Abstract
Angelman syndrome (AS) is a severe neurological disorder caused by a deficiency of ubiquitin protein ligase E3A (Ube3a), but the pathophysiology of the disease remains unknown. We now report that in the brains of AS mice in which the maternal Ube3a allele is mutated (m-) and the paternal allele is potentially inactivated by imprinting (p+) (Ube3a m-\p+) the mitochondria are abnormal and exhibit a partial oxidative phosphorylation (OXPHOS) defect. Electron microscopy of the hippocampal region of the Ube3a m-\p+ mice (n=6) reveals small, dense mitochondria with altered cristae, relative to wild-type littermates (n=6) and reduced synaptic vesicle density. The specific activity of OXPHOS complex III is reduced in whole brain mitochondria in Ube3a m-\p+ (n=5) mice versus wild-type littermates (n=5). Therefore, mitochondrial dysfunction may contribute to the pathophysiology of Angelman Syndrome.
Keywords: Angelman syndrome, Mitochondrial complexes, Synaptic vesicle, Electron microscopy, Ube3a deficient mouse
Introduction
Angelman syndrome (AS) is clinically characterized by seizures, microcephaly, motor dysfunction, mental retardation and mild dysmorphic features. Based on molecular analyses, there are four known genetic/epigenetic mechanisms causing AS: (i) deletions of maternal chromosome region 15q11-q13 involving the ubiquitin-protein ligase E3A (UBE3A) critical region, (ii) paternal uniparental disomy (UPD) of this region, (iii) imprinting center defects (IDs), and (iv) mutations in the UBE3A gene [15]. UBE3A expression differs in the maternal and paternal alleles because of different methylation patterns (imprinting). The paternal silencing of the UBE3A gene occurs in a brain region-specific manner, the maternal allele almost exclusively being expressed in the hippocampus and cerebellum [10]. UBE3A encodes an 865-amino acid protein E6-associated protein (E6-AP), which acts as a cellular ubiquitin ligase which creates a covalent linkage (e.g., the “ligase” function) between a 76-amino acid, ubiquitin molecule and its target protein to create a polyubiquitylated substrate [15]. The AS associated ubiquitin ligase localized to neuronal synapse, nucleus and to presynaptic and postsynaptic compartments in cultured hippocampal neurons, and maternal deficiency resulted in abnormal dendritic spine morphology [6]. Ubiquitination is critical for proteasomal degradation of proteins and plays a role in numerous cellular processes including regulation of the cell cycle, signal transduction, synaptic plasticity and transcription [2, 12].
Mitochondria are central to many cellular functions, including the generation of ATP and intracellular Ca2+ homeostasis. They are also responsible for the formation of reactive oxygen species (ROS) as well as for triggering cell death [24]. Brain neurons, liver, heart and skeletal muscles in particular rely on mitochondria because of their high levels of activity and need for energy. Mitochondrial dysfunction is involved in the pathophysiology of many neurological diseases [17].
A mouse model of AS has been created by the inactivation of the Ube3a gene. Ube3a is highly expressed in hippocampal neurons and Purkinje cells [1] and in mouse tissues in which the maternal allele is mutated (m-) and the wild type paternal allele (p+) is inactivated by imprinting, we would expect a near complete lack of Ube3a protein level in Ube3a m-/p+ tissues. Since hippocampal and Purkinje cells have the highest expression of Ubea3a, we might predict that these cells would have the greatest sensitivity to the Ube3a m-\p+ genotype.
Ube3a m-\p+ mice have been observed to exhibit defects in hippocampal long-term potentiation (LTP) [14] and in cerebellar motor function [11]. The hippocampal LTP defect of Ube3a m-\p+ mice has been linked to increased inhibitory phosphorylation of αCaMKII in the calcium/calmodulin kinase type 2 (CaMKII) signaling pathway [25], and the neurological deficits of Ube3a m-\p+ can be rescued through a mutation in CaMKII that prevents its inhibitory phosphorylation [23].
Hippocampal mitochondria of Ube3a m-\p+ mice are small and dense with altered cristae, and brain complex III activity is reduced. Synaptic vesicle density is also reduced. The mitochondria are an important system for modulating cytosolic calcium. Therefore, our data suggest that some of the characteristics of the mouse phenotype, and perhaps of human AS, may also implicate a mitochondrial etiology.
Materials and methods
Mouse model for AS
Ube3a deficient mice were developed as described previously [14]. Maternal deficient heterozygous mice (m-\p+), which represent the AS disease condition and wild-type littermates were kindly provided by Drs Jiang and Beaudet (Baylor College of Medicine, Houston, TX).
Electron Microscopy
Wild-type littermates (n=6) and m-/p+ mice (n=6) at 3-4 months of age were perfused with the solution of 4% paraformaldehyde plus 0.1% glutaraldehyde in 0.1M PB. These treated brains were further fixed in the same solution for 2-4 h. 100 μm sections were cut with a vibratom, then collected in PBS and fixed in 1% glutaraldehyde over night. Tissue samples were fixed in 1% Osmium for 1 h at 4°C and serially dehydrated in ethanol, which were embedded in Eponate 12 resin at 65°C for 24-36 h. Ultrathin (60∼80 nm) sections were cut with a diamond knife. Sections were stained in 1% uranyl acetate for 30 min, followed by lead citrate for 7-10 min. Sections were examined with a Philips CM10 transmission electron microscope. Electron micrographs were taken with a Gatan UltraScan US1000 digital camera.
Mitochondrial isolation and OXPHOS enzyme analysis
Mitochondria were isolated from the liver, heart, skeletal muscle (vastus lateralis), whole brain, and hippocampi and cerebellum separately by homogenization and differential centrifugation as described before [22]. Mitochondrial enzyme complex activities were determined by OXPHOS enzyme assays using our standard protocols [22].
Western blotting
Protein expression levels of the mitochondrial complexes I-IV were determined by Western blotting analyses. Equal amount of proteins were separated on SDS-PAGE gels or native gels. Proteins were then transferred to nitrocellulose membranes, incubated in blocking buffer (5% dry milk in 1xPBS-T), and treated with primary antibodies for the mitochondrial complexes: microprofile WB kit (MitoSciences, Eugene, OR), and porin (Molecular Probes, Eugene, OR). Appropriate secondary antibody (anti-mouse) was used. As appropriate, mitochondrial porin level was determined for each blot to verify equal protein loading.
The synaptic vesicle protein was quantified in whole brain by reaction of brain protein Western blots with synaptophysin (SYP) antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The goat synaptophysin antibody binding was detected by reaction with rabbit antigoat secondary antibody. Protein loading levels were verified by actin staining.
Data analysis
Digital electron micrographs were analyzed using Adobe Photoshop and NIH Image. In order to analyze full synaptic features, the selected synapses met the following three criteria: (i) a widened synaptic cleft, (ii) presence of a cluster of synaptic vesicles associated with the presynaptic membrane and (iii) the postsynaptic membrane. Mean synaptic vesicle numbers within 0.4 × 0.2 μm2 of the presynaptic membrane were calculated. Vesicles touching the presynaptic membrane were included in the estimate of docked vesicles. The size of mitochondria within 2.6 × 2.6 μm2 of area was assessed in the CA1 region of the hippocampi. Six different areas of each section were reviewed with a total of 6 sections studied. Kolmogorov-Smirnov two sample test was used to assess differences in values of m-\p+ and wild-type littermates. Measurements of mitochondrial enzyme activities were repeated 3 times for each test on each animal. The unpaired student’s t test was used to evaluate differences in values of m-\p+ and wild-type littermates. Data represent mean ± SD. An analysis with a value of p≤0.05 was considered to be statistically significant.
Results
Ultrastructural analysis of the neuronal mitochondria of hippocampal CA1 region in the Ube3a deficient mice
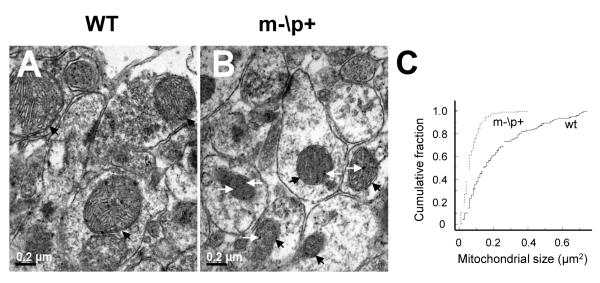
To clarify ultrastructural properties of hippocampal neurons, electron microscopic images were obtained from the Ube3a deficient mice. At low magnification (×2200), we calculated the numbers of mitochondria located in the 10.4×10.4 μm2 area of each section (data not shown). There were 85-95 mitochondria within 10.4×10.4 μm2 of area (0.78-0.88/μm2) in wild-type littermates and m-\p+ mice, indicating no obvious change in the number of mitochondria. To ensure that the orientation of sections did not have an effect on the results, we made 6 serial sections and analyzed mitochondrial sizes located in 2.6×2.6 μm2 in hippocampal CA1 neurons at a high magnification (× 8900). Morphologically the Ube3a m-\p+ mouse mitochondria were compressed, with disturbances of mitochondrial cristae. They also contain multiple small, electron-dense spheroids within the mitochondrial matrix when compared to their wild-type littermates (Fig. 1 A and B). We found that the mean size of mitochondria was significantly smaller in m-\p+ mice versus wild-type littermates (Fig. 1 C, p<0.001, Kolmogorov-Smirnov two sample test). Curves were constructed from 142 mitochondria in wild-type littermates, 227 mitochondria in the m-\p+ mice. Our analysis revealed that the Ube3a deficiency is associated with mitochondria in small size.
Figure 1. Ube3a deficiency results in morphological changes of mitochondria in mouse hippocampal CA1 neurons.
Electron micrographs of wild-type littermate (WT, A) and m-\p+ mouse (B). Magnification: ×8900. Black arrows show mitochondria and white arrows indicate small, electron- dense spheroids within mitochondria. C) Cumulative probability curves of a distribution of mitochondrial sizes in the m-\p+ mice and their WT. X-axis indicates mitochondrial size and Y-axis indicates cumulative fraction.
Enzyme activities of the brain mitochondrial complexes in the Ube3a deficient mice
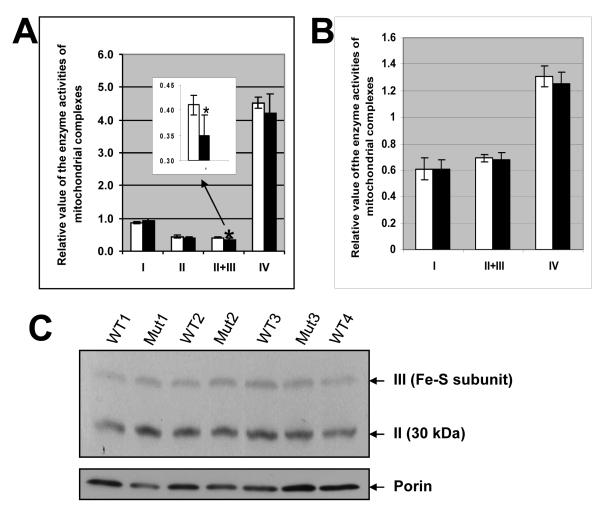
To determine if the morphological alterations of the brain mitochondria could be correlated with perturbations of mitochondrial function, we measured the enzyme activities of complexes I, II, II+III and IV in the brain from the Ube3a m-\p+ mice and wild-type littermates. Mitochondrial matrix citrate synthase (CS) was used to normalize the enzyme activities of mitochondrial complexes. We observed that the activity of brain mitochondrial complexes II+III activity was significantly reduced in the m-\p+ mice (0.35±0.04, n=5) when compared to wild-type littermates (0.41±0.02, n=5) (p<0.05, Student t-test), whereas the activities of brain complexes I and IV were unchanged (Fig. 2A). We also assessed the individual activity of complex II and found that the brain complex II was not changed in the m-\p+ mice. Therefore, the defect appears to be associated with respiratory complex III in the Ube3a deficient mice. Interestingly, the skeletal muscle in the m-\p+ mice possibly showed induction of mitochondrial complex IV activity from 2.63±0.24 (wild-type littermates) to 3.07±0.33 (m-\p+ mice) (p=0.05, Student t-test; n=5/group; data not shown), whereas muscle complexes I and II+III activities were unchanged. There were no significant differences in the activities of complexes I, II+III and IV in the liver and heart tissues (data not shown).
Figure 2. Ube3a deficiency leads to decrease in enzyme activities of brain mitochondrial complex III.
The enzyme activities of the brain (A) and hippocampal (B) mitochondrial complexes. An inserted chart in A is enlarged for complexes II+III. The data were normalized with citrate synthase. Measurements were repeated 3 times for each test on each animal. * p<0.05 (Student t test, M±SD). Empty bars indicate WT; Black bars indicate the m-\p+ mice. C) Western blotting analysis of mitochondrial complexes II and III is shown in upper panel, and porin staining in lower panel. The arrows indicate complexes II, III and porin on the right, and genotypes of mice are shown above.
To further define the regions of the brain that accounted for the overall complex III defect, we also assayed the enzyme complexes in mitochondria isolated from the hippocampi and cerebellums from the Ube3a m-\p+ mice. The activities of hippocampal (Fig. 2B) and cerebellar (data not shown) mitochondrial complexes I, II+III and IV were not significantly different from controls, indicating that the mitochondrial enzyme defect was more pronounced in the cortex of 3 to 4 month old animals.
To determine the protein expression levels of the brain OXPHOS complexes, we separated the complexes using blue native gel electrophoresis and transferred the complexes to filters by Western blotting. The filters were then reacted with either a cocktail of 5 monoclonal antibodies that allows detection of mitochondrial complexes I, II, III, IV and V, or an anticomplex III antibody alone. Our Western blots revealed that the relative levels of the OXPHOS complexes were not significantly altered in the Ube3a m-\p+ mice when comparing to wild-type littermates (Fig. 2 C).
Analyses of chemical synapses in hippocampal CA1 region of the Ube3a deficient mice
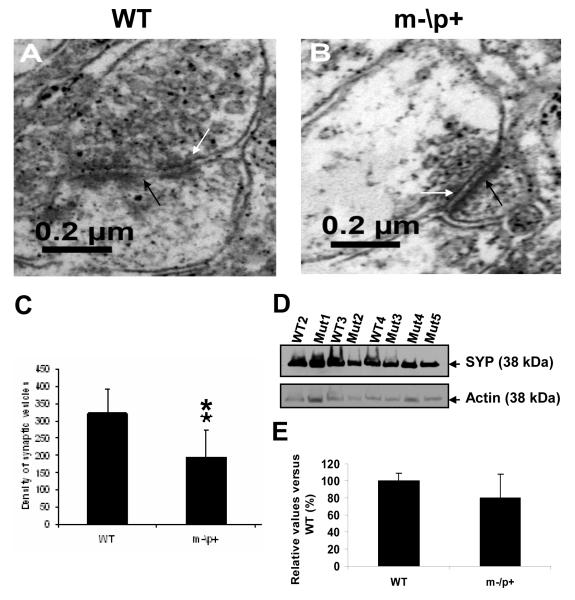
A previous study has shown deficits in hippocampal LTP in the Ube3a deficient mice, but ultrastructural evidence of pre- or postsynaptic membranes was not studied [14]. We employed electron microscopy to characterize the ultrastructure of the hippocampal CA1 area in the m-\p+ mice and wild-type littermates. Analysis of the hippocampal CA1 region at higher magnification (× 8900) demonstrated the presence of chemical synapses with many abnormal features in the m-\p+ mice including a cluster of vesicles embedded in a loosely organized matrix of electron dense material, associated with a cleft of 10-25 nm separating a pre- and postsynaptic process (Fig. 3 A and B). Electron dense material was present within the cleft and was also concentrated on the intracellular surface of the opposing postsynaptic membrane. The majority of vesicles located in the synaptic terminals, ranging 20-45 nm in diameter, are observed in wild-type littermates and m-\p+ mice (Fig. 3 A and B). There are a remarkably lower density of synaptic vesicles in m-/p+ mice (n=45 synapses) when compared to the wild-type littermates (n=25 synapses), (Fig. 3C, P<0.01, Student t test, M±SD). This result suggested that reduction of synaptic vesicle density in the hippocampi of Ube3a deficient mice may be also implicated in the pathogenesis of AS.
Figure 3. Ube3a deficiency results in reduction of synaptic vesicle density in hippocampal CA1 neurons.
Electron micrographs of chemical synapses in WT (A) and m-\p+ mouse (B). Magnification: ×8900. C) Analysis of synaptic vesicle density. ** p<0.01 (Student t test, M±SD). D) Western blotting analysis using synaptophysin (SYP) (upper panel) and an actin-specific antibody (lower panel) in the m-\p+ mice and their WT brains. The arrows indicate SYP and actin on the right. E) Densitometic analysis of SYP expression in the m-\p+ mice and their WT brains. Y-axis indicates relative values of SYP vs WT (%) and X-axis indicates genotypes of mice.
The expression levels of the synaptic vesicle protein, synaptophysin, were also tested in the brain of the Ube3a m-\p+ mice by Western blots. The relative protein levels in the Ube3a m-\p+ mice (n=5) were downregulated by 21% when comparing to the wild-type littermates (n=3) (Fig. 3D and E). Such the changed synaptic vesicle protein levels were not statistically different between the Ube3a m-\p+ mice and wild-type littermates (Student t test, p> 0.05, M±SD) due to the variability of the Ube3a m-\p+ mice. But there were remarkable reductions of the protein levels in 3 out 5 Ube3a m-\p+ mice.
Discussion
Using mice heterozygous for a null mutation in the Ube3a gene, in which the mutant gene was contributed by the mother, we have observed a partial mitochondrial respiratory complex III defect in the brains of Ube3a m-\p+ mice. This mitochondrial enzyme defect is associated with abnormal brain mitochondrial morphology and alteration in the synaptic vesicle density.
While the complex III defect was significantly reduced for the whole brain, it was not significantly reduced when the hippocampus or cerebellum mitochondria were assayed separately. This may be explained by the observation that the paternal allele of Ube3a is strongly inactivated in hippocampal and cerebellar neurons [11,14]. Therefore, during development, those cells in which the paternal allele was fully inactivated would become complex III null, which is incompatible with life. Hence, this subset of cells would be lost prior to birth, resulting in the Angelman Syndrome phenotype. It is well established that different types of differentiated cells have different mitochondrial physiologies and express different sets of mitochondrial genes, including different isoforms of the same protein [24]. Therefore, it is likely that some neurons and various classes of glial cells would have normal or only partially reduce the expression of the p+ allele. Therefore, in the hippocampus and cerebellum where the p+ allele inactivation would be most complete, all cells undergoing p+ inactivation would be lost during development resulting in the AS phenotype. The remaining cells would thus not be subject to p+ inactivation, such that the specific activity of the mitochondria of the remaining cells would be maximal. By contrast, in other parts of the brain where p+ inactivation would be partial, the partial complex III defect would be more apparent and thus detectable with the methods currently available. Even relatively mild alterations in mitochondrial function can result in mitochondrial morphological changes. Hence, it would be expected that a 50% reduction in complex III in the Ube3a m-\p+ mice would result in morphological alterations in the brain mitochondria that we observed.
The mechanism by which a defect in Ube3a could cause a mitochondrial defect is unknown. The reduced enzyme activities of the brain mitochondrial complexes in the Ube3a deficient mice may be due to inactivation of the enzymes causing impairment of the electron transport chain. Oxidative damage could potentially preferentially affect the stability and function of the complexes containing iron-sulfur clusters (complexes II+III) in the Ube3a deficient mice brain. However, it is clear that ubiquitination is important in maintaining the structural and functional integrity of mitochondria in yeast [8, 21]. Moreover, the mitochondrial ubiquitin ligase MARCH-V is important in regulating the mitochondrial dynamics of Drp1 and Fis1 in mammalian cells [16, 20] and the human deubiquitinating enzyme ubiquitin-specific protease 30 (USP30) is embedded in the mitochondrial outer membrane and participates in the maintenance of mitochondrial morphology [19]. Therefore, partial defects in Ube3a could have either primary or secondary consequences for the maintenance of mitochondrial complex III. Since the Ube3a protein E6-AP has been shown to interact with Drosophila High-wire which regulated synaptic growth, alteration in ubiquitin biochemistry could also explain the aberrations in synaptic morphology [3], acting either through the mitochondria or by an independent pathway.
It is becoming increasingly clear that alterations in mitochondrial function accompany epigenomic alterations. One of the best examples is Rett syndrome (RS), which shares many characteristics with AS [13], and is the result of mutations in the X-linked MeCP2 gene. RS patients have been documented to have brain mitochondrial structural and OXPHOS complex defects [7, 4] and mice in which the MeCP2 gene is inactivated have alterations in complex III gene expression and uncoupled mitochondrial [18].
The hippocampus which resides in the temporal lobe has a low seizure threshold [26]. Yashiro et al [27] found that the Ube3a deficient mice had impaired synaptic plasticity in experience-dependent neocortical development, demonstrating Ube3a plays a critical role in the modifications of neuronal circuits. This is consistent with the observed reduction in spine density and synaptic current frequency of the pyramidal neurons of the cortex [27, 5]. The reduction of synaptic vesicle density in the hippocampus may also be the result of a maturational delay associated with the Ube3a deletion and possibly contributes to the impaired neuronal circuits and also to pathophysiology of AS.
Therefore, while we lack sufficient depth of understanding of the biology of the mitochondrion to know the full nature of the interactions between the nucleus and mitochondrion, the similarities in clinical phenotypes that have been observed between patients with mitochondrial disease [24] and patients with diseases of the epigenome [9] are sufficiently similar as to suggest a common pathophysiological mechanism. If this proves to be true, then approaches that are being developed for mitochondrial disease therapies may be help for the more complex epigenomic diseases.
Acknowledgments
The authors thank Dr Beaudet for providing the Ube3a deficient mice. This research was supported by the Rare Diseases Clinical Research Consortia (USA), 5 U54 RR019478-05 and an RDCRN postdoctoral trainee award (H.S.), R01AR050236 (V.E.K.), California Regenerative Medicine Predoctoral Fellowship TI-00008 (W.F.), the National Institute of Health (USA) grants NS21328, NS41850, AG13154, AG24373 and AG16573 (D.C.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- [2].Ciechanover A, Schwartz AL. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- [4].Cornford ME, Philippart M, Jacobs B, Scheibel AB, Vinters HV. Neuropathology of Rett syndrome: case report with neuronal and mitochondrial abnormalities in the brain. J Child Neurol. 1994;9:424–431. doi: 10.1177/088307389400900419. [DOI] [PubMed] [Google Scholar]
- [5].Crozier RA, Wang Y, Liu CH, Bear MF. Deprivation-induced synaptic depression by distinct mechanisms in different layers of mouse visual cortex. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1383–1388. doi: 10.1073/pnas.0609596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Human molecular genetics. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- [7].Eeg-Olofsson O, al-Zuhair AG, Teebi AS, Daoud AS, Zaki M, Besisso MS, Al-Essa MM. Rett syndrome: a mitochondrial disease? J Child Neurol. 1990;5:210–214. doi: 10.1177/088307389000500311. [DOI] [PubMed] [Google Scholar]
- [8].Escobar-Henriques M, Westermann B, Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. The Journal of cell biology. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- [10].Glenn CC, Deng G, Michaelis RC, Tarleton J, Phelan MC, Surh L, Yang TP, Driscoll DJ. DNA methylation analysis with respect to prenatal diagnosis of the Angelman and Prader-Willi syndromes and imprinting. Prenat Diagn. 2000;20:300–306. [PubMed] [Google Scholar]
- [11].Heck DH, Zhao Y, Roy S, LeDoux MS, Reiter LT. Analysis of cerebellar function in Ube3a-deficient mice reveals novel genotype-specific behaviors. Human molecular genetics. 2008;17:2181–2189. doi: 10.1093/hmg/ddn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hochstrasser M. Ubiquitin-dependent protein degradation. Annual review of genetics. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- [13].Jedele KB. The overlapping spectrum of rett and angelman syndromes: a clinical review. Seminars in pediatric neurology. 2007;14:108–117. doi: 10.1016/j.spen.2007.07.002. [DOI] [PubMed] [Google Scholar]
- [14].Jiang Y.-h., Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman Ubiquitin Ligase in Mice Causes Increased Cytoplasmic p53 and Deficits of Contextual Learning and Long-Term Potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- [15].Jiang YH, Beaudet AL. Human disorders of ubiquitination and proteasomal degradation. Current opinion in pediatrics. 2004;16:419–426. doi: 10.1097/01.mop.0000133634.79661.cd. [DOI] [PubMed] [Google Scholar]
- [16].Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. The Journal of cell biology. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Keating DJ. Mitochondrial dysfunction, oxidative stress, regulation of exocytosis and their relevance to neurodegenerative diseases. Journal of neurochemistry. 2008;104:298–305. doi: 10.1111/j.1471-4159.2007.04997.x. [DOI] [PubMed] [Google Scholar]
- [18].Kriaucionis S, Paterson A, Curtis J, Guy J, Macleod N, Bird A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Molecular and cellular biology. 2006;26:5033–5042. doi: 10.1128/MCB.01665-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nakamura N, Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Molecular biology of the cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO reports. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Neutzner A, Youle RJ. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. The Journal of biological chemistry. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- [22].Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods in enzymology. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- [23].van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, de Avila Freire R, Jiang Y.-h., Elgersma Y, Weeber EJ. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of [alpha]CaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10:280–282. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- [24].Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual review of genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Weeber EJ, Jiang Y-H, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, Christian JM, Mirnikjoo B, Silva A, Beaudet AL, Sweatt JD. Derangements of Hippocampal Calcium/Calmodulin-Dependent Protein Kinase II in a Mouse Model for Angelman Mental Retardation Syndrome. J. Neurosci. 2003;23:2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yaari Y, Beck H. “Epileptic neurons” in temporal lobe epilepsy. Brain pathology (Zurich, Switzerland) 2002;12:234–239. doi: 10.1111/j.1750-3639.2002.tb00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, Weinberg RJ, Ehlers MD, Philpot BD. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009 doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]