cause protein damage) (Cuervo et al., 1999; Cuervo et al., 1995; Kiffin et al., 2004). Under these conditions, the selectivity of CMA allows the removal of altered proteins without affecting neighboring functional ones. CMA has been described only in mammalian cells, and the LAMP-2A variant is not conserved in other species (yeast, flies, zebra fish), supporting that CMA was acquired late in evolution. Alterations in CMA underlie the pathogenesis of different neurodegenerative disorders such as Parkinson's disease, and certain forms of tauopathies (Cuervo et al., 2004; Martinez-Vicente et al., 2008; Wang et al., 2009). A decrease in CMA activity, induced by the pathogenic protein that accumulates in the affected neurons, renders these cells susceptible to numerous stressors and often precipitates cell death. In addition, CMA contributes to the removal of some pathogenic forms of huntingtin, the protein that accumulates in Huntington's disease (Thompson et al., 2009). CMA pathology is not restricted to neurons, as decreased CMA activity is behind the kidney hypertrophy that presents as a common complication of diabetic patients (Sooparb et al., 2004) and has also been described in certain lysosomal storage disorders such as mucolipidosis type IV (Venugopal et al., 2009).
4.2.2. Changes with age in the autophagic system
Decreased macroautophagic activity with age has been described in different mammalian tissues (Cuervo, 2008; Cuervo et al., 2005). In fact, malfunctioning of macroautophagy in livers of old rodents was described even before a complete molecular characterization of this pathway was available. In this early work, it was shown that although cargo sequestration by the double membrane vesicles still takes place in the old tissues, the elimination of these vesicles by fusion with the lysosomal compartment was severely impaired (Fig. 3) (Terman, 1995). In addition, as in the case of the proteasome, cross-linked products that reach the lysosome through their sporadic fusion with the double membrane vesicles, exert a potent inhibitory effect on lysosomal proteolysis by directly inducing undesired posttranslational modifications in the enzymatic machinery and also by changing the properties of the intralysosomal milieu, normally optimal for functioning of these enzymes (Brunk and Terman, 2002; Terman and Brunk 1998). Incomplete degradation of cargo inside lysosomes leads to the accumulation of undegraded products that through further modification and cross-linking give rise to the autofluorescent pigment known as lipofuscin, an often used marker of aging. Studies on the regulation of macroautophagy, also in liver of old rodents, identified alterations in the hormonal regulation of this process (Bergamini and Kovacs, 1990). Thus, it was known that insulin and glucagon exert opposite inhibitory and stimulatory effects, respectively, on macroautophagy. In old organisms the stimulatory effect of glucagon is compromised mostly due to increased basal signaling activity from the insulin receptor with age (Fig. 3) (Bergamini et al., 1994; Marino et al., 1998). This insulin-independent signaling seems upregulated in response to the oxidative stress characteristic of old organisms. Caloric restriction and treatment with lipogenic agents are able to prevent the dysregulation of macroautophagy in old rodents, although the mechanisms by which they exert their action is still subject of investigation (Cavallini et al., 2001; Donati et al., 2004).
Now that the different components that participate in the macroautophagic process have been characterized, decrease of some macroautophagy markers have been described in different tissues of rodents and in the aging human brain (Shibata et al., 2006). However, as in the case of the proteasome, the macroautophagic defect seems to be also tissue-dependent (Wohlgemuth et al., 2007).
In the case of CMA, we have found that the activity of this autophagic pathway decreases in almost all tissues of old rodents and in senescent human fibroblasts in culture (Cuervo and Dice, 2000; Dice, 1982). A step-by-step comparative analysis of CMA in livers of young and old rodents revealed a decrease with age in binding and lysosomal translocation of the substrate proteins, due to progressively lower levels of the CMA receptor at the lysosomal membrane with age (Cuervo and Dice, 2000). Changes with age in the lipid composition of the lysosomal membrane are likely behind the instability of LAMP-2A in old organisms (Kiffin et al., 2007).
4.2.3. Modulating autophagy in the pursuit of longevity
The first genetic connection between autophagy and aging was established in worms. The lifespan extension observed in worms mutant for the equivalent of the insulin-signaling pathway, such as daf-2, is drastically reduced when macroautophagy genes are knocked down (Hars et al., 2007; Melendez et al., 2003). Functional macroautophagy has also proven necessary to attain the maximal lifespan extension mediated by mutations in other genes such as the nutrient-sensor TOR (Hansen et al., 2008), the tumor suppressor protein p53 (Tavernarakis et al., 2008) and even the lifespan prolonging effect of Sirt-1 (Morselli et al., 2010). Inversely, all these factors have also shown to exert a regulatory role on macroautophagy both in invertebrates and in mammals (Crighton et al., 2006; Lee et al., 2008). Other factors that promote longevity in invertebrates, such as the Foxo family of Forkhead transcription factors, also upregulate macroautophagy (Salih and Brunet, 2008). Activation of macroautophagy is a common feature of all the long-lived mutant worms, including the models for dietary restriction (Hansen et al., 2008; Hars et al., 2007; Melendez et al., 2003; Tóth et al., 2008). In fact, the role of macroautophagy in lifespan extension mediated by caloric restriction has also been genetically confirmed in C. elegans (Jia and Levine, 2007). An aspect that has been origin of controversy is whether or not mutation of essential autophagic genes decreases lifespan. Although some of the studies in worms have failed to see this shortening (Jia and Levine, 2007; Melendez et al., 2003), studies by other groups in worms and flies seem to support that functional macroautophagy is necessary to preserve normal lifespan (Juhasz et al., 2007; Morck and Pilon, 2006; Simonsen et al., 2007).
Highlighting the importance in proteostasis maintenance, recent work in flies has shown that expression of specific autophagy genes prolongs lifespan and provides resistance to particular stressors (Simonsen et al., 2007). Interventions using chemical modulators of macroautophagy have gained considerable interest during this last year as a result of the publication of marked increase in lifespan and enhance health-span in mice treated with rapamycin, a well-characterized inhibitor of mTOR (Harrison et al., 2009). However, it is premature to attribute the beneficial effect observed with the rapamycin treatment to the stimulatory effect of this drug on macroautophagy, because inhibition of this major cellular kinase should also affect other cellular functions such as mRNA translation, ribosome biogenesis, angiogenesis, mitochondrial metabolism, adipogenesis and cellular growth (Dennis et al., 2001). Future studies should aim at dissecting the specific contribution of each of these pathways to the increase in life- and health-span in these mice. In addition, identification of other chemical modulators more specific for autophagy should also help to elucidate the role of autophagy in mammalian longevity. On this respect, recent studies with spermidine, a natural polyamine, have shown promising autophagy-dependent effects on yeast, flies and worms longevity and on mouse health-span (Eisenberg et al., 2009).
Regarding CMA, the lack of information on this pathway in species commonly used for genetic manipulations such as flies and worms, has not permitted to generate genetic evidence linking this autophagic pathway with longevity. However, to determine the contribution of declined CMA to aging, our laboratory has recently prevented the age-dependent decrease of CMA in liver through genetic manipulations in mice (Zhang and Cuervo, 2008). Activation of an exogenously added copy of the CMA receptor in transgenic mice when its endogenous levels start to decrease prevents the decline in CMA activity in the old animals. Livers of mice with preserved CMA function show better cellular homeostasis, improved ability to respond to the cell damage induced by different stressors and their liver function is comparable to that of young mice (Zhang and Cuervo, 2008). This is the first evidence in mammals that preventing the decline in autophagic activity slows down cellular aging and preserves organ function. Interestingly, the activity of other quality control mechanisms, such as macroautophagy and the UPS, is also improved in these animals (Zhang and Cuervo, 2008).
5. Concluding remarks and the many open paths ahead
Protein quality control is essential for proper cellular function and in the orchestration of an efficient cellular response to stress. Growing evidence supports that functional decline of the different components of the proteostasis network is one of the essential factors that contribute to cellular and organism aging. The better understanding of the molecular basis and regulation of the quality control systems inside cells has been the fuel behind the numerous biochemical and genetic connections recently established between protein homeostasis and longevity. The future of this field is challenging but exciting. Although individual studies in preferred experimental systems have offered a good overall view of the proteostasis network and its malfunctioning in aging, there is still a broad lack of knowledge on the specific tissue-dependent characteristics in the functioning of this network and on how networks from different organ and tissues cross-talk in a whole organism. The components of the quality control mechanisms are highly conserved through evolution, however, species-specific differences should be expected. For example, CMA is absent, or at least does not have the same components in unicellular organisms and invertebrates. Why this proteolytic mechanism appears late in evolution and the repercussions that this late appearance has on the functioning of the other mechanisms of quality control remains unknown. In the same context, it is obviously naïve considering that the different chaperones and proteolytic systems act in a completely autonomic manner. Cross-talk between chaperone families and among the different autophagic and non-autophagic pathways has been already demonstrated in different model systems. In fact, this coordinated function could explain why a small improvement in one of the components of the quality control in old organisms has such dramatic beneficial effects. How aging affects this cross-talk and whether functional asynchronism could be behind defective quality control in old organisms remain to be elucidated. Particular pressure should be placed in the development of genetic models in which the impairment of the quality control mechanisms can be exerted late in life, as these models will better mimic the aging conditions and should allow analyzing the effect of the dysfunction once compensatory mechanisms can no longer take place. Design of better chemical modulators of the different components of the proteostasis network is also essential for the development of future anti-aging interventions based on the manipulations of the quality control mechanisms. Despite these limitations, correction of the defects in protein homeostasis in aging organism represents one of the more promising avenues in the fight against the aging process.
Acknowledgements
Work in our laboratory is supported by NIH grants from NIA (AG021904, AG031782), NIDKK (DK041918), NINDS (NS038370), a Glenn Foundation Award and a Hirsch/Weill-Caulier Career Scientist Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarraberes F, Terlecky S, Dice J. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altomare K, Greco V, Bellizzi D, Berardelli M, Dato S, DeRango F, Garasto S, Rose G, Feraco E, Mari V, Passarino G, Franceschi C, De Benedictis G. The allele (A)(−110) in the promoter region of the HSP70-1 gene is unfavorable to longevity in women. Biogerontology. 2003;4:215–220. doi: 10.1023/a:1025182615693. [DOI] [PubMed] [Google Scholar]
- Ambra R, Mocchegiani E, Giacconi R, Canali R, Rinna A, Malavolta M, Virgili F. Characterization of the hsp70 response in lymphoblasts from aged and centenarian subjects and differential effects of in vitro zinc supplementation. Exp Gerontol. 2004;39:1475–1484. doi: 10.1016/j.exger.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini E, Del Roso A, Gori Z, Masiello P, Masini M, Pollera M. Endocrine and amino acid regulation of liver macroautophagy and proteolytic function. Am J Physiol. 1994;266:G118–122. doi: 10.1152/ajpgi.1994.266.1.G118. [DOI] [PubMed] [Google Scholar]
- Bergamini E, Kovacs J. Exploring the age-related changes in hormone-regulated protein breakdown by the use of a physiologic model of stimulation of liver autophagy. In: Segal H, Rothstein M, Bergamini E, editors. Protein metabolism in Aging. Modern Aging Research; Wiley-Liss, New York: 1990. pp. 361–370. [Google Scholar]
- Broadley SA, Hartl FU. Mitochondrial stress signaling: a pathway unfolds. Trends Cell Biol. 2008;18:1–4. doi: 10.1016/j.tcb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583:2647–2653. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Brunk U, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Rad Biol Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001b;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Petropoulos I, Friguet B. Age-related alterations of proteasome structure and function in aging epidermis. Exp Gerontol. 2000;35:767–777. doi: 10.1016/s0531-5565(00)00136-4. [DOI] [PubMed] [Google Scholar]
- Carrard G, Dieu M, Raes M, Toussaint O, Friguet B. Impact of ageing on proteasome structure and function in human lymphocytes. Int J Biochem Cell Biol. 2003;35:728–739. doi: 10.1016/s1357-2725(02)00356-4. [DOI] [PubMed] [Google Scholar]
- Cavallini G, Donati A, Gori Z, Pollera M, Bergamini E. The protection of rat liver autophagic proteolysis from the age-related decline co-varies with the duration of anti-ageing food restriction. Exp Gerontol. 2001;36:497–506. doi: 10.1016/s0531-5565(00)00224-2. [DOI] [PubMed] [Google Scholar]
- Chiang H, Terlecky S, Plant C, Dice J. A role for a 70 kDa heat shock protein in lysosomal degradation of intracellular protein. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Petropoulos I, Franceschi C, Friguet B, Gonos ES. Fibroblast cultures from healthy centenarians have an active proteasome. Exp Gerontol. 2000;35:721–728. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Stratford FL, Trougakos IP, Friguet B, Rivett AJ, Gonos ES. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J Biol Chem. 2003;278:28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Hildebrand H, Bomhard E, Dice J. Direct lysosomal uptake of alpha2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–545. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Knecht E, Terlecky S, Dice J. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and Aging: keeping that old broom working. Trends Genetic. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2009 doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. J. [DOI] [PubMed] [Google Scholar]
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- Deretic V. Links between autophagy, innate immunity, inflammation and Crohn's disease. Dig Dis. 2009;27:246–251. doi: 10.1159/000228557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice J. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- Dice J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Dice J. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- Donati A, Cavallini G, Carresi C, Gori Z, Parentini I, Bergamini E. Anti-aging effects of anti-lipolytic drugs. Exp Gerontol. 2004;39:061–067. doi: 10.1016/j.exger.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Ehrenfried JA, Evers BM, Chu KU, Townsend CM, Jr., Thompson JC. Caloric restriction increases the expression of heat shock protein in the gut. Ann Surg. 1996;223:592–597. doi: 10.1097/00000658-199605000-00015. discussion 597-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Frohlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Esser C, Alberti S, Hohfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim Biophys Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Fargnoli J, Kunisada T, Fornace AJ, Jr., Schneider EL, Holbrook NJ. Decreased expression of heat shock protein 70 mRNA and protein after heat treatment in cells of aged rats. Proc Natl Acad Sci U S A. 1990;87:846–850. doi: 10.1073/pnas.87.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009;19:323–328. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friguet B, Szweda LI. Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett. 1997;405:21–25. doi: 10.1016/s0014-5793(97)00148-8. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Gavilan MP, Pintado C, Gavilan E, Jimenez S, Rios RM, Vitorica J, Castano A, Ruano D. Dysfunction of the unfolded protein response increases neurodegeneration in aged rat hippocampus following proteasome inhibition. Aging Cell. 2009;8:654–665. doi: 10.1111/j.1474-9726.2009.00519.x. [DOI] [PubMed] [Google Scholar]
- Ghazi A, Henis-Korenblit S, Kenyon C. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proc Natl Acad Sci U S A. 2007;104:5947–5952. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;18:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DM, Xu L, Drake VJ, Oberley LW, Oberley TD, Moseley PL, Kregel KC. Aging reduces adaptive capacity and stress protein expression in the liver after heat stress. J Appl Physiol. 2000;89:749–759. doi: 10.1152/jappl.2000.89.2.749. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic L, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–310. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hars E, Qi H, Ryazanov A, Jin S, Cai L, Hu C, Liu L. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- Hawse JR, Hejtmancik JF, Horwitz J, Kantorow M. Identification and functional clustering of global gene expression differences between age-related cataract and clear human lenses and aged human lenses. Exp Eye Res. 2004;79:935–940. doi: 10.1016/j.exer.2004.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydari AR, Wu B, Takahashi R, Strong R, Richardson A. Expression of heat shock protein 70 is altered by age and diet at the level of transcription. Mol Cell Biol. 1993;13:2909–2918. doi: 10.1128/mcb.13.5.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydari AR, You S, Takahashi R, Gutsmann-Conrad A, Sarge KD, Richardson A. Age-related alterations in the activation of heat shock transcription factor 1 in rat hepatocytes. Exp Cell Res. 2000;256:83–93. doi: 10.1006/excr.2000.4808. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hussain SG, Ramaiah KV. Reduced eIF2alpha phosphorylation and increased proapoptotic proteins in aging. Biochem Biophys Res Commun. 2007;355:365–370. doi: 10.1016/j.bbrc.2007.01.156. [DOI] [PubMed] [Google Scholar]
- Hutt DM, Powers ET, Balch WE. The proteostasis boundary in misfolding diseases of membrane traffic. FEBS Lett. 2009;583:2639–2646. doi: 10.1016/j.febslet.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 Forms Two Distinct Phosphatidylinositol 3-Kinase Complexes with Mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahngen JH, Lipman RD, Eisenhauer DA, Jahngen EG, Jr., Taylor A. Aging and cellular maturation cause changes in ubiquitin-eye lens protein conjugates. Arch Biochem Biophys. 1990;276:32–37. doi: 10.1016/0003-9861(90)90006-k. [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1-10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul SC, Yaguchi T, Taira K, Reddel RR, Wadhwa R. Overexpressed mortalin (mot-2)/mthsp70/GRP75 and hTERT cooperate to extend the in vitro lifespan of human fibroblasts. Exp Cell Res. 2003;286:96–101. doi: 10.1016/s0014-4827(03)00101-0. [DOI] [PubMed] [Google Scholar]
- Kaula SC, Reddelb RR, Sugiharac T, Mitsuia Y, Wadhwac R. Inactivation of p53 and life span extension of human diploid fibroblasts by mot-2. FEBS Lett. 2000;474:159–164. doi: 10.1016/s0014-5793(00)01594-5. [DOI] [PubMed] [Google Scholar]
- Keller JN, Huang FF, Markesbery WR. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience. 2000 b;98:149–156. doi: 10.1016/s0306-4522(00)00067-1. [DOI] [PubMed] [Google Scholar]
- Kiffin R, Christian C, Knecht E, Cuervo A. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kopito R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:523–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Kourie JI, Henry CL. Protein aggregation and deposition: implications for ion channel formation and membrane damage. Croatian Medical Journal. 2001;42:359–374. [PubMed] [Google Scholar]
- Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. Biochim Biophys Acta. 2009;1793:1404–1412. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev Cell. 2007;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow G. Stress response and aging in Caenorhabditis elegans. Results Probl Cell Differ. 2000;29:131–148. doi: 10.1007/978-3-540-48003-7_7. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended lifespan conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M. Heat shock transcription factor activation and hsp72 accumulation in aged skeletal muscle. Cell Stress Chaperones. 2000;5:45–51. doi: 10.1043/1355-8145(2000)005<0045:HSTFAA>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M, Tanguay RM. Diminished heat shock response in the aged myocardium. Cell Stress Chaperones. 1996;1:251–260. doi: 10.1379/1466-1268(1996)001<0251:dhsrit>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini M, Lapalombella R, Canaider S, Farina A, Monti D, De Vescovi V, Morellini M, Bellizzi D, Dato S, De Benedictis G, Passarino G, Moresi R, Tesei S, Franceschi C. Heat shock response by EBV-immortalized B-lymphocytes from centenarians and control subjects: a model to study the relevance of stress response in longevity. Exp Gerontol. 2004;39:83–90. doi: 10.1016/j.exger.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Marino M, Dolfi C, Paradiso C, Cavallini G, Masini M, Gori Z, Pollera M, Trentalance A, Bergamini E. Age-dependent accumulation of dolichol in rat liver: is tissue dolichol a biomarker of aging? Journals of Gerontology Series A-Biological Sciences & Medical Sciences. 1998;53:B87–93. doi: 10.1093/gerona/53a.2.b87. [DOI] [PubMed] [Google Scholar]
- Marques C, Guo W, Pereira P, Taylor A, Patterson C, Evans PC, Shang F. The triage of damaged proteins: degradation by the ubiquitin-proteasome pathway or repair by molecular chaperones. FASEB J. 2006;20:741–743. doi: 10.1096/fj.05-5080fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Kaushik S, Massey A, Mazzulli J, Mosharov E, Hodara R, Fredenburg R, Wu D, Follenzi A, Dauer W, Przedborski S, Ischiropoulos H, Lansbury P, Sulzer D, Cuervo A. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Archiv. B. Cell Pathology. 1981;36:219–234. doi: 10.1007/BF02912068. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Min JN, Whaley RA, Sharpless NE, Lockyer P, Portbury AL, Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28:4018–4025. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SA, Lopez A, Richardson A, Pahlavani MA. Effect of age and dietary restriction on expression of heat shock protein 70 in rat alveolar macrophages. Mech Ageing Dev. 1998;104:59–73. doi: 10.1016/s0047-6374(98)00052-9. [DOI] [PubMed] [Google Scholar]
- Morck C, Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Battistini S, Zhang P, Tanguay RM. Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J Biol Chem. 2004a;279:43382–43385. doi: 10.1074/jbc.C400357200. [DOI] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004b;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, Michaud M, Madeo F, Tavernarakis N, Kroemer G. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6 doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Lardeux BR, Adams CE. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. Journal of Biological Chemistry. 1988;263:2506–2512. [PubMed] [Google Scholar]
- Mounier N, Arrigo AP. Actin cytoskeleton and small heat shock proteins: how do they interact? Cell Stress Chaperones. 2002;7:167–176. doi: 10.1379/1466-1268(2002)007<0167:acashs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ. Longevity and heat stress regulation in Caenorhabditis elegans. Mech Ageing Dev. 2003;124:43–48. doi: 10.1016/s0047-6374(02)00168-9. [DOI] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Master M, Zhu Y, Pack AI. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J Neurosci. 2008;28:6539–6548. doi: 10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cariomyocytes in the basal state and in response to hemodynamic stress. Nature Medicine E pub. 2007 doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Navon A, Ciechanover A. The 26 S proteasome: from basic mechanisms to drug targeting. J Biol Chem. 2009;284:33713–33718. doi: 10.1074/jbc.R109.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. Endocytosis of lipid-anchored proteins: excluding GEECs from the crowd. J Cell Biol. 2009;186:457–459. doi: 10.1083/jcb.200907119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. Journal of Biological Chemistry. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Nollen EA, Brunsting JF, Roelofsen H, Weber LA, Kampinga HH. In vivo chaperone activity of heat shock protein 70 and thermotolerance. Mol Cell Biol. 1999;19:2069–2079. doi: 10.1128/mcb.19.3.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss JE, Choksi KB, DeFord JH, Papaconstantinou J. Decreased enzyme activities of chaperones PDI and BiP in aged mouse livers. Biochem Biophys Res Commun. 2008;365:355–361. doi: 10.1016/j.bbrc.2007.10.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K. 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J Biol Chem. 1999;274:23787–23793. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlavani MA, Harris MD, Moore SA, Weindruch R, Richardson A. The expression of heat shock protein 70 decreases with age in lymphocytes from rats and rhesus monkeys. Exp Cell Res. 1995;218:310–318. doi: 10.1006/excr.1995.1160. [DOI] [PubMed] [Google Scholar]
- Pandey KN. Ligand-mediated endocytosis and intracellular sequestration of guanylyl cyclase/natriuretic peptide receptors: role of GDAY motif. Mol Cell Biochem. 2009 doi: 10.1007/s11010-009-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz Gavilan M, Vela J, Castano A, Ramos B, del Rio JC, Vitorica J, Ruano D. Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol Aging. 2006;27:973–982. doi: 10.1016/j.neurobiolaging.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Pickart C, Cohen R. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Rabek JP, Boylston WH, 3rd, Papaconstantinou J. Carbonylation of ER chaperone proteins in aged mouse liver. Biochem Biophys Res Commun. 2003;305:566–572. doi: 10.1016/s0006-291x(03)00826-x. [DOI] [PubMed] [Google Scholar]
- Rattan SI, Ali RE. Hormetic prevention of molecular damage during cellular aging of human skin fibroblasts and keratinocytes. Ann N Y Acad Sci. 2007;1100:424–430. doi: 10.1196/annals.1395.047. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Renkawek K, Bosman GJ, Gaestel M. Increased expression of heat-shock protein 27 kDa in Alzheimer disease: a preliminary study. Neuroreport. 1993;5:14–16. doi: 10.1097/00001756-199310000-00003. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ruotolo R, Grassi F, Percudani R, Rivetti C, Martorana D, Maraini G, Ottonello S. Gene expression profiling in human age-related nuclear cataract. Mol Vis. 2003;9:538–548. [PubMed] [Google Scholar]
- Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU. Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16412–16418. doi: 10.1073/pnas.182426899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper W, Hoozemans JJ. Endoplasmic reticulum protein quality control in neurodegenerative disease: the good, the bad and the therapy. Curr Med Chem. 2009;16:615–626. doi: 10.2174/092986709787458506. [DOI] [PubMed] [Google Scholar]
- Scrofano MM, Shang F, Nowell TR, Jr., Gong X, Smith DE, Kelliher M, Dunning J, Mura CV, Taylor A. Calorie restriction, stress and the ubiquitin-dependent pathway in mouse livers. Mech Ageing Dev. 1998a;105:273–290. doi: 10.1016/s0047-6374(98)00097-9. [DOI] [PubMed] [Google Scholar]
- Shama S, Lai CY, Antoniazzi JM, Jiang JC, Jazwinski SM. Heat stress-induced life span extension in yeast. Exp Cell Res. 1998;245:379–388. doi: 10.1006/excr.1998.4279. [DOI] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Shibatani T, Nazir M, Ward WF. Alteration of rat liver 20S proteasome activities by age and food restriction. J Gerontol A Biol Sci Med Sci. 1996;51:B316–322. doi: 10.1093/gerona/51a.5.b316. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Finley KD. Linking lysosomal trafficking defects with changes in aging and stress response in Drosophila. Autophagy. 2007;3:499–501. doi: 10.4161/auto.4604. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kolvraa S, Bross P, Jensen UB, Gregersen N, Tan Q, Knudsen C, Rattan SI. Reduced heat shock response in human mononuclear cells during aging and its association with polymorphisms in HSP70 genes. Cell Stress Chaperones. 2006;11:208–215. doi: 10.1379/CSC-184R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitte N, Huber M, Grune T, Ladhoff A, Doecke WD, Von Zglinicki T, Davies KJ. Proteasome inhibition by lipofuscin/ceroid during postmitotic aging of fibroblasts. FASEB J. 2000;14:1490–1498. doi: 10.1096/fj.14.11.1490. [DOI] [PubMed] [Google Scholar]
- Sloan LA, Fillmore MC, Churcher I. Small-molecule modulation of cellular chaperones to treat protein misfolding disorders. Curr Opin Drug Discov Devel. 2009;12:666–681. [PubMed] [Google Scholar]
- Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Masternak MM, Kopchick JJ, Conover CA, Bartke A, Miller RA. Endocrine regulation of heat shock protein mRNA levels in long-lived dwarf mice. Mech Ageing Dev. 2009;130:393–400. doi: 10.1016/j.mad.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Khazaeli AA, Curtsinger JW. Chaperoning extended life. Nature. 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- Terman A. The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology. 1995;41:319–325. doi: 10.1159/000213753. [DOI] [PubMed] [Google Scholar]
- Terman A, Brunk U. Lipofuscin- Mechanisms of formation and increase with age. APMIS. 1998;106:265–276. doi: 10.1111/j.1699-0463.1998.tb01346.x. [DOI] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, JG OS-R, Khashwji H, Lukacsovich T, Zhu YZ, Lau AL, Massey A, Hayden MR, Zeitlin SO, Finkbeiner S, Green KN, Laferla FM, Bates G, Huang L, Patterson PH, Lo DC, Cuervo AM, Marsh JL, Steffan JS. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Tóth M, Sigmond T, Borsos E, Barna J, Erdélyi P, Takács-Vellai K, Orosz L, Kovács A, Csikós G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- True HL. The battle of the fold: chaperones take on prions. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Tsirigotis M, Zhang M, Chiu RK, Wouters BG, Gray DA. Sensitivity of mammalian cells expressing mutant ubiquitin to protein-damaging agents. J Biol Chem. 2001;276:46073–46078. doi: 10.1074/jbc.M109023200. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008;4:1076–1078. doi: 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal B, Mesires N, Kennedy J, Curcio-Morelli C, Laplante JM, Dice JF, Slaugenhaupt SA. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–353. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 2007;21:2672–2682. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Kaur K, Aida S, Yaguchi T, Kaul Z, Hirano T, Taira K, Kaul SC. Identification and characterization of molecular interactions between mortalin/mtHsp70 and HSP60. Biochem J. 2005;391:185–190. doi: 10.1042/BJ20050861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Walker GA, White TM, McColl G, Jenkins NL, Babich S, Candido EPM, Jonhson TE, Lithgow GJ. Heat shcok protein accumulation is upregulated in a long-lived mutant of Caenorhabditis elegans. J Gerontol. 2001;56:B281–B287. doi: 10.1093/gerona/56.7.b281. [DOI] [PubMed] [Google Scholar]
- Wang C, Li Q, Redden DT, Weindruch R, Allison DB. Statistical methods for testing effects on “maximum lifespan”. Mech Ageing Dev. 2004;125:629–632. doi: 10.1016/j.mad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA, Tetrault JM, Dellavalle RP, Lindquist SL. A new method for manipulating transgenes: engineering heat tolerance in a complex, multicellular organism. Curr Biol. 1993;3:842–853. doi: 10.1016/0960-9822(93)90218-d. [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM, Jr., Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmus MM, Boelens WC, Otte-Holler I, Kamps B, de Waal RM, Verbeek MM. Small heat shock proteins inhibit amyloid-beta protein aggregation and cerebrovascular amyloid-beta protein toxicity. Brain Res. 2006;1089:67–78. doi: 10.1016/j.brainres.2006.03.058. [DOI] [PubMed] [Google Scholar]
- Willis MS, Schisler JC, Portbury AL, Patterson C. Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc Res. 2009;81:439–448. doi: 10.1093/cvr/cvn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn WA., Jr. Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 2007;10:281–292. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Fukumoto K, Murakami T, Harada S, Hosono R, Wadhwa R, Mitsui Y, Ohkuma S. Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. 2002;516:53–57. doi: 10.1016/s0014-5793(02)02470-5. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Stanhill A, Yang Y, Zhang Y, Haynes CM, Xu CF, Neubert TA, Mor A, Philips MR, Ron D. Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:7094–7099. doi: 10.1073/pnas.0707025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]