Abstract
Genetic variants in the brain-derived neurotrophic factor (BDNF) gene, predominantly the functional Val66Met polymorphism, have been associated with risk of bipolar disorder and other psychiatric disorders. However, not all studies support these findings, and overall the evidence for BDNF association with disease risk is weak. As differences in population genetic structure between patient samples could cause discrepant or spurious association results, we investigated this possibility by carrying out population genetic analyses of the BDNF genomic region. Substantial variation was detected in BDNF coding region SNP allele and haplotype frequencies between 58 global populations, with the derived Met allele of Val66Met ranging from 0–72% frequency across populations. FST analyses to assess diversity in the HapMap populations determined that the Val66Met FST value was at the 99.8th percentile among all SNPs in the genome. As the BDNF population genetic differences may be due to local selection, we performed the long-range haplotype (LRH) test for selection using 68 SNPs spanning the BDNF genomic region in 12 European-derived pedigrees. Evidence for positive selection was found for a high frequency Val-carrying haplotype, with a relative extended haplotype homozygosity (REHH) value above the 99th percentile compared to HapMap data (P=4.6 ×10−4). In conclusion, we observed considerable BDNF allele and haplotype diversity among global populations and evidence for positive selection at the BDNF locus. These phenomena can have a profound impact on detection of disease susceptibility genes and must be considered in gene association studies of BDNF.
Keywords: selection, diversity, gene association, single-nucleotide polymorphism, haplotype, susceptibility locus
Introduction
Brain-derived neurotrophic factor (BDNF) is a key neurotrophic factor important for neuronal development, survival, and plasticity1,2. A non-synonymous polymorphism in BDNF that leads to a valine to methionine change at position 66 of the proBDNF protein, referred to as Val66Met, has been shown to have an important role in neuronal and cognitive function3–10. BDNF is implicated as a genetic risk factor for several psychiatric disorders. We previously reported overrepresentation of the Val allele of the Val66Met polymorphism and lower frequency of a Met-carrying haplotype in BP patients11. Val66Met association with BP has received mixed support in subsequent studies (Table S1). A meta-analysis we performed using data from 14 case-control and parent-proband trio studies detected modest but statistically significant evidence for BDNF association with BP (P = 0.004)12. BDNF variants have also been associated with other psychiatric disorders including obsessive-compulsive disorder13–15, schizophrenia16–19, psychosis20, major depression21, anxiety22, and eating disorders23–26. Lack of association has also been reported for many of these disorders, however, raising the question of whether BDNF is in fact a disease susceptibility gene.
One explanation for discrepant association findings is differences in population genetic structure between patient samples. Isolation between human populations over generations has resulted in genetic differentiation marked by allele frequency and linkage disequilibrium (LD) differences. Thus, a single population may not accurately reflect variation in other populations, and within the same population some members may not be representative of others. Such differences could have a profound impact on detection of disease risk genes because association studies examine a fraction of gene polymorphisms, and rely extensively on LD between markers and susceptibility alleles to detect disease associations. Furthermore, if LD patterns vary dramatically between groups, population specific haplotypes will arise that could explain conflicting association results. Thus, assessment of the distribution of polymorphic markers and haplotypes worldwide is necessary to accurately interpret disease association data from different patient populations.
We therefore performed a population genetic study of the BDNF genomic region to determine whether population dynamics could explain the inconsistent psychiatric association findings. Our data provide evidence for substantial population diversity at the BDNF locus, as well as two putative selection events acting on BDNF alleles, that have significant implications for disease association studies of this gene.
Materials and Methods
Subjects
The study received approval from all appropriate Institutional Review Boards. DNA from the following collections was utilized: 12 multiplex pedigrees (93 individuals) obtained from the Centre d’Etudie Polymorphisme Humain (CEPH) repository27,28, the HGDR-CEPH Human Genome Diversity Cell Line Panel consisting of 1064 individuals from 57 worldwide populations29, and 31 parent-child trios from the Yoruba people of Ibadan, Nigeria. The CEPH sample overlaps by 50% the HapMap European (CEU) sample30.
SNP Genotyping
SNPs were selected from the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/), from our previous study11, and from the former Celera Discovery System database. Genotyping was performed by Sequenom MassARRAY (San Diego, CA) mass spectrometry as described previously11. SNP genotype data was subjected to the following quality control criteria prior to analyses: 1) >86% of attempted genotypes were successful (average 97% CEPH, >99% HGDR panel, 98% Yoruban), 2) founder alleles were in Hardy-Weinberg equilibrium (goodness of fit test P>0.05), 3) <2% of transmitted alleles had Mendelian inheritance errors, and 4) minor allele frequency >2%.
Haplotype Genealogy and Global Diversity
Twelve SNPs within the BDNF coding region were genotyped in the HGDR-CEPH Human Diversity Panel, 31 Yoruban parent-child trios, and 12 CEPH pedigrees. SNP identifiers are provided in Table S2. Maximum-likelihood expectation (MLE) haplotypes comprising the SNPs were reconstructed using an expectation maximization (EM) algorithm using data from all populations together to minimize artifact separation of the populations. SNP ancestral alleles were predicted from consensus genotypes from chimpanzee, gorilla, and orangutan DNA. Haplotype frequencies were determined for Africa, Asia, and Europe continental groups using all chromosomes that could be definitively classified into one of the groups (364 African, 444 Asian, and 418 European chromosomes).
The Sweep program (P. Varilly, B. Fry, and P. Sabeti; http://www.broad.mit.edu/mpg/sweep/) was used to create BDNF haplotype genealogical trees for the three major continental regions using all classifiable chromosomes. A map of global diversity of BDNF haplotypes was created by displaying pie charts representing the calculated haplotype frequencies for each population on a world map using the Diaspora software program (B. Fry, unpublished software). Fixation index (FST) values to assess population differentiation were calculated using Weir-Hill unbiased estimator31 for SNPs that were polymorphic in at least one population in the HapMap Phase II (release 21) dataset32. Population comparisons were performed by calculating FST values separately for the HapMap European (CEU), African (Yoruban - YRI), and Asian (Chinese and Japanese – CHB/JPT) populations. Significance of the FST values was determined by comparison to HapMap data.
Long-Range Haplotype Test
The Long-Range Haplotype (LRH) test33 was used to examine evidence for recent positive selection at BDNF. Genotype data were utilized from 68 SNPs (Table S2) spanning 500kb of the BDNF genomic region genotyped in 12 CEPH pedigrees, as well as genotype data from the International HapMap Project data release 1630. Haplotype blocks were defined according to Gabriel et al. 200234. MLE haplotypes were reconstructed using an EM algorithm. The LRH test detects evidence of an allele’s recent and rapid rise in prevalence in a population, based on disparity in the allele age estimated from its high frequency (characteristic of an old allele) and its long-range LD (characteristic of a young allele). Long-range association was measured by extended haplotype homozygosity (EHH)33. For a population of individuals sharing allele ‘t’, EHH at a distance ‘x’ from the locus is the probability that two random chromosomes carrying the allele are identical by descent for the entire interval from the locus to ‘x’. Correction for local variation in recombination rates was performed using the relative EHH (REHH), the factor by which EHH decays on the tested allele compared to all other alleles at a locus. The REHH and EHH were corrected by matching genetic distances of the loci using recombination rates from HapMap data. Decay of the extended ancestral haplotype on which the tested allele arose was visualized using the Bifurcator program (B. Fry; http://benfry.com/bifurcator/).
Results
Global diversity of BDNF SNPs
Analysis of BDNF diversity focused on the Val66Met polymorphism (rs6265) and 11 other SNPs within the BDNF coding region that together encompass the region containing all BDNF variants reported to be associated with BP (Val66Met, a functional repeat polymorphism located 1kb upstream of exon 535, 36, and a coding region haplotype11). All 12 SNPs had sizeable allele frequencies differences between Europe, Asia, and Sub-Saharan Africa continental groups. Notably, the derived Met allele of the Val66Met polymorphism (hereafter referred to as Met66) increased in frequency from 0.55% to 19.9% and 43.6% frequency in Sub-Saharan Africa, Europe, and Asia, respectively. Subsequent analysis of SNP distributions in each of 58 global populations identified considerable diversity in allele frequencies (Table S3), corroborating the trend observed in the continental groups. The Met66 frequency varied widely from 0–72% among populations (Figure 1), being virtually absent in all Sub-Saharan African and some American indigenous populations.
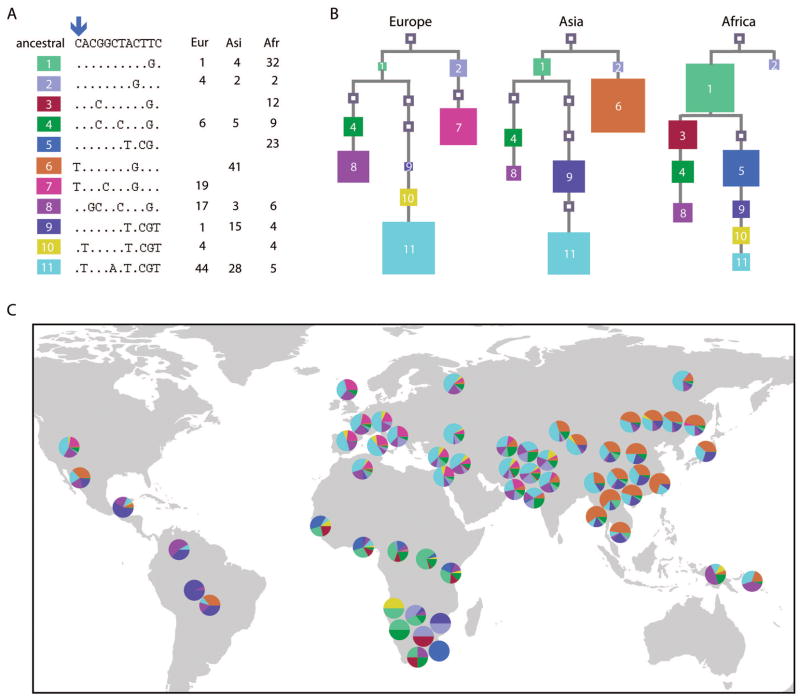
Figure 1.
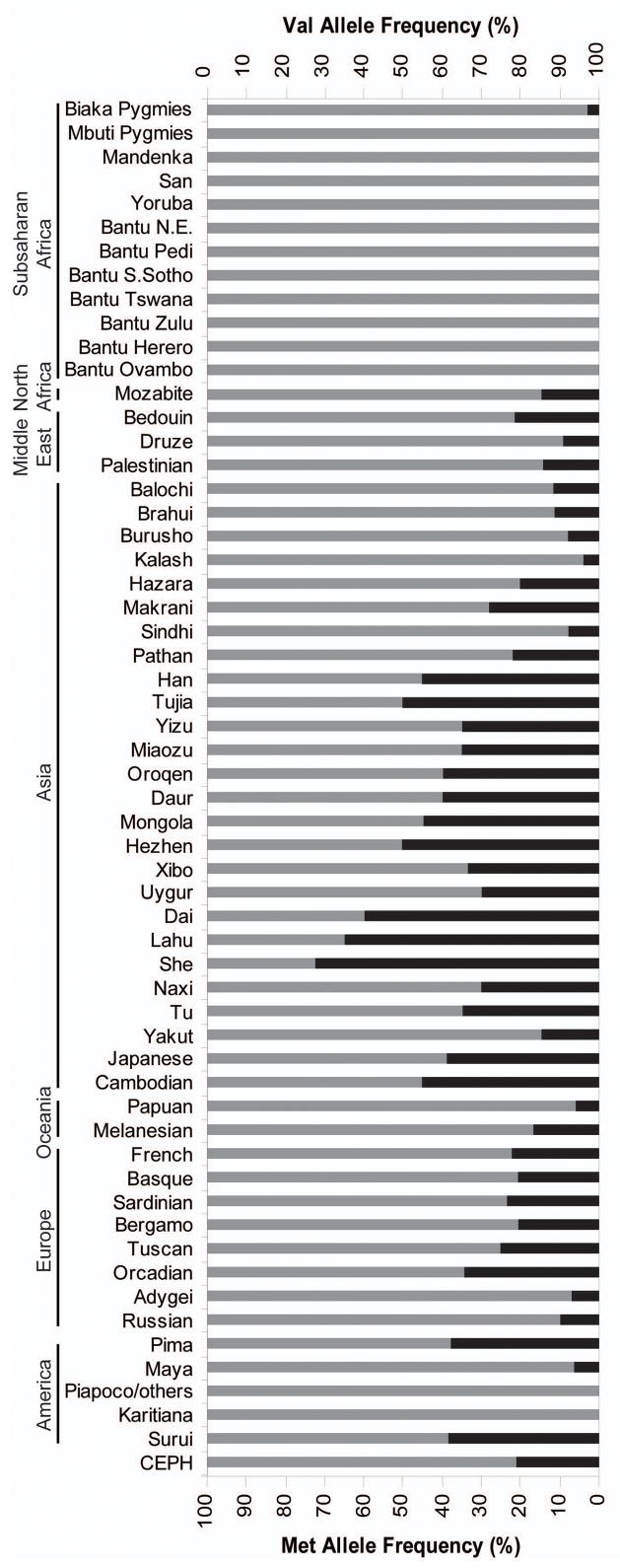
BDNF Val66Met allele frequencies in global populations. Frequencies of the Val66 allele (gray) and Met66 allele (black) are shown for each of the 57 populations in the HGDR-CEPH Human Diversity Panel and for CEPH pedigrees.
Population diversity of Val66Met was supported by comparison of FST values from the International HapMap Project. FST analyses detect population genetic differences that originated after the divergence of human populations, roughly 50,000 to 75,000 years ago37. Comparison of FST values of all non-synonymous SNPs in the genome, including Val66Met, to the distribution of all HapMap SNPs (synonymous and non-synonymous) determined that the Val66Met FST value (0.149) was in the 99.8th percentile and ranked 25th among >11,000 non-synonymous SNPs in the genome. This polymorphism also had high FST values when comparing the HapMap European, Asian (Chinese and Japanese treated as a single population), and Yoruban populations to each other (European vs. Asian, FST = 0.186, 86th percentile; European vs. Yoruban, FST = 0.168, 75th percentile; Asian vs. Yoruban, FST = 0.439, 94th percentile).
Global diversity of BDNF haplotypes
We subsequently examined population diversity of haplotypes comprised of the 12 BDNF SNPs. Eleven haplotypes observed above 2% frequency in at least one of the Europe, Asia, and Sub-Saharan Africa continental groups had clear frequency differences between the three groups (Figure 2A and Table S3). Met66 (“T” allele) was present on two haplotypes each exclusive to Asia and Europe (haplotype-6 and -7, respectively), and was not present on any Sub-Saharan African haplotypes. Haplotype-7 corresponds to the Met66-carrying haplotype we previously found associated with BP11. BDNF haplotype genealogical trees were constructed for each of the continental groups to clarify the population genetic structure (Figure 2B). The number and length of branches in the three trees were comparable, indicating similar history and mutational distance from the ancestral haplotype. An exception was the branch that contains haplotypes-6 or -7 in the Asian or European trees, respectively, which was shorter in the African tree due to having only one haplotype (haplotype-2). Haplotype-6 was found to be ancestral to haplotype-7, differing only in the allele of rs2049045 (of the SNPs we examined), with haplotype-6 having the ancestral “G” allele and haplotype-7 having the derived “C” allele (Figure 2A).
Figure 2.
Global diversity and genealogy of BDNF haplotypes. (A) BDNF haplotypes comprised of 12 coding region SNPs observed in European, Asian, and Sub-Saharan African continental groups. Val66Met/rs6265 is indicated by an arrow (C = Val, T = Met). Shown is the ancestral haplotype (top) and eleven haplotypes above 2% frequency in at least one of the continental groups (dots = ancestral alleles, nucleotide letters = derived alleles), with frequencies shown to the right. (B) BDNF haplotype genealogical trees in European, Asian, and Sub-Saharan African groups. Trees are rooted at the predicted ancestral haplotype (square at top) and each haplotype is shown as a square of size proportional to frequency and branch length proportional to mutational distance. Unfilled squares indicate unobserved haplotypes that are necessary links. (C) BDNF haplotype frequencies in global populations are indicated by pie diagrams placed relative to population geographic locations.
Examination of BDNF haplotypes among the global populations identified considerably different haplotype structure and frequencies between populations, but generally similar patterns within a geographic group (Figure 2C and Table S3). Of the Met66-carrying haplotypes, haplotype-6 ranged from 2–70% frequency in Asian, Oceania, and some Central and South American populations, whereas haplotype-7 was at 2–29% frequency in European, North African, Middle Eastern, and some Asian (solely Pakistani) populations. All together, these data further point toward considerable diversity of BDNF polymorphisms among worldwide populations.
Evidence for positive selection of haplotypes spanning the BDNF region
The observed population genetic differences in BDNF alleles may be due to local selection, which can have large effects on LD and thereby impact association studies. We therefore looked for evidence of selection across the BDNF genomic region within European-derived CEPH pedigrees using the Long-Range Haplotype (LRH) test33. This method detects evidence for more recent positive selection (<30,000 years old) compared to the FST analyses above. The LRH test identifies variants that appear to have recently arisen in a population, based on the extent of the surrounding ancestral chromosome, but that are prevalent in the population, suggesting natural selection may have caused a rapid rise in the allele’s frequency.
We examined 68 SNPs across a 500kb region on chromosome 11 spanning the BDNF, LIN7C, LGR4, and CCDC34 genes. A common 45% frequency haplotype located 5′ of BDNF had evidence for an extended ancestral haplotype, as measured by relative extended haplotype homozygosity (REHH) (see Methods). The long-range haplotype extending from this haplotype covers the entire genomic region examined (Figure S1A). The haplotype carries the Val66 allele and corresponds to haplotype-11 in Figure 2. Further evidence for selection of the BDNF haplotype was obtained by comparison of its REHH and frequency to the empirical distribution of chromosome 11 haplotypes from the HapMap European (CEU) data30. The haplotype REHH value was above the 99th percentile when compared to 1,652 chromosome 11 haplotypes with comparable frequencies (40–45%) (Figure S1B; uncorrected P = 4.6 ×10−4). This result indicates that the haplotype has more extensive LD given its frequency than expected under neutrality, which is suggestive of selection at the BDNF genomic region. Selection on this Val66-carrying haplotype is unlikely to explain the high Val66Met FST values above, since the elevated Met allele frequency outside of Africa appears to drive the FST result.
Discussion
Our study to gain insight into the potential impact of population diversity on association studies of BDNF revealed several lines of evidence suggesting the BDNF genomic region is not neutrally evolving. First, prominent differences were found among global populations in frequencies of the Val66Met variant and BDNF haplotypes. The derived Met66 allele ranged considerably from 0–72% frequency across populations, and exists on different population-specific haplotypes in Europeans and Asians. This substantial global diversity was supported by FST analyses in the HapMap populations, in which the Val66Met FST value was above the 99th percentile in the distribution of all SNPs in the genome. This putative selection signal appears to be driven by the derived Met allele and reflects its rise in frequency across Sub-Saharan African, European, and Asian populations. Second, the presence of young polymorphisms at high frequency in European-derived populations is indicative of a more recent selection event. A long-range haplotype detected at the BDNF locus carries the Val allele of Val66Met, and thus appears to be a different selection event than the Met allele signal identified in FST analyses. These data suggest that selection may have been operating on more than one BDNF allele or haplotype. It is important to note, however, that due to extensive LD, the long-range haplotype extends throughout the entire region examined, spanning BDNF, LIN7C, LGR4, and CCDC34, and it is therefore not possible to distinguish from which gene this selection event originates. Given the critical functions of BDNF in neuronal survival and plasticity1,2, selection may increase the prevalence of new variants that increase fitness by altering any of these functions.
The global diversity of Met66 and its rapid rise across Sub-Saharan African, European, and Asian populations is intriguing given its seemingly detrimental effect on neuronal and cognitive function4–10. In addition, Met66 is at lower frequency in BP and other psychiatric patients compared to controls (except in eating disorders), rather than at higher frequency as expected given its apparently deleterious function. As BDNF has diverse roles in neural development, cell survival, and synaptic plasticity1,2, however, it is possible that Met66 has pleiotropic effects, some detrimental and some beneficial. As discussed by Lipsky and Marini2, downregulation of synaptic plasticity by Met66, particularly in excitatory glutamatergic circuits, may be protective against psychiatric disorders, despite Met66 being associated with reduced cognitive performance4, 6–10.
The observed population genetic differences in BDNF have implications for interpreting the conflicting association literature for psychiatric disorders. If Val66Met is a true risk variant, its presence on different haplotypic backgrounds may produce different association patterns in different populations, which could account for the inconsistent association signals between studies. Alternatively, if Val66Met does not have a direct role in disease, but rather is in LD with the true associated allele to varying degrees in different populations, inconsistent association findings with Val66Met could also result due to population differences in LD patterns. It will be crucial to consider population genetic information when carrying out future disease association studies of this locus.
In conclusion, we have uncovered substantial global diversity and evidence for positive selection of the BDNF Val66Met polymorphism and BDNF region haplotypes. The unusual genetic diversity at the locus may explain the varying results between previous association studies using different patient populations. Further studies that take into account the population diversity in the BDNF genomic region are required to elucidate whether this gene has a role in major mental illness.
Supplementary Material
Supplementary information is available at the Molecular Psychiatry website.
Acknowledgments
We thank Dr. Manuel Ferreira for analytical assistance, and Dr. Roy Perlis, Dr. James Potash, Dr. Melvin McInnis, and Dr. J. Raymond De Paulo for valuable comments on the manuscript. The HGDR-CEPH Human Genome Diversity Cell Line Panel DNA was obtained from David Reich, M.D.. This study was supported by NIMH grant R01 MH062137, postdoctoral fellowships from the Canadian Institutes for Health Research (TLP) and the Charles A. King Trust, Bank of America, Co-Trustee (JBF), Young Investigator awards from the National Alliance on Schizophrenia and Depression (TLP, JBF), and a Burroughs Wellcome Career Award in the Biomedical Sciences (PCS).
References
- 1.Lu B, Pang PT, Woo NH. The yin and yang of neurotrophin action. Nat Rev Neurosci. 2005 Aug;6(8):603–614. doi: 10.1038/nrn1726. [DOI] [PubMed] [Google Scholar]
- 2.Lipsky RH, Marini AM. Brain-derived neurotrophic factor in neuronal survival and behavior-related plasticity. Annals of the New York Academy of Sciences. 2007 Dec;1122:130–143. doi: 10.1196/annals.1403.009. [DOI] [PubMed] [Google Scholar]
- 3.Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004 May 5;24(18):4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003 Jan 24;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006 Oct 6;314(5796):140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, et al. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003 Jul 30;23(17):6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dempster E, Toulopoulou T, McDonald C, Bramon E, Walshe M, Filbey F, et al. Association between BDNF val66 met genotype and episodic memory. Am J Med Genet B Neuropsychiatr Genet. 2005 Apr 5;134(1):73–75. doi: 10.1002/ajmg.b.30150. [DOI] [PubMed] [Google Scholar]
- 8.Tan YL, Zhou DF, Cao LY, Zou YZ, Wu GY, Zhang XY. Effect of the BDNF Val66Met genotype on episodic memory in schizophrenia. Schizophr Res. 2005 Sep 15;77(2–3):355–356. doi: 10.1016/j.schres.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Rybakowski JK, Borkowska A, Czerski PM, Skibinska M, Hauser J. Polymorphism of the brain-derived neurotrophic factor gene and performance on a cognitive prefrontal test in bipolar patients. Bipolar Disord. 2003 Dec;5(6):468–472. doi: 10.1046/j.1399-5618.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 10.Rybakowski JK, Borkowska A, Skibinska M, Szczepankiewicz A, Kapelski P, Leszczynska-Rodziewicz A, et al. Prefrontal cognition in schizophrenia and bipolar illness in relation to Val66Met polymorphism of the brain-derived neurotrophic factor gene. Psychiatry Clin Neurosci. 2006 Feb;60(1):70–76. doi: 10.1111/j.1440-1819.2006.01462.x. [DOI] [PubMed] [Google Scholar]
- 11.Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G, et al. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Mol Psychiatry. 2002;7(6):579–593. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- 12.Fan J, Sklar P. Genetics of bipolar disorder: focus on BDNF Val66Met polymorphism. In: Foundation N, editor. Novartis Foundation Symposium 289: Growth Factors and Psychiatric Disorders. Wiley; Chichester: 2008. pp. 60–73.pp. 87–93. [DOI] [PubMed] [Google Scholar]
- 13.Hall D, Dhilla A, Charalambous A, Gogos JA, Karayiorgou M. Sequence variants of the brain-derived neurotrophic factor (BDNF) gene are strongly associated with obsessive-compulsive disorder. American journal of human genetics. 2003 Aug;73(2):370–376. doi: 10.1086/377003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso P, Gratacos M, Menchon JM, Saiz-Ruiz J, Segalas C, Baca-Garcia E, et al. Extensive genotyping of the BDNF and NTRK2 genes define protective haplotypes against obsessive-compulsive disorder. Biological psychiatry. 2008 Mar 15;63(6):619–628. doi: 10.1016/j.biopsych.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Hemmings SM, Kinnear CJ, Van der Merwe L, Lochner C, Corfield VA, Moolman-Smook JC, et al. Investigating the role of the brain-derived neurotrophic factor (BDNF) val66met variant in obsessive-compulsive disorder (OCD) World J Biol Psychiatry. 2008;9(2):126–134. doi: 10.1080/15622970701245003. [DOI] [PubMed] [Google Scholar]
- 16.Muglia P, Vicente AM, Verga M, King N, Macciardi F, Kennedy JL. Association between the BDNF gene and schizophrenia. Mol Psychiatry. 2003 Feb;8(2):146–147. doi: 10.1038/sj.mp.4001221. [DOI] [PubMed] [Google Scholar]
- 17.Nanko S, Kunugi H, Hirasawa H, Kato N, Nabika T, Kobayashi S. Brain-derived neurotrophic factor gene and schizophrenia: polymorphism screening and association analysis. Schizophr Res. 2003 Aug 1;62(3):281–283. doi: 10.1016/s0920-9964(02)00349-3. [DOI] [PubMed] [Google Scholar]
- 18.Neves-Pereira M, Cheung JK, Pasdar A, Zhang F, Breen G, Yates P, et al. BDNF gene is a risk factor for schizophrenia in a Scottish population. Mol Psychiatry. 2005 Feb;10(2):208–212. doi: 10.1038/sj.mp.4001575. [DOI] [PubMed] [Google Scholar]
- 19.Qian L, Zhao J, Shi Y, Zhao X, Feng G, Xu F, et al. Brain-derived neurotrophic factor and risk of schizophrenia: an association study and meta-analysis. Biochem Biophys Res Commun. 2007 Feb 16;353(3):738–743. doi: 10.1016/j.bbrc.2006.12.121. [DOI] [PubMed] [Google Scholar]
- 20.Rosa A, Cuesta MJ, Fatjo-Vilas M, Peralta V, Zarzuela A, Fananas L. The Val66Met polymorphism of the brain-derived neurotrophic factor gene is associated with risk for psychosis: evidence from a family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2006 Mar 5;141(2):135–138. doi: 10.1002/ajmg.b.30266. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher J, Jamra RA, Becker T, Ohlraun S, Klopp N, Binder EB, et al. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biological psychiatry. 2005 Aug 15;58(4):307–314. doi: 10.1016/j.biopsych.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Lang UE, Hellweg R, Kalus P, Bajbouj M, Lenzen KP, Sander T, et al. Association of a functional BDNF polymorphism and anxiety-related personality traits. Psychopharmacology (Berl) 2005 Jun;180(1):95–99. doi: 10.1007/s00213-004-2137-7. [DOI] [PubMed] [Google Scholar]
- 23.Ribases M, Gratacos M, Fernandez-Aranda F, Bellodi L, Boni C, Anderluh M, et al. Association of BDNF with restricting anorexia nervosa and minimum body mass index: a family-based association study of eight European populations. Eur J Hum Genet. 2005 Apr;13(4):428–434. doi: 10.1038/sj.ejhg.5201351. [DOI] [PubMed] [Google Scholar]
- 24.Ribases M, Gratacos M, Fernandez-Aranda F, Bellodi L, Boni C, Anderluh M, et al. Association of BDNF with anorexia, bulimia and age of onset of weight loss in six European populations. Hum Mol Genet. 2004 Jun 15;13(12):1205–1212. doi: 10.1093/hmg/ddh137. [DOI] [PubMed] [Google Scholar]
- 25.Ribases M, Gratacos M, Armengol L, de Cid R, Badia A, Jimenez L, et al. Met66 in the brain-derived neurotrophic factor (BDNF) precursor is associated with anorexia nervosa restrictive type. Mol Psychiatry. 2003 Aug;8(8):745–751. doi: 10.1038/sj.mp.4001281. [DOI] [PubMed] [Google Scholar]
- 26.Dmitrzak-Weglarz M, Skibinska M, Slopien A, Szczepankiewicz A, Rybakowski F, Kramer L, et al. BDNF Met66 allele is associated with anorexia nervosa in the Polish population. Psychiatr Genet. 2007 Aug;17(4):245–246. doi: 10.1097/YPG.0b013e3280991229. [DOI] [PubMed] [Google Scholar]
- 27.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d’etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990 Mar;6(3):575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 28.Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, Manion F, et al. A comprehensive human linkage map with centimorgan density. Cooperative Human Linkage Center (CHLC) Science. 1994 Sep 30;265(5181):2049–2054. doi: 10.1126/science.8091227. [DOI] [PubMed] [Google Scholar]
- 29.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, et al. A human genome diversity cell line panel. Science. 2002 Apr 12;296(5566):261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 30.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005 Oct 27;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 32.The International HapMap Consortium. Second Phase of HapMap project completed. Pharmacogenomics. 2007 Nov;8(11):1489–1491. [Google Scholar]
- 33.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002 Oct 24;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 34.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002 Jun 21;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 35.Proschel M, Saunders A, Roses AD, Muller CR. Dinucleotide repeat polymorphism at the human gene for the brain-derived neurotrophic factor (BDNF) Hum Mol Genet. 1992 Aug;1(5):353. doi: 10.1093/hmg/1.5.353-a. [DOI] [PubMed] [Google Scholar]
- 36.Okada T, Hashimoto R, Numakawa T, Iijima Y, Kosuga A, Tatsumi M, et al. A complex polymorphic region in the brain-derived neurotrophic factor (BDNF) gene confers susceptibility to bipolar disorder and affects transcriptional activity. Mol Psychiatry. 2006 Jul;11(7):695–703. doi: 10.1038/sj.mp.4001822. [DOI] [PubMed] [Google Scholar]
- 37.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, et al. Positive natural selection in the human lineage. Science. 2006 Jun 16;312(5780):1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is available at the Molecular Psychiatry website.