Abstract
A b-type heme is conserved in membrane-bound complex II enzymes (SQR, succinate-ubiquinone reductase). The axial ligands for the low spin heme b in Escherichia coli complex II are SdhC His84 and SdhD His71. E. coli SdhD His71 is separated by 10 residues from SdhD Asp82 and Tyr83 which are essential for ubiquinone catalysis. The same His-10x-AspTyr motif dominates in homologous SdhD proteins, except for Saccharomyces cerevisiae where a tyrosine is at the axial position (Tyr-Cys-9x-AspTyr). Nevertheless, the yeast enzyme was suggested to contain a stoichiometric amount of heme, however, with the Cys ligand in the aforementioned motif acting as heme ligand. In this report, the role of Cys residues for heme coordination in the complex II family of enzymes is addressed. Cys was substituted to the SdhD-71 position and the yeast Tyr71Cys72 motif also recreated. The Cys71 variant retained heme, although it was high spin, while the Tyr71Cys72 mutant lacked heme. Previously the presence of heme in S. cerevisiae was detected by a spectral peak in fumarate-oxidized, dithionite-reduced mitochondria. Here it is shown that this method must be used with caution. Comparison of bovine and yeast mitochondrial membranes shows that fumarate induced reoxidation of cytochromes in both SQR and the bc1 complex (ubiquinol-cytochrome c reductase). Thus, this report raises a concern about the presence of low spin heme b in S. cerevisiae complex II.
Keywords: Succinate dehydrogenase, complex II, cytochrome b, heme, Saccharomyces cerevisiae
1. Introduction
Succinate-quinone reductase (SQR, complex II), a membrane-bound enzyme, is part of the mitochondrial and bacterial electron transport chain. During aerobic metabolism, SQR oxidizes succinate to reduce quinone to quinol. SQRs normally consist of four subunits that comprise a soluble succinate dehydrogenase domain and a trans-membrane quinone reductase domain. The soluble domain is comprised of the SdhA subunit with a covalently attached FAD cofactor and the dicarboxylate binding site, and the SdhB subunit that coordinates three distinct iron-sulfur centers [1]. The hydrophobic domain consists of the transmembrane spanning SdhC and SdhD subunits that also coordinate the low spin hexa-coordinated heme b [1–3]. The quinone-binding and heme binding sites are composed of amino acid residues from the SdhB, SdhC, and SdhD subunits. X-ray structures of E. coli, chicken, and pig SQR, as well as, for several homologous fumarate reductase complexes have become available in recent years [4–12]. A substantial sequence and structural homology of the catalytic dehydrogenase fragment throughout the complex II family of proteins has allowed insight into the catalytic mechanism of succinate/fumarate interconversion [7]. By contrast, before the structural data on Complex II became available, the lack of sequence homology in the SdhC and SdhD subunits caused concern as to the overall catalytic mechanism of quinone reduction among complex II enzymes. Therefore, it was gratifying that comparison of the x-ray structures of E. coli and mitochondrial SQRs revealed that the position of key catalytic residues involved in heme b coordination and quinone reduction are absolutely conserved [8–13]. As a result, E. coli SQR emerged as a most convenient model for studying the molecular mechanism of complex II enzymes due to high levels of protein expression and simple genetic manipulation.
In the SdhD subunit, three residues that are critical for SQR assembly and activity are located on helix II of the protein (Fig. 1A). Using E. coli SdhD numbering, one finds that SdhD His71 is positioned in the middle of the transmembrane segment at a kink of a helix and provides one of the axial ligands for the b heme. SdhD His71 is separated by 10 residues from SdhD Asp82 Tyr83 which are part of the same helix and essential for ubiquinone catalysis [9, 10, 13]. This same motif His-10x-AspTyr dominates in SdhD homologous proteins (Fig. 2). In spite of recent progress, the role of the heme b in complex II enzymes is still not completely elucidated.
Figure 1.
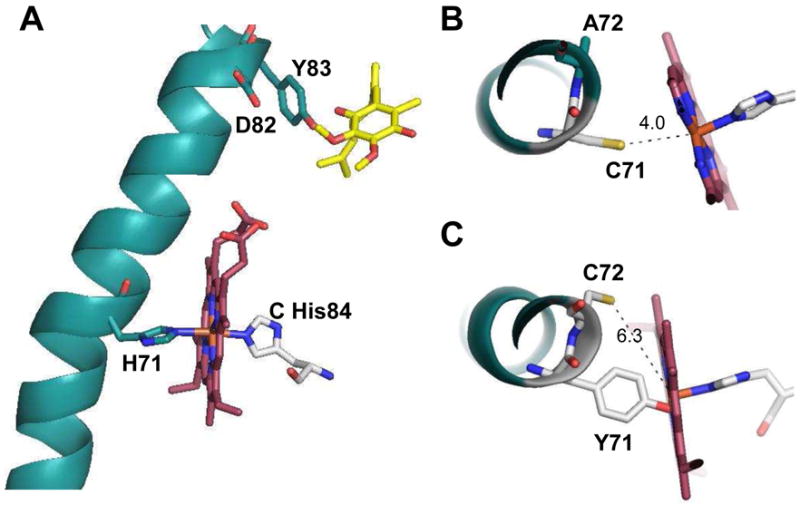
Crystal structure of helix II of the E. coli SdhD subunit with heme b and ubiquinone (Protein data bank code 1NEK). A. Side chains of the residues that form a conservative motif His-10x-AspTyr and their interaction with heme b (magenta) and ubiquinone (yellow) are shown. Computer simulation: B. Top view at the heme b with His 71 substituted to a cysteine residue. C. Simulation of the double mutant: His71 substituted with Tyr and Ala72 with Cys. The figure was prepared using PyMOL (DeLano Scientific LLC, Palo Alto, CA). The mutated side chain rotamers were chosen manually based on the best alignment with original amino acid positions.
Figure 2.
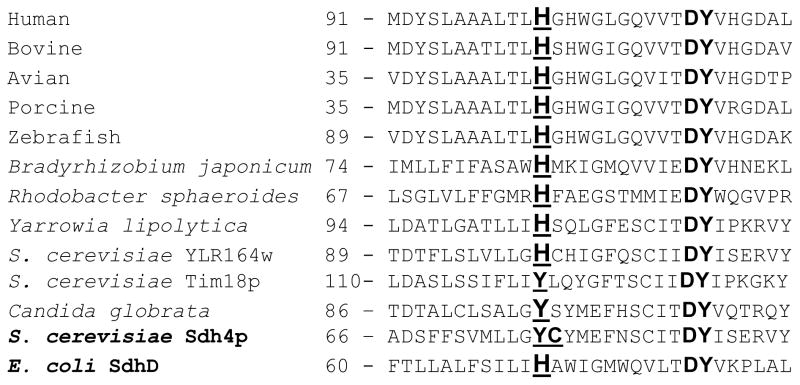
Comparison of the amino acid sequence of the SdhD homologous subunits from various species. Conservative positions for the His-10x-AspTyr are highlighted in bold and the His axial ligand or comparable position in yeast is underlined, as is the Cys residue in yeast Sdh4p that was proposed to ligand the heme [19].
It was recently demonstrated that E. coli SQR could be assembled as an active membrane-bound complex without the heme b when one of the axial histidine ligands for the heme was substituted with a tyrosine [14]. It is of interest why the heme b is evolutionarily conserved in complex II enzymes even though a Tyr could substitute to provide alternative stabilization for the transmembrane domain. Nature has provided a potential example of such possibility. The yeast Saccharomyces cerevisiae has a Tyr rather than a His (Fig. 2) in the canonical motif in the Sdh4p protein, the homologue of the E. coli SdhD subunit [15]. It should be noted that in the S. cerevisiae genomic database two other Sdh4p homologous genes (YLR164w and YOR297c) are found. The predicted YLR164w and YOR297c protein sequences are 52% and 74%, respectively, similar to Sdh4p. Interestingly, YLR164w has a His residue in the position homologous to SdhD His71 in E. coli SdhD (Fig. 2). The YOR297c gene encodes Tim18p [16], a subunit of the TIM22 import complex of the mitochondrial inner membrane and has a Tyr in the position similar to Tyr77 in Sdh4p. In addition, a hypothetical SdhD protein from the yeast Candida globrata genome is reported to have a Tyr-10x-AspTyr motif sequence (genebank: CAA86683.1).
An early report in an abstract suggested that S. cerevisiae complex II does not contain heme, consistent with the lack of a His at the canonical His-residue position in SdhD, and a very low heme b/FAD ratio of 0.2 [17]. More recent studies, however, have suggested for the yeast a stoichiometric heme b to FAD content in the mitochondrial membrane [18]. Additional mutagenesis [19, 20] and molecular modeling studies [21] identified the axial heme ligands in S. cerevisiae complex II as His106 of Sdh3p (SdhC homologue) and Cys78 of Sdh4p. A Cys ligand for the heme b is unprecedented in the complex II family of enzymes. Since in the yeast Sdh4p sequence Tyr77 is in the position usually occupied by a His in other complex II family members, it was apparently not considered that the heme b could be a high spin penta-coordinated heme and thus Cys78 was suggested as the sixth axial ligand to the heme iron [21]. Mutagenesis studies, in the yeast, where both modeled axial heme ligands were mutated by Ala showed that the heme is not critical for SQR assembly and activity [22]. This is similar to what was found for E. coli SQR, though in the E. coli SdhD H71Y mutant the hydrogen-bonding interaction of SdhC H84 and SdhD Y71 was suggested to play a role for stability of the protein [14].
The overwhelming majority of protein-derived hexa-coordinated low spin heme irons have either dual histidine or histidine/methionine axial ligands [23–25 and Refs. therein]. Cysteine is found as an axial ligand in high spin penta-coordinated hemes in cytochromes P450, NO synthase, and chloroperoxidases. Tyrosine is also found as an axial ligand in penta-coordinated heme of catalase. Attempts to substitute one of the bis-histidine axial ligands with Cys in flavocytochrome b2 resulted in a drop in the Em by ~250 mV compared to wild-type protein and loss of axial coordination, thus transforming the protein into a high spin heme [26]. Therefore, the suggested Cys and His heme b coordination in S. cerevisiae SQR [21] would be a unique case among low spin hexa-coordinated hemes. The suggestion that the yeast SQR contains a Cys-coordinated heme also challenges the importance of the structural conservation of the His-10x-AspTyr motif and the possibility to form a stable active protein without heme by proteins with a Tyr-10x-AspTyr sequence [14]. The important role of Sdh4p Tyr89 for ubiquinone catalysis in yeast SQR [27] indicates the relevance of the conservative motif for S. cerevisiae SQR.
This communication addresses the role of a Cys residue in E. coli SdhD for heme b coordination. A single substitution of SdhD His71 to Cys was constructed and also a double mutant converting SdhD His71 to Tyr and SdhD Ala72 to Cys in order to mimic the TyrCys motif found in yeast Sdh4p (Fig. 1B and C). Both of the E. coli mutant enzymes were overexpressed allowing their detailed characterization within isolated membranes. Similar to previous observations in the SdhD H71Y mutant [14], the double mutant assembles within the membrane but without heme, and it retains the ability to reduce quinone. The substitution of SdhD His71 to Cys resulted in protein that retained penta-coordinated heme b indicating that Cys is not able to provide coordination for the heme in E. coli SQR even in its optimal structural position. Comparison of the heme spectral environment in the bacterial, mammalian, and yeast SQRs also provided insight into the heme b coordination in S. cerevisiae complex II.
2. Materials and Methods
2.1 Strains and plasmids
E. coli strain DW35 (ΔfrdABCD, sdhC::kan) which was used as the host for expression of wild-type and mutant forms of SQR have been previously described [28]. Plasmid pFAS (PFRDsdhC+D+A+B+) was used for expression of SQR enzymes [29]. S. cerevisiae strain D273-10B was used for mitochondrial membranes isolation.
2.2 Mutagenesis
Mutations were accomplished using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) following manufacturer protocols and using appropriate mutagenic primers for a SdhD H71C (5′ GCGCTGTTTTCTATCTTGATCTGTGCCTGGATCGGCATGTG 3′) and the double mutant Sdh D H71Y/A72C (5′ CGCTGTTTTCTATCTTGATCTATTGCTGGATCGGCATGT GGCAGG 3′). The underlined nucleotide indicates the site of the mutation. All mutations were verified by sequencing the AarI and BstBI restriction fragment and subcloned back into pFAS. All cloning procedures were performed by methods previously described [28, 29].
2.3 Growth conditions and membrane and enzyme purification
E. coli DW35 harboring the appropriate plasmid was grown under microaerophilic conditions in Terrific Broth medium as previously described [30]. Isolation of SQR [30] and membrane fractions [31] was performed according to previously published methods. Bovine submitochondrial particles (SMP) were prepared according to standard procedure [32]. Yeast were grown on YPGal medium at 30 °C and harvested at stationary phase. Yeast spheroplasts were obtained by using Zymolyase 20T (MP Biomedicals, Solon, Ohio) as previously described [33]. In order to obtain mitochondrial membranes, spheroplasts in the presence of protein inhibitors (Complete Protease inhibitor, Roche Diagnostics) were disrupted by an Avestin cell homogenizer (Avestin Inc., Ottawa, Canada). The disrupted cells were separated by sedimentation (25 min at 38,000 × g) and mitochondrial membranes were collected by centrifugation (75 min at 130,000 × g).
2.4 Measurement of enzyme activity
Enzymatic activity was assayed as previously described [34]. For enzyme activation, SQR membranes were diluted to 5 mg protein/ml in 30 mM BTP (Bis-Tris-Propane, pH 7.0), 0.1 mM EDTA, 3 mM malonate, and incubated for 20 min at 30 °C. Activated enzyme was then stored on ice for the duration of the experiment. The standard assay medium contained 50 mM BTP (pH 8.0), 0.1 mM EDTA, 3 mM potassium cyanide, 10 mM succinate (30 °C). Succinate oxidation using phenazine ethosulfate (PES) or ubiquinone-1 (UQ1) as reoxidizing substrates, was monitored by the reduction of dichlorophenol indophenol (DCIP) ε600=21.8 mM−1cm−1.
2.4 Analytical methods
Optical spectra were recorded at 25 °C with an Agilent 8453 diode array spectrophotometer. The protoheme IX content of cytochrome b was determined by the pyridine hemochromogen assay using an extinction coefficient for the wavelength pair 556.5–540 nm = 23.98 mM−1cm−1 [35]. The heme d content was determined from the reduced minus “as isolated” difference spectrum, with the ε (628–607 nm) = 10.8 mM−1 cm−1 [36]. FAD content was determined as previously described [37]. Protein concentration was determined by the Biuret method.
3. Results and Discussion
3.1 Kinetic properties of the mutants
Two SdhD mutant variants of E. coli SQR were constructed. The first single substitution converted the axial His ligand (SdhD His71) to a Cys residue (H71C), whereas the double mutant converted His71 to Tyr and Ala72 to Cys (SdhD Y71C72) a sequence that mimics the yeast Sdh4p sequence. The SdhD H71Y single substitution had been made previously [14]. Cells harboring the mutated plasmids were grown under microaerophilic conditions on rich medium, and the cellular membranes were isolated. The purified cell membranes were a rich brown in color indicating the presence of heterotetrameric SQR enzymes. Table 1 shows the kinetic parameters of the succinate-oxidase reactions catalyzed by wild-type and the mutant SQR variants. All of the mutants demonstrated significant rates of succinate oxidation with ubiquinone or the artificial electron acceptor PES. Both the H71C and Y71C72 mutants showed higher PES or UQ1 reductase activities than that shown by the previously described H71Y mutant which is known to lack heme [14]. The H71C single substitution retains 43% of ubiquinone reductase activity compared to wild-type SQR (Table 1), however, comparing the ratio of UQ1 reductase to PES reductase activity (0.66) shows that the quinone reductase activity in this mutant is impaired to a greater extent than its succinate-oxidase activity measured with PES. The Y71C72 double mutant shows significant improvement in its activity compared to either single substitutions at His71 (Table 1). Nevertheless, there is a somewhat greater impairment of quinone reductase activity compared to PES reductase activity as noted in the UQ/PES ratio in Table 1. In the double mutant the ubiquinone reductase activity is 2-fold higher than in the H71Y mutant. All three mutants demonstrate a minor effect on UQ1 binding as evidenced by the 4- to 5-fold higher Km for Q1 compared to wild-type SQR.
Table 1.
Kinetic parameters of succinate-oxidase reaction of membranes enriched with E. coli SQR.
| membranes | Succinate:PES s−1 per FAD | Succinate:UQ1 s−1 per FAD | Km UQ1 μM | UQ1/PES ratio |
|---|---|---|---|---|
| Wt SQR | 98 | 102 | 2.5 | 1.04 |
| H71Y | 37 | 34 | 10 | 0.92 |
| H71C | 65.5 | 43.5 | 12 | 0.66 |
| 71Y72C | 99 | 72 | 13 | 0.72 |
Previous substitutions of the SdhD axial His71 ligand destabilized the mutant proteins to different extents [14, 29]. The SdhD H71Q mutant was suitable for enzyme purification, whereas the SdhD H71Y substitution was very unstable in the presence of even low concentrations of detergent [14, 30]. This sensitivity to detergents holds true for either the SdhD H71C or Y71C72 double mutant; that is, the succinate-ubiquinone reductase activity of both mutants is completely eliminated in the presence of as little as 0.001% (w/v) detergent, a concentration at least 9-fold lower than the critical micelle concentration (CMC) of the detergents used; i.e., Triton X-100, (CMC 0.017%); C12E9 (Thesit), (CMC0.009%); or dodecylmaltoside (DDM, CMC 0.027%) all had the same effect. This loss of quinone-reductase activity is rapidly followed by a decrease of succinate-PES reductase activity which is indicative of a loss of protein integrity of the overall SdhCDAB complex. Therefore, it was not possible to further purify the mutants by the use of detergents (data not shown).
3.2 Spectral properties and heme b content in purified membranes
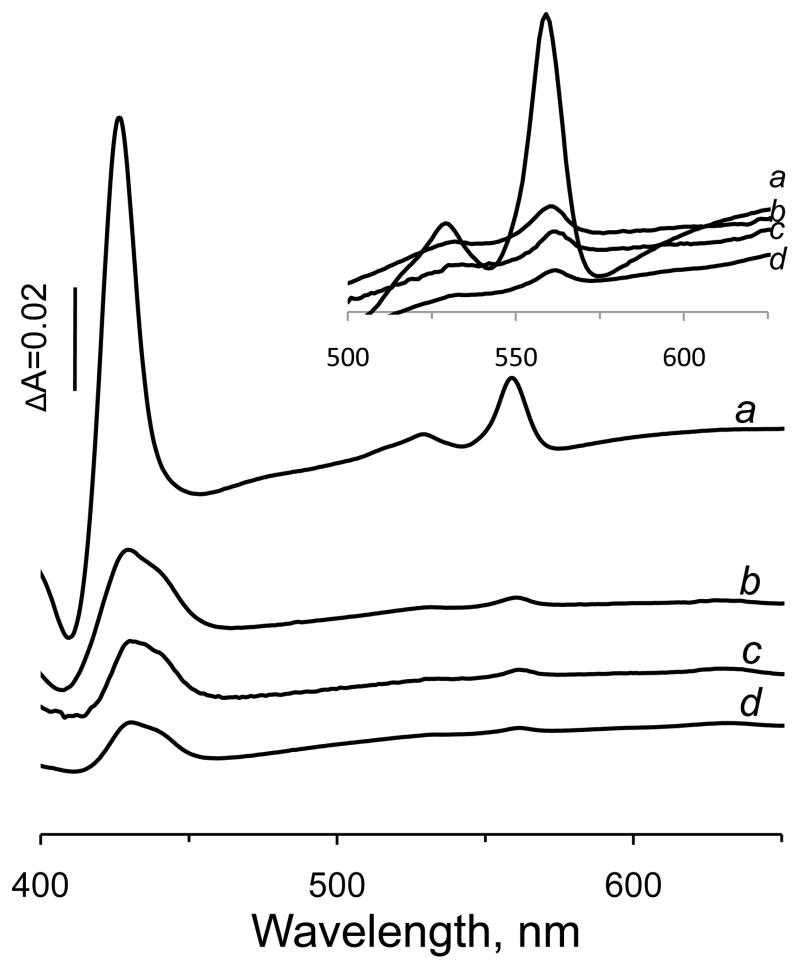
Figure 3 demonstrates dithionite reduced minus oxidized optical spectra of isolated E. coli membranes and Table 2 indicates the content of FAD and heme b in the membranes. The presence of a low spin heme b in the wild-type SQR membrane is evident by the significant absorbance at the Soret and α-regions (Fig. 3). The stoichiometric molar ratio of heme b to FAD shown in Table 2 confirms that wild-type SQR contains one heme b per one covalently bound FAD. Membranes isolated from E. coli cells lacking SQR/QFR proteins show small absorbances attributed to hemes from non-SQR membrane-bound proteins in their difference spectrum [30]. Cytochrome bd is a quinol oxidase from E. coli which is optimally expressed under microaerophilic growth conditions. It is a heterodimeric membrane-bound complex that contains one low spin heme b558, and two high spin hemes, b559 and d [36]. The difference spectrum of DW35 membranes lacking SQR and QFR proteins exhibits a typical absorbance characteristic of cytochrome b at 560 nm and cytochrome d at 628–649 nm [30]. The total heme b content in DW35 membranes is estimated at 0.5 nmol per mg protein, and part of it (0.16 nmol per mg protein) is attributed to the bd quinol oxidase based on the heme d content (0.08 mol per mg protein) (Table 2) [14, 30]. The remainder of the heme b remaining in the DW35 membranes can be attributed to other heme complexes expressed in E. coli [38] under the microaerophilic growth conditions used in this study. Notably, membranes with high expression levels of wild-type SQR are significantly depleted of other heme containing proteins, including bd-oxidase. We have previously shown that membranes containing the SdhD H71Y protein do not contain heme attributed to SQR [14]. Moreover, SdhD H71Y membranes show a low ratio of total heme per FAD of 0.22 (Table 1). Similar to the single H71Y mutant, the membranes with the double substitution (H71Y/A72C) show the presence of covalently-bound FAD without an increased content of heme b that gives a low heme/FAD ratio of 0.2 (Table 2). Taken together these data clearly indicated that analogous to the SdhD H71Y single variant, the double mutant is assembled without heme. The computer simulated substitutions of Y71/C72 in the E. coli SQR structure (Fig. 1C) predict that the side chain of Tyr in the SdhD 71 position would be within hydrogen bonding distance from the imidazole ring of SdhD His84. This suggests that formation of this H-bond affords the preservation of the structural organization of the hydrophobic domain of SQR and prevents the incorporation of the heme moiety. In this model Cys in the SdhD 72 position is not able to act as an axial heme ligand.
Figure 3.
Absorption difference (dithionite-reduced minus oxidized) spectra of E. coli DW35 membranes harboring SQR mutant protein. The spectra were determined at 25°C in a 1 ml cuvette containing 1 mg/ml membrane protein in 50 mM potassium phosphate (pH 7.0), 0.1 mM EDTA. Solid sodium dithionite was added to reduce the membrane suspensions. Trace a – wild type SQR; trace b- SdhD H71C; trace c – SdhD H71Y; trace d – SdhD Y71/C72. Insert shows the magnification of the α region of the same spectra.
Table 2.
Cofactor contents of membranes enriched with E. coli SQR.
| Membrane source | Heme b nmol/mg protein | FAD nmol/mg protein | Heme b/FAD ratio | Heme d nmol/mg protein |
|---|---|---|---|---|
| Wt SQR | 3.50 | 3.40 | 1.0 | 0.01 |
| H71Y | 0.58 | 2.60 | 0.22 | 0.08 |
| H71C | 1.93 | 2.48 | 0.78 | 0.03 |
| 71Y72C | 0.42 | 2.08 | 0.20 | 0.06 |
| DW35 | 0.50 | 0 | nd | 0.08 |
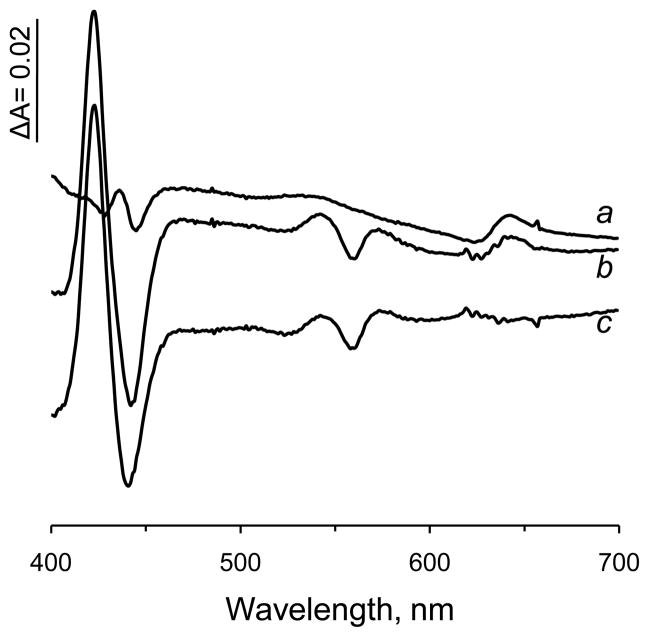
In this study we have also probed whether cysteine could provide the sixth coordination for the heme iron when introduced at the canonical SdhD His71 position. Table 2 shows a substantial increase of total heme content to 1.93 nmol per mg protein in the membranes harboring the SdhD H71C mutant. The difference spectrum of the SdhD H71C membranes, however, shows a less pronounced increase of the amplitude at the Soret band thus indicating altered spectral properties of the heme b within the SdhD H71C mutant membranes. We had previously observed that changes in protein-derived axial ligands transformed the heme iron state from low spin to high spin (i.e., from hexa-coordinated to penta-coordinated) in the SdhD H71Q mutant, which also affected the optical properties of the heme [30]. In the SdhD H71Q mutant, the high spin heme b was detected by changes in optical spectra upon carbon monoxide binding to the dithionite reduced protein and by EPR spectroscopy. Therefore, we examined the ability of CO to react with dithionite-reduced SdhD H71C membranes (Fig. 4). Because of the presence of bd-oxidase, DW35 membranes show the characteristic spectrum for high spin cytochrome d in the 642-622 nm range upon CO binding to the dithionite reduced membranes. In the SdhD H71C membranes, CO induced significant spectral changes with maximum at 423 nm, and minima at 441 and 560 nm. Based on the amplitude at the 642-622 nm region we subtracted the spectrum of the control DW35 membranes from the H71C membranes. The resulting double difference spectrum (Fig. 4) represents the spectrum of a CO-coordinated heme b within the His71Cys mutant enzyme and is very similar to the spectrum of SdhD H71Q. These data clearly indicate that a Cys residue at the SdhD His71 position is unable to substitute for His in providing the sixth coordination site for the heme b (Fig. 1B). We also observed that the heme b in the SdhD H71C membranes was not reducible by succinate which is consistent with a lowered Em for high spin hemes in E. coli SQR mutants [29]. Similar experiments in changing the bis-histidine heme ligand His-66 to Cys in the soluble flavocytochrome b2 from S. cerevisiae resulted in a protein with a ~250 mV lower Em with indications that the remaining His heme-iron ligand was displaced [26]. Comparison of the SdhD H71C mutant membranes with previously described SdhC H84L and SdhD H71Q mutants show that they have similar spectral properties and interaction with CO. These data are all consistent with the interpretation that the SdhD H71C variant contains a penta-coordinated high spin heme.
Figure 4.
Effect of carbon monoxide on the absorption difference spectra of isolated E. coli membranes. The spectra were determined as described in the legend for Fig 3. DW35 control membranes (trace a) and membranes with the SdhD H71C mutant (trace b) were reduced with dithionite and the spectra recorded. The suspensions in the cuvette were then treated with CO gas for 1 min and the spectra recorded. The CO-reduced minus dithionite-reduced difference spectra are shown. Trace c - Double difference spectrum represents subtraction of background bd-oxidase from DW35 control membranes (trace b minus trace a) to show the CO induced spectrum of the H71C mutant.
3.3 Comparison of heme b mutants of E. coli SQR with eukaryotic complex II
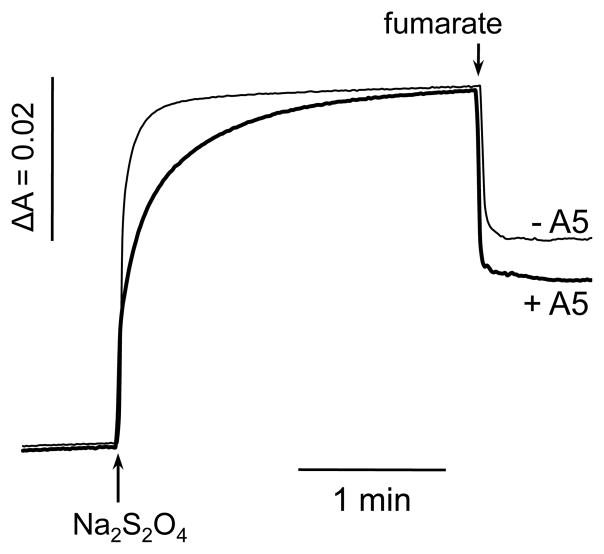
In this work we utilized E. coli SQR as a model of complex II to address the role of cysteine residues in protein derived coordination for heme b. The cysteine substitutions were introduced at two positions in the SdhD subunit: first; to substitute axial His71 and second; to replace Ala72 to create the double mutant Y71C72 that mimics the sequence in the Sdh4p subunit of S. cerevisiae SQR. Cysteine as an axial ligand for a heme moiety is typically found in high spin hemes, and the His-Cys coordination proposed for the heme b from S. cerevisiae SQR would make it unique among this cytochrome family. The data presented here on the E. coli double mutant shows that assembly of catalytically active membrane-bound complex without heme b does not agree with previous studies on S. cerevisiae SQR that show a stoichiometric FAD to heme b content in the yeast complex. It should be stressed that S. cerevisiae SQR has never been isolated except for a brief abstract suggesting that it lacked heme [17]. Otherwise all analytical measurements including the examination of heme b content in numerous yeast mutants were done in either isolated yeast mitochondria or membranes with wild-type expression levels of SQR and therefore in the presence of other heme b containing proteins found in the inner mitochondrial membrane [18–22]. The quantitative tool used for S. cerevisiae SQR heme b determination came from publications focusing on yeast complex III. These studies in turn were based on the original observation by Hatefi and Galante who showed that b heme in isolated dithionite-reduced bovine SQR is reoxidized by fumarate [39]. Bruel et al., suggested that the SQR cytochrome b in yeast membranes is the only heme b that is totally oxidized by fumarate upon addition to dithionite reduced membranes [40]. The original data on isolated bovine SQR, however, demonstrated that fumarate induced about 50% of heme b reoxidation compared to ubiquinone [40]. Figure 5 demonstrates that fumarate also causes about 40% reoxidation of dithionite-reduced isolated E. coli SQR. Dithionite rapidly reduces heme b in the presence of added UQ1 (trace a), and in the “as isolated” protein (data not shown). Addition of a potent Q-site inhibitor atpenin A5 [10, 41] significantly slows the heme b reduction (trace b), while fumarate induces about the same level of the heme b reoxidation. These data show that ubiquinone mediated heme reduction by dithionite (even when an excess of dithionite was used) is in agreement with previous observations that heme b is in fast electron equilibrium with the quinone pool [42]. Importantly, atpenin A5 does not prevent heme b reoxidation by fumarate.
Figure 5.
Reoxidation of dithionite-reduced isolated E. coli SQR by fumarate. SQR 2.4 μM was reduced by solid sodium dithionite in 1 ml of 50 mM of potassium phosphate (pH 6.5) with 30 μM UQ1 present and subsequent addition of 20 μl of 1 M fumarate. 2 μM atpenin A5 was added before the reduction with dithionite. Absorbance changes were recorded at 558 nm.
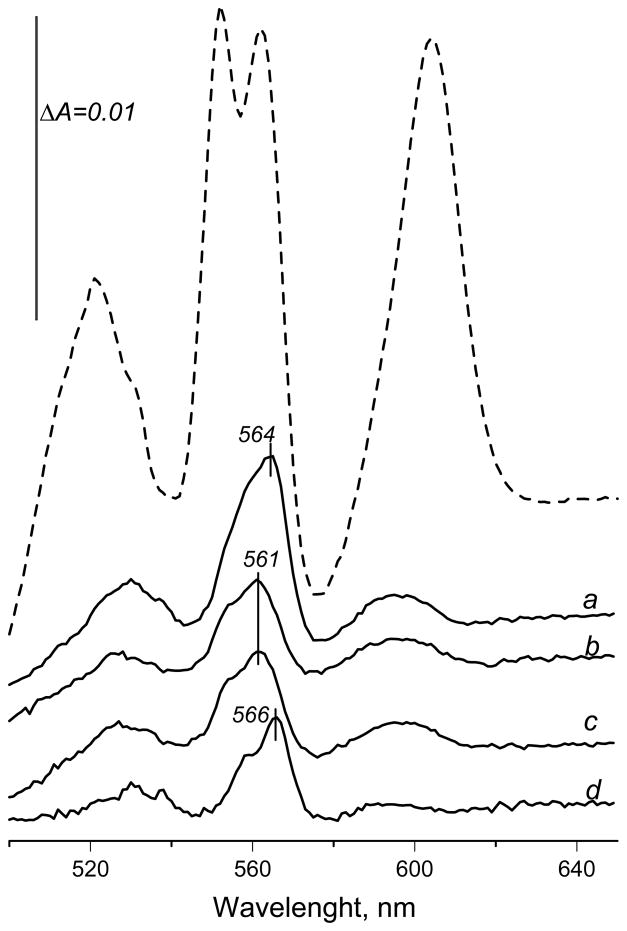
Complex II is a much more efficient succinate:ubiquinone reductase, but it is capable of catalyzing the reverse reaction of ubiquinol oxidation by fumarate [29, 43]. If redox equilibrium between the ubiquinone pool and Q-binding sites of the membrane bound complexes is faster than ubiquinone reduction with dithionite, one may expect that upon addition of fumarate to the dithionite-reduced membranes the b-hemes within complex III may be also partially reoxidized via ubiquinone. Atpenin A5, a specific and potent inhibitor for bovine, E. coli [10, 41] and yeast membrane-bound SQR (data not shown), is a useful reagent to examine fumarate reoxidation of the b hemes in isolated membrane fractions. This concentration of atpenin A5 is also sufficient to completely block activity in both E. coli and bovine SQR. Thus, the redox status of the cytochromes in isolated membranes in the presence of dithionite/fumarate was examined by using specific ubiquinone-binding site inhibitors for SQR and the bc1 complex in bovine submitochondrial particles (SMP). Figure 6 (trace a) shows that the fumarate induced reoxidation of the dithionite-reduced SMP and a difference spectrum demonstrates an asymmetrical peak with a maximum at 564 nm and an amplitude of almost one-third of that compared to the dithionite reduced membranes (Fig. 6, dashed line). When the reaction medium was supplemented with either atpenin A5 (trace b), or specific complex III Q-site inhibitors antimycin plus myxothiazol (trace c), it was observed that dithionite minus fumarate spectra are virtually identical and show a peak with maximum at 561 nm and an amplitude about half of that without the inhibitors. Based on the fact that atpenin A5 does not prevent heme b from reoxidation by fumarate (Fig. 5) this latter peak can be assigned to SQR heme b. The spectral changes that are due to presence of the inhibitors (trace d; trace a minus trace b) show an asymmetrical peak with maximum at 566 nm, characteristic for the low potential heme b566 of the bc1 complex, and a shoulder at 561 nm most likely due to a smaller amplitude of high potential heme b562 [44]. In agreement with Oyedotun and Lemire [18] the fumarate induced changes are linked to complex II activity, and malonate does not cause any changes in the dithionite reduced spectra (data not shown). Therefore, when fumarate is used to reoxidize dithionite-reduced bovine SMP, hemes b in both SQR and complex III are partially reoxidized and contribute to the observed spectral changes.
Figure 6.
Reoxidation of the dithionite reduced bovine submitochondrial particles by fumarate. SMP (0.8 mg protein/ml) were suspended in 1 ml of 0.25 M sucrose, 10 mM potassium phosphate (pH 6.5), 2 mM potassium cyanide, and 0.1% of dodecyl maltoside. Routinely 25 μM K3Fe(CN)6 was added to ensure complete oxidation of the cytochromes. Dashed line shows a spectrum of completely reduced membrane 2 minutes after a few grains of solid sodium dithionite was added. 20 ul of 1 M fumarate was added to the cuvette and the spectra recorded and the dithionite reduced minus fumarate oxidized spectra are shown; 2% correction for the dilution was made to the dithionite reduced spectrum before subtraction. The following additions were made before addition of dithionite: trace a – none; trace b – 2 μM atpenin A5, trace c – 5 μM antimycin and 10 μM myxothiazol. Trace d represents a double difference spectrum after trace b was subtracted from trace a.
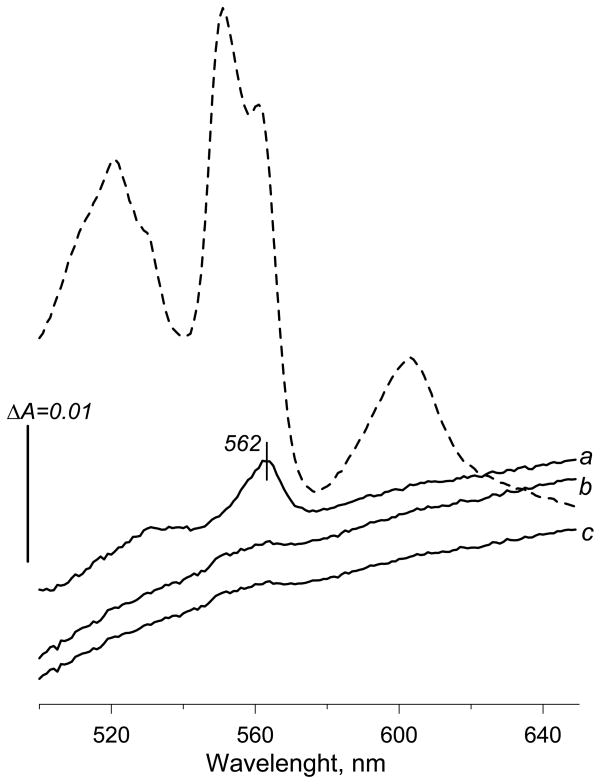
Next, mitochondrial membranes were isolated from S. cerevisiae strain (D273-10B) that was originally used for determination of the amino terminal sequence for subsequent cloning of Sdh4p subunit [15]. Succinate-UQ reductase activity in membrane fractions from yeast was not affected by the addition of 0.1% DDM, however, the activity was completely blocked by the addition of 2 μM atpenin A5. When fumarate was added to the dithionite reduced yeast membranes a peak with maximum at 562 nm, similar to the previously described [18], was observed (Fig. 7 trace a). In the presence of either atpenin A5 (trace b) or antimycin with myxothiazol (trace c), addition of fumarate did not induce any significant changes in the spectrum of the dithionite reduced membranes (Fig. 7). It should be pointed out that the low potential heme b in the isolated yeast bc1 complex exhibits an 563.5–564 nm absorption making the shoulder at 558 nm much less noticeable [45], suggesting it might be misrecognized and attributed to complex II heme upon reoxidation by fumarate. It is also important to stress that excess of dithionite was used to maintain anaerobiosis and high reduction levels of the respiratory enzymes in the membrane, and under these conditions the data (Fig. 6 and 7) demonstrate that reoxidation of complex III is tightly coupled via the ubiquinone pool with fumarate reduction by SQR. Hence the fumarate inducible spectral changes of dithionite-reduced membranes cannot be used to unambiguously determine specific spectral features of the SQR heme b, as the cytochromes of the bc1 complex significantly contribute to the fumarate induced difference spectrum. In addition, heme b of SQR is not completely reoxidized by fumarate in the presence of dithionite in both bovine and E. coli SQR in spite of the differences in the Em of the hemes. The method relies entirely on the catalytic abilities of the enzyme to reduce fumarate in the reaction that requires the association of dehydrogenase catalytic fragment with the transmembrane domain [1, 2, 30, 46], both of which may be affected by introduced mutations. Substitution of amino acid residues lining the heme binding cavity or axial ligands for the iron atom often result in significant alterations of the spectral properties of the heme [30], (and as seen in the H71C mutant in this study). Consequently, the typical absorption coefficient cannot be applied for heme b quantification in such mutants.
Figure 7.
Reoxidation of dithionite-reduced S. cerevisiae mitochondrial membranes by fumarate. The experiment was performed as described in the legend for Fig. 6. Yeast membranes were suspended at concentrations 1.8 mg protein/ml. The dashed line represents a spectrum of dithionite reduced membranes before addition of fumarate. Trace a – no additions; trace b – 2 μM atpenin A5, trace c – 5 μM antimycin and 10 μM myxothiazol.
Interestingly, yeast SQR active in quinone-reductase activity can be extracted from mitochondrial membranes by the common non-ionic detergents (Triton X-100, C12E9, DDM) that caused a complete loss of succinate-ubiquinone reductase activity in the E. coli SdhD Y71 and Y71/C72 mutants described in this study. It should be noted, however, that initial attempts to purify yeast SQR with the aforementioned detergents and ion exchange chromatography results in the rapid loss of ubiquinone-reductase activity. This finding is similar to that of E. coli SQR which contains a pentacoordinated heme [30] which also rapidly inactivates during analysis. The E. coli SQR SdhD Y71 and Y71/C72 mutants do not contain heme (Table 2), but the additional stability to non-ionic detergents noted for the yeast enzyme complex may be afforded by the 29 amino acid C-terminal extension of the Sdh4p protein compared to E. coli and mammalian SdhD. The C. globrata homologue of the Sdh4p protein also contains the C-terminal extension. Perhaps the original method based on the use of bile salt detergents and ammonium sulfate precipitation where active yeast SQR depleted of heme b was found [17] might provide stability to maintain interaction of the hydrophilic (SdhAB) and hydrophobic (SdhCD) subunits and thus allow retention of quinone reductase activity.
In conclusion, there appears to be a concern about the experiments that lead to the interpretation regarding the presence of heme b in S. cerevisiae SQR [15, 17–22]. The data presented in this manuscript show that heme b determination using spectral changes induced by addition of fumarate to dithionite-reduced membranes is not a reliable analytical tool. In the E. coli SQR model system we have shown that Cys at either axial position-71 or in a double mutant (Y71C72) is not able to act as a heme ligand. In the case of the single substitution mutant the heme is retained in a high spin state. When tyrosine is present at the axial position, E. coli SQR forms a membrane-bound protein without heme even when a cysteinyl residue is introduced next to tyrosine resembling the S. cerevisiae SQR sequence. In addition, recent structural analysis of the bis-histidine coordinated membrane-bound ferri hemes defined a four-helix bundle as a frequent characteristics of these proteins [25 and Refs. therein]. The histidine ligands occur opposite each other on the two diametrically opposed helices. The two non-ligand-bearing helices of the bundle also obey this symmetry and have a conserved small residue, usually glycine or serine/threonine, where the edge of the heme ring makes contact with the helix backbone [25]. Surprisingly, in yeast the Sdh3p subunit (genbank: CAA52088), the homolog of E. coli SdhC, the conserved glycine is replaced by a bulky leucine. It was suggested that a four helix bundle formed by the Sdh3p and Sdh4p subunits may not bind heme [25], although computational modeling [21] of the yeast enzyme suggests it is possible.
Therefore, further characterization of the isolated yeast protein is evidently needed before a final conclusion about the structural composition of the yeast complex II can be made. The abstract report [17], which also suggested a heme-less yeast SQR, may be consistent with findings observed for the Y71C72 E. coli SQR mutant reported in this communication. Alternatively, studies with E. coli SQR high spin heme mutants showed impaired protein stability [30], and if yeast SQR retains penta-coordinated heme b this would suggest it could be readily lost during purification attempts consistent with detergent sensitivity and previous suggestions [17] that the enzyme lacked heme. It would also complicate detection of its spectral signatures in a complex membrane environment.
Acknowledgments
This work was supported by National Institutes of Health Grant GM61606 and by the Department of Veterans Affairs, Office of Research and Development, Biomedical Laboratory Research Division. We thank Dr. Eugenia Mileykovskaya for providing the yeast strain.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cecchini G. Function and structure of complex II of the respiratory chain. Annu Rev Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- 2.Hägerhäll C. Succinate:quinone oxidoreductases variation on a conserved theme. Biochim Biophys Acta. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- 3.Cecchini G, Schröder I, Gunsalus RP, Maklashina E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim Biophys Acta. 2002;1553:140–157. doi: 10.1016/s0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 4.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 5.Lancaster CR, Kröger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- 6.Taylor P, Pealing SL, Reid GA, Chapman SK, Walkinshaw MD. Structural and mechanistic mapping of a unique fumarate reductase. Nat Struct Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- 7.Reid GA, Miles CS, Moysey RK, Pankhurst KL, Chapman SK. Catalysis in fumarate reductase. Biochim Biophys Acta. 2000;1459:310–315. doi: 10.1016/s0005-2728(00)00166-3. [DOI] [PubMed] [Google Scholar]
- 8.Huang LS, Shen JT, Wang AC, Berry EA. Crystallographic studies of the binding of ligands to the dicarboxylate site of complex II, and the identity of the ligand in the “oxaloacetate inhibited” state. Biochim Biophys Acta. 2006;1757:1073–1083. doi: 10.1016/j.bbabio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 10.Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Ōmura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate:ubiquinone oxidoreductase): A mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006;281:7309–7316. doi: 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- 11.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rhao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA. 3-Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem. 2006;281:5965–5972. doi: 10.1074/jbc.M511270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006;281:32310–32317. doi: 10.1074/jbc.M607476200. [DOI] [PubMed] [Google Scholar]
- 14.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. An Escherichia coli succinate dehydrogenase variant lacking the heme b. Proc Natl Acad Sci USA. 2007;104:18007–18012. doi: 10.1073/pnas.0707732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullis BL, Lemire BD. Isolation and characterization of the Saccharomyces cerevisiae SDH4 gene encoding a membrane anchor subunit of succinate dehydrogenase. J Biol Chem. 1994;269:6543–6549. [PubMed] [Google Scholar]
- 16.Kerscher O, Sepuri NB, Jensen RE. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol Biol Cell. 2000;11:103–116. doi: 10.1091/mbc.11.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilling RJ, Baldwin T, Palmer G. The characterization of highly purified complex II from Baker’s yeast. Fed Proc. 1982;41:896. [Google Scholar]
- 18.Oyedotun KS, Lemire BD. The Saccharomyces cerevisiae succinate-ubiquinone reductase contains a stoichiometric amount of cytochrome b562. FEBS Lett. 1999;442:203–207. doi: 10.1016/s0014-5793(98)01657-3. [DOI] [PubMed] [Google Scholar]
- 19.Oyedotun KS, Lemire BD. The Saccharomyces cerevisiae succinate-ubiquinone oxidoreductase. Identification of Sdh3p amino acid residues involved in ubiquinone binding. J Biol Chem. 1999;274:23956–23962. doi: 10.1074/jbc.274.34.23956. [DOI] [PubMed] [Google Scholar]
- 20.Oyedotun KS, Yau PF, Lemire BD. Identification of the heme axial ligands in the cytochrome b562 of the Saccharomyces cerevisiae succinate dehydrogenase. J Biol Chem. 2004;279:9432–9439. doi: 10.1074/jbc.M311877200. [DOI] [PubMed] [Google Scholar]
- 21.Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem. 2004;279:9424–9431. doi: 10.1074/jbc.M311876200. [DOI] [PubMed] [Google Scholar]
- 22.Oyedotun KS, Sit CS, Lemire BD. The Saccharomyces cerevisiae succinate dehydrogenase does not require heme for ubiquinone reduction. Biochim Biophys Acta. 2007;1767:1436–1445. doi: 10.1016/j.bbabio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Anderson JLR, Chapman SK. Ligand probes for heme proteins. Dalton Trans. 2005;1:13–24. doi: 10.1039/b413046d. [DOI] [PubMed] [Google Scholar]
- 24.Rydberg P, Sigfridsson E, Ryde U. On the role of the axial ligand in heme proteins: a theoretical study. J Biol Inorg Chem. 2004;9:203–223. doi: 10.1007/s00775-003-0515-y. [DOI] [PubMed] [Google Scholar]
- 25.Berry EA, Walker FA. Bis-histidine-coordinated hemes in four-helix bundles: how the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J Biol Inorg Chem. 2008;13:483–498. doi: 10.1007/s00775-008-0372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mowat CG, Miles CS, Munro AW, Cheesman MR, Quaroni LG, Reid GA, Chapman SK. Changing the heme ligation in flavocytochrome b2: substitution of histidine-66 by cysteine. J Biol Inorg Chem. 2000;5:584–592. doi: 10.1007/s007750000141. [DOI] [PubMed] [Google Scholar]
- 27.Silkin Y, Oyedotun KS, Lemire BD. The role of Sdh4p Tyr-89 in ubiquinone reduction by the Saccharomyces cerevisiae succinate dehydrogenase. Biochim Biophys Acta. 2007;1767(2):143–50. doi: 10.1016/j.bbabio.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Westenberg DJ, Gunsalus RP, Ackrell BAC, Sices H, Cecchini G. Escherichia coli fumarate reductase frdC and frdD mutants: identification of amino acid residues involved in catalytic activity with quinones. J Biol Chem. 1993;268:815–822. [PubMed] [Google Scholar]
- 29.Maklashina E, Berthold DA, Cecchini G. Anaerobic expression of Escherichia coli succinate dehydrogenase: The functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J Bacteriol. 1998;180:5989–5996. doi: 10.1128/jb.180.22.5989-5996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maklashina E, Rothery RA, Weiner JH, Cecchini G. Retention of heme in axial ligand mutants of succinate:ubiquinone oxidoreductase (complex II) from Escherichia coli. J Biol Chem. 2001;276:18968–18976. doi: 10.1074/jbc.M011270200. [DOI] [PubMed] [Google Scholar]
- 31.Rothery RA, Seime AM, Spiers AMC, Maklashina E, Schröder I, Gunsalus RP, Cecchini G, Weiner JH. Defining the Qp-site of Escherichia coli fumarate reductase (FrdABCD) by site-directed mutagenesis, fluorescence quench titrations, and EPR spectroscopy. FEBS J. 2005;272:313–326. doi: 10.1111/j.1742-4658.2004.04469.x. [DOI] [PubMed] [Google Scholar]
- 32.Fessenden JM, Racker E. A-particles and P-particles. Methods Enzymol. 1967;10:194–197. [Google Scholar]
- 33.Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 34.Maklashina E, Cecchini G. Comparison of catalytic activity and inhibitors of quinone reactions of succinate dehydrogenase (succinate-ubiquinone oxidoreductase) and fumarate reductase (menaquinol-fumarate oxidoreductase) from Escherichia coli. Arch Biochem Biophys. 1999;369:223–232. doi: 10.1006/abbi.1999.1359. [DOI] [PubMed] [Google Scholar]
- 35.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 36.Borisov V, Arutyunyan AM, Osborne JP, Gennis RB, Konstantinov AA. Magentic circular dichroism used to examine the interaction of Escherichia coli cytochrome bd with ligands. Biochemistry. 1999;38:740–750. doi: 10.1021/bi981908t. [DOI] [PubMed] [Google Scholar]
- 37.Singer TP, Salach J, Hemmerich P, Ehrenberg A. Flavin Peptides. Methods Enzymol. 1971;18:416–427. [Google Scholar]
- 38.Gennis RB, Stewart V. Respiration, Escherichia coli and Salmonella. In: Neidhardt FC, editor. Cellular and Molecular Biology. Vol. 1. American Society for Microbiology Press; Washington, D. C: 1996. pp. 217–261. [Google Scholar]
- 39.Hatefi Y, Galante YM. Isolation of cytochrome b560 from complex II (succinate-ubiquinone oxidoreductase) and its reconstitution with succinate dehydrogenase. J Biol Chem. 1980;255:5530–5537. [PubMed] [Google Scholar]
- 40.Bruel C, di Rago JP, Slonimski PP, Lemesle-Meunier B. Role of the evolutionarily conserved cytochrome b tryptophan 142 in the ubiquinol oxidation catalyzed by the bc1 complex in the yeast Saccharomyces cerevisia. J Biol Chem. 1995;270:22321–22328. doi: 10.1074/jbc.270.38.22321. [DOI] [PubMed] [Google Scholar]
- 41.Miyadera H, Shiomi K, Ui H, Yamaguchi Y, Masuma R, Tomada H, Miyoshi H, Osanai A, Kita K, Ōmura S. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase) Proc Natl Acad Sci USA. 2003;100:473–477. doi: 10.1073/pnas.0237315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson RF, Hille R, Shinde SS, Cecchini G. Electron transfer within complex II. Succinate:ubiquinone oxidoreductase of Escherichia coli. J Biol Chem. 2005;280:33331–33337. doi: 10.1074/jbc.M506002200. [DOI] [PubMed] [Google Scholar]
- 43.Grivennikova VG, Gavrikova EV, Timoshin AA, Vinogradov AD. Fumarate reductase activity of bovine heart succinate-ubiquinone reductase. New assay system and overall properties of the reaction. Biochim Biophys Acta. 1993;1140:282–292. doi: 10.1016/0005-2728(93)90067-p. [DOI] [PubMed] [Google Scholar]
- 44.Trumpower BL. Function of the iron-sulfur protein of the cytochrome b-c1 segment in electron-transfer and energy-conserving reactions of the mitochondrial respiratory chain. Biochim Biophys Acta. 1981;639:129–155. doi: 10.1016/0304-4173(81)90008-2. [DOI] [PubMed] [Google Scholar]
- 45.Siedow JN, Power S, de la Rosa FF, Palmer G. The preparation and characterization of highly purified, enzymatically active complex III from baker’s yeast. J Biol Chem. 1978;253:2761–2769. [PubMed] [Google Scholar]
- 46.Ackrell BAC, Ball MB, Kearney EB. Peptides from complex II active in reconstitution of succinate-ubiquinone reductase. J Biol Chem. 1980;255:2761–2769. [PubMed] [Google Scholar]