Summary
MLL is involved in chromosomal rearrangements that generate fusion proteins with deregulated transcriptional activity. The mechanisms of MLL fusion protein-mediated transcriptional activation are poorly understood. Here we show MLL interacts directly with the Polymerase Associated Factor complex (PAFc) through sequences flanking the CxxC domain. PAFc interacts with RNA polymerase II and stimulates post-translational histone modifications. PAFc augments MLL and MLL-AF9 mediated transcriptional activation of Hoxa9. Conversely, knock down of PAFc disrupts MLL fusion protein-mediated transcriptional activation and MLL recruitment to target loci. PAFc gene expression is down regulated during hematopoiesis and likely serves to regulate MLL function. Deletions of MLL that abolish interactions with PAFc also eliminate MLL-AF9 mediated immortalization indicating an essential function for this interaction in leukemogenesis.
Highlights
PAFc interacts with the pre-CxxC and RD2 domains of MLL
Transcriptional activity of MLL and MLL fusion proteins is stimulated by PAFc
MLL fusion protein mediated transformation is dependent upon interaction with PAFc
PAFc expression is coordinately down regulated during myeloid differentiation
Significance
Translocations involving the MLL gene create potent oncogenic fusion proteins that account for up to 80% of infant acute leukemias, underscoring the importance of understanding the molecular mechanisms driving oncoprotein function. Using biochemical approaches we have identified direct physical interaction of the PAF complex (PAFc) with sequences invariably retained in MLL fusion proteins. At a molecular level we show PAFc is required for full MLL fusion protein mediated transcriptional activation and for proper recruitment of MLL to target genes. This study demonstrates that PAFc is an essential co-factor for MLL fusion proteins that is necessaryin vivo for cell immortalization and consequently may serve as an attractive new therapeutic target for leukemias with MLL rearrangements.
Introduction
MLL is a histone methyltransferase containing a C terminal SET domain that methylates histone H3 lysine 4, a mark commonly associated with gene activation (Milne et al., 2002; Nakamura et al., 2002; Strahl et al., 1999). MLL is required for normal embryonic development through proper maintenance of patterns of Hox gene expression (Yu et al., 1995). MLL also plays a central role in regulating hematopoetic stem cell self-renewal and progenitor expansion (Jude et al., 2007; McMahon et al., 2007). The protein is of particular biomedical importance as rearrangements involving MLL located at chromosome 11q23 are one of the most common genetic alterations in human leukemia. While the mechanisms are likely to differ, all rearranged forms of MLL positively up regulate expression of HOX genes including HOXA9 and the HOX cofactor MEIS1, which has been shown to be critical for transformation (Armstrong et al., 2002; Ayton and Cleary, 2003; Kumar et al., 2004). The most common MLL rearrangements are balanced translocations that account for up to 80% of infant acute leukemia and approximately 5–10% of adult acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL) cases (Aplan, 2006; Hess, 2004). The resulting oncogenic fusion proteins fuse approximately 1400 N-terminal amino acids of MLL in frame to one of over 60 different translocation partners (Krivtsov and Armstrong, 2007). The breakpoint cluster region in MLL invariably includes the CxxC domain and adjacent RD2 region in MLL fusion proteins, but deletes the downstream PHD fingers and SET domain (Zhang and Rowley, 2006). Because the SET domain is lost during rearrangement, MLL fusion proteins activate transcription through mechanisms dependent on the translocation partner. The most common of these fusion proteins involve translocations between MLL and nuclear translocation partners including AF9, AF4 and ENL among others that interact with a complex of proteins termed ENL Associated Proteins (EAPs) or a closely related complex called AEP for AF4 family/ENL family/P-TEFb complex (Yokoyama et al.). In addition to the most common MLL translocation partners, EAP includes the histone H3 lysine 79 methyltransferase hDOT1L and transcription elongation factors CDK9 and CyclinT1 (collectively known as pTEFb) (Krivtsov et al., 2008; Mueller et al., 2007).
Several structural domains have been identified in MLL and MLL fusion proteins that are required for transcriptional activity. Identifying these domains and their molecular interactions is important because of their promise for therapeutic targeting. For example, interaction of N-terminal sequences of MLL in a trimolecular complex with Menin and LEDGF is required for targeting the fusion protein to chromatin and for leukemogenicity (Caslini et al., 2007; Yokoyama and Cleary, 2008; Yokoyama et al., 2005). The DNA methyltransferase homology region or CxxC domain of MLL binds nonmethylated CpG islands and protects against DNA methylation (Ayton et al., 2004; Erfurth et al., 2008). Point mutations that block DNA binding by this region also block immortalization (Ayton et al., 2004).
In addition to the CxxC domain itself, regions immediately adjacent to the CxxC domain also appear to be important for MLL function. In the study by Bach et. al. involving domain swaps between MLL1 and MLL2, the CxxC domain along with flanking sequences of MLL1 were essential for immortalization by MLL fusion proteins (Bach et al., 2009). In particular immortalization was dependent on MLL sequences between aa 1149 and 1154 (the so called “pre CxxC domain) as well as sequences in the adjacent basic “post CxxC” or RD2 region between aa 1298 and 1337 (Ayton et al., 2004; Bach et al., 2009).
Increasing evidence shows that both histone H3 lysine 4 (H3K4) methylation, which is mediated by MLL among other methyltransferases, and histone H3 lysine 79 (H3K79) methylation mediated by DOT1L, which is recruited by MLL fusion proteins, is dependent on histone H2B mono-ubiquitination. The Polymerase Associated Factor complex (PAFc) plays a critical role in mediating H2B ubiquitination as well as promoting H3K4 and H3K79 methylation (Dover et al., 2002; Krogan et al., 2003; Ng et al., 2003; Sun and Allis, 2002). PAFc is composed of five subunits in mammals including PAF1, LEO1, CDC73, CTR9 and WDR61 (Rozenblatt-Rosen et al., 2005; Zhu et al., 2005a).
The role of PAFc in transcriptional regulation is beginning to be better defined. PAFc was originally identified as a protein complex in yeast that associates with RNA polymerase II (RNAP II) (Wade et al., 1996). The complex associates with both initiating (Ser5-phosphorylated) and elongating (Ser2-phosphorylated) RNAPII (Pokholok et al., 2002). Increasing evidence suggests that in mammals many genes controlling development, including the Hox genes, are regulated at the level of transcriptional elongation (Chopra et al., 2009). At such “pause prone” promoters, RNA polymerase II (RNAPII) interacts with two complexes that inhibit transcriptional elongation, Negative Elongation Factor (NELF) and DRB sensitivity inducing factor (DSIF). NELF is a complex of four subunits that binds to the unphosphorylated RNAPII C terminal domain (CTD) and induces proximal promoter pausing. DSIF is composed of a heterodimer of hSpt4 and hSpt5 that plays a dual role in both transcriptional activation and repression. hSpt5 contains six copies of the sequence G-S-R/Q-T-P (the C terminal repeats or CTR), which is similar in sequence to the RNAPII CTD Y-S-P-T-S-P-S. When the CTR is unphosphorylated, DSIF represses transcriptional elongation. As an early step in transcriptional activation, pTEF-b phosphorylates both the Pol II CTD as well as the hSpt5 CTR. CTD phosphorylation releases NELF. Importantly phosphorylation of the hSpt5 CTR in DSIF also promotes recruitment of PAFc (Liu et al., 2009).
Once recruited, PAFc then has two critical activities. One is the recruitment of the E2 ubiquitin ligase RAD6 and the E3 ubiquitin ligase BRE1 (in mammals hBRE1A/RNF20 and BRE1B/RNF40), which is mediated through either a direct or indirect interaction between PAF1 and BRE1 (Kim et al., 2009; Pavri et al., 2006). Importantly, the interaction with PAF1 not only recruits BRE1 and RAD6 but also stimulates the ubiquitinating activity of the heterocomplex for histone H2B lysine 120. In yeast this is followed by recruitment of the proteosomal ATPases Rpt4 and Sug1 (Rpt6) and methylation by the yeast MLL complex homolog COMPASS, which is recruited by interaction with monoubiquitinated histone H2B via the Cps35 subunit as well as through PAF1 (Krogan et al., 2003; Lee et al., 2007). However, the human homolog of Cps35, WDR82, doesn’t interact with MLL complexes (Wu et al., 2008), suggesting that MLL is recruited by alternative mechanisms.
In this study, we investigated the importance of PAFc in MLL transcriptional activation and MLL fusion protein transformation.
Results
PAFc interacts with the CxxC-RD2 domain of MLL
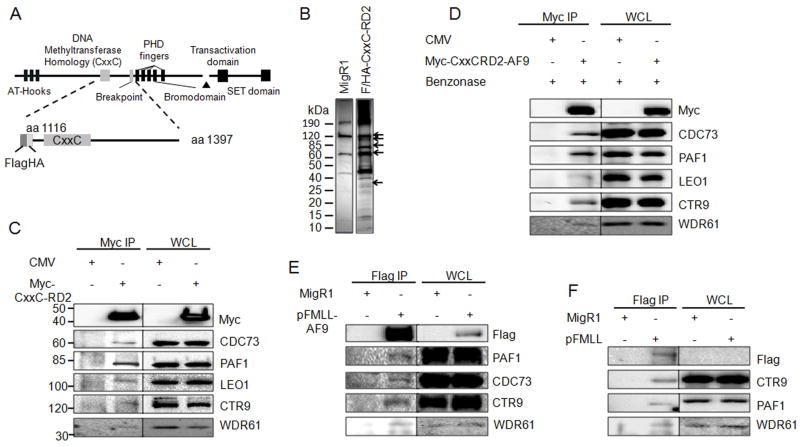
In order to identify proteins that associate with MLL CxxC-RD2, we expressed epitope tagged portions of this region transiently in human embryonic kidney 293 cells (Figure 1A). Flag-tagged CxxC-RD2 including a nuclear localization signal (NLS) was immunoprecipitated from transiently transfected 293 cells using M2 anti-Flag agarose beads. An “empty” expression vector with Flag epitope tag and NLS was also subjected to immunoprecipitation as a non-specific immunoprecipitation control. Coeluted proteins were resolved by SDS-PAGE (Figure 1B) and analyzed by mass spectroscopy. Multiple peptides corresponding to subunits of PAFc were identified with high probability including CTR9, LEO1, PAF1, CDC73 and WDR61 that correlated with silver stained bands at 133 kDa, 105 kDa, 75 kDa, 64 kDa and 34 kDa, respectively (Figure 1B). Each of the five PAFc subunits identified by mass spectrometry (PAF1, CDC73, CTR9, LEO1 and WDR61) was confirmed to co-immunoprecipitate with Myc-tagged CxxC-RD2 by western blotting in 293 cells (Figure 1C). To exclude the possibility of a DNA-mediated MLL-PAFc interaction and confirm that the PAFc interaction is preserved in the context of a fusion protein, immunoprecipitations were repeated with Myc-tagged CxxC-RD2-AF9 in 293 cells following Benzonase treatment (Figure S1A). Immunoprecipitation was preserved in the presence of Benzonase indicating the interaction is not DNA dependent (Figure 1D). Furthermore, PAFc co-immunoprecipitated with Myc-CxxC-RD2-AF9 (Figure 1D), as well as, CxxC-RD2 (Figure S1B). These experiments were repeated with transfection of an expression vector for Flag-tagged full-length MLL or Flag-tagged MLL-AF9 into 293 cells followed by immunoprecipitation and western blotting (Figure 1E, 1F). We also confirmed the PAFc interaction by co-immunoprecipitation of CDC73 with MLL-ENL in the KOPN8 cell line (Figure S1C). Together, these experiments show the MLL-PAFc interaction is maintained both in the context of full length (MLLN) as well as in the context of a leukemogenic MLL fusion protein (Figure 1E, 1F, S1C).
Figure 1. MLL binds to the PAF complex in a DNA independent manner.
A) Schematic diagram of the full length MLL protein with key domains indicated. The Flag/HA tagged CxxC-RD2 MLL fragment used for immunoprecipitation is shown below. The first and last amino acids of the protein fragments are indicated. B) MigR1 and Flag/HA-tagged CxxC-RD2 was expressed in 293 cells and immunoprecipitated. Immunoprecipitates were analyzed by SDS-PAGE and visualized by silver staining. Arrows indicate bands at the predicted molecular weights of the PAF complex components. C) Immunoprecipitation and western blot of the PAF complex following immunoprecipitation of Myc tagged CxxC-RD2 or Myc tag control from transiently tranfected 293 cells. D) The experiment described in C was repeated after treatment of the lysate with benzonase to digest DNA indicating the PAFc interaction with CxxC-RD2 is DNA-independent. Myc tagged CxxC-RD2-AF9 but not the Myc tag alone interacts with PAFc subunits showing that PAFc binding to CxxC-RD2 is maintained in the presence of the AF9 translocation partner. E and F) A full length flag tagged MLL-AF9 fusion protein (E) or Flag tagged full length MLL (F) was expressed in 293 cells and immunoprecipitated. The PAF complex stably associated with full length MLL fusion protein and MLL as indicated by western blot. (See also Figure S1)
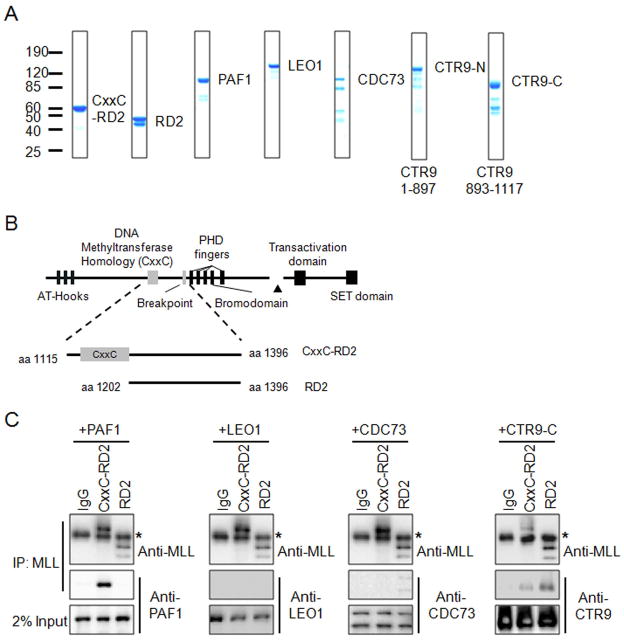
To determine if the interaction between MLL and PAFc is direct and to identify the PAFc subunit(s) involved, we bacterially expressed and purified the MLL CxxC-RD2 region (amino acids 1115 – 1396) and the RD2 region alone (amino acids 1202 – 1396) for in vitro pull-down experiments with bacterially expressed PAF1, LEO1, CDC73 and CTR9 (expressed as N and C terminus proteins termed CTR9-N and CTR9-C) (Figure 2A and 2B). These in vitro immunoprecipitations were performed with either CxxC-RD2 or RD2 with MLL antibodies after incubation with individual components of PAFc. Strong interaction of PAF1 with CxxC-RD2 but not the RD2 region was detected indicating an interaction between PAF1 and amino acids 1115 and 1201 of MLL (Figure 2C). These findings are consistent with our in vivo immunoprecipitation experiments in which PAFc co-immunoprecipitated with a small fragment of MLL N-terminal to the CxxC domain (amino acids 1115 – 1154) (Figure 3, see below). We also detected a second interaction between CTR9-C and both the MLL CxxC-RD2 and RD2 regions (Figure 2C). In addition a weak association of CDC73 with RD2 was also detected. These interactions were confirmed by reciprocal MBP pull down of CDC73, CTR9-N and CTR9-C, which showed that the MLL CxxC-RD2 and RD2 regions specifically associated with CTR9-C, along with a weak interaction detected between RD2 and CDC73 (Figure S2).
Figure 2. PAF1 and CTR9 bind directly to the CxxC-RD2 region of MLL.
A) Coomassie blue staining of bacterially purified His-MOCR tagged CxxC-RD2, RD2, PAF1, LEO1, and His-MBP tagged CDC73, CTR9-N and CTR9-C. The amino acids of CTR9-N and CTR9-C are indicated. B) Schematic diagram of MLL and bacterially purified CxxC-RD2 and RD2 regions. Starting and ending amino acids are indicated. C) Immunoprecipitations performed with bacterially purified recombinant CxxC-RD2 or RD2 and PAF complex components. Individual PAF components were incubated with either CxxC-RD2 or RD2 and immunoprecipitated with MLL antibodies. PAF components and immunoprecipitated MLL fragments were detected with the indicated antibodies by western blot. Asterisk denotes detection of the IgG heavy chain. (See also Figure S2)
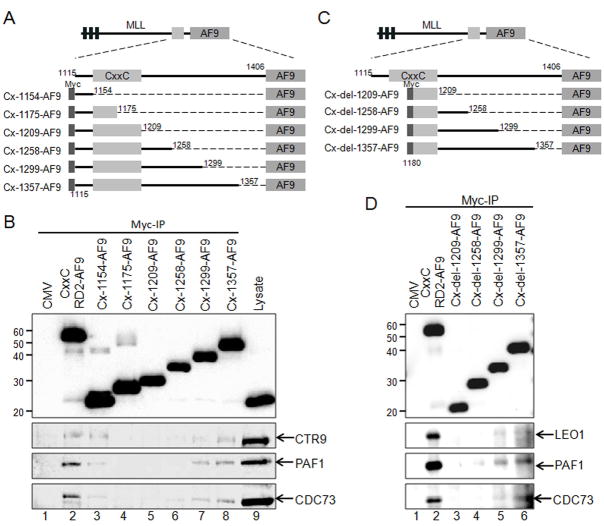
Figure 3. Amino Acids within the RD2 region are necessary for MLL interaction with the PAF complex.
A) Schematic of Myc-tagged CxxC-RD2-AF9 constructs made with serial deletions of the RD2 region. The first and last MLL amino acid retained in the expression constructs are indicated. All constructs include the AF9 fusion partner at the C-terminus and Myc tag at the N-terminus. The Cx-1154-AF9 through Cx-1357-AF9 constructs are named according to the last MLL residue retained in the fusion protein. B) Myc-tagged RD2 deletion constructs described in A were expressed in 293 cells followed by immunoprecipitation. The Myc-tagged MLL deletion proteins and associated PAF complex components were detected by western blot with the indicated antibodies. C) Schematic of a second set of deletion constructs made through the RD2 region that also delete the pre-CxxC domain. Cx-del-1209-AF9 through Cx-del-1357-AF9 were made by deleting amino acids 1115 through 1179 of MLL. The amino acids included in each construct are indicated. D) Myc tagged constructs described in C were expressed in 293 cells, immunoprecipitated and detected as described in B.
We then performed a series of deletion experiments to further map the MLL residues that participate in the MLL-PAFc interaction. Expression vectors for Myc-tagged CxxC-RD2-AF9 deletion mutants spanning the RD2 region of MLL (Figure 3A, Cx-1154-AF9 through Cx-1357-AF9) were transiently transfected into 293 cells and tested for PAFc interaction. These experiments reveal a sharp decrease in the MLL-PAFc interaction when C terminal deletions were made past amino acid 1299 (Figure 3B, compare lanes 6 and 7). To overcome the residual low level binding of PAFc with proteins deleted at amino acids 1209 or amino acids 1258, believed to be the result of pre CxxC interaction (Figure 2), we repeated this experiment with a set of deletion constructs that begin with MLL amino acid 1180 thereby deleting the proximal site of PAFc interaction (Figure 3C, Cx-del-1209-AF9 through Cx-del-1357-AF9). These experiments showed PAFc interaction with MLL is completely eliminated with deletions beyond amino acid 1299 (Figure 3D, compare lanes 4 and 5). Together, our data suggest the MLL-PAFc interaction is multivalent involving residues of MLL both in the pre CxxC domain as well as the RD2 region. Furthermore, the binding of PAFc by both pre and post CxxC domains is consistent with the solution structure of the MLL CxxC domain (Protein Database structure, 2J2S) (Allen et al., 2006), which shows the DNA binding CxxC domain coordinates two zinc atoms thereby bringing the pre and post CxxC regions into close opposition (Figure 8D).
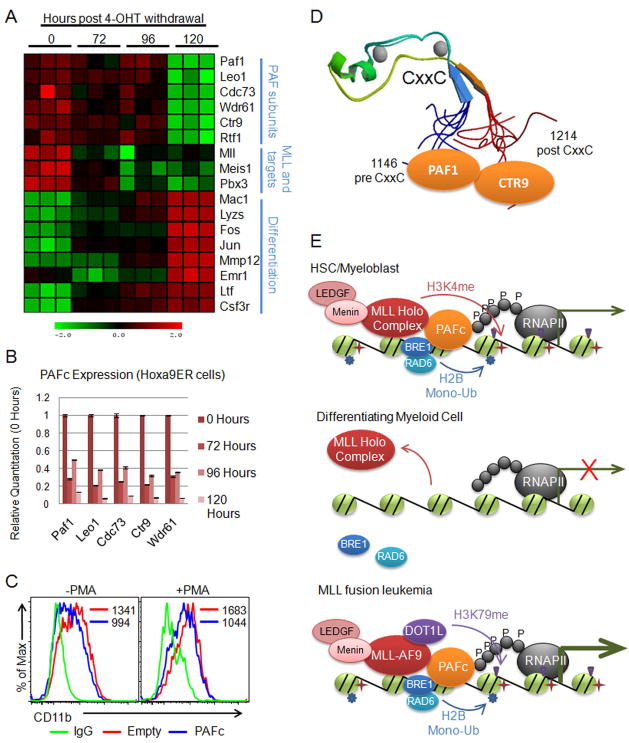
Figure 8. PAFc is down regulated during differentiation of hematopoietic cells.
A) Heat map generated from expression array data collected from differentiation of the Hoxa9ER cell line. Data is shown in triplicate for each time point following tamoxifen withdrawal. Labels indicate components of the PAF complex, MLL and MLL targets, and genes associated with myeloid cell differentiation. B) Expression of PAF components were verified using qPCR by separately differentiating the Hoxa9ER cell line by tamoxifen withdrawal. Time points indicate hours post tamoxifen withdrawal that RNA was collected. Expression for each component is shown relative to 0 hours. Error bars indicate +/− SD. C) THP-1 cells were transfected with an equal mixture of PAFc expression vectors or empty vectors along with a GFP vector at a 5:1 ratio (PAFc/Empty:GFP). Half were treated with PMA to induce differentiation. Surface expression of CD11b was monitored by FACS in the GFP positive gated cell population to track differentiation. Mean fluorescence values are shown for each sample. D) Structure of the CxxC domain and flanking sequences (PDB code: 2JYI). Solution structure of MLL CxxC region shows the flanking sequences of the CxxC domain brought into close juxtaposition creating a binding surface for both PAF1 and CTR9 of the PAF complex. E) Model for myeloid cell differentiation showing (top) PAFc dependent recruitment of MLL to target genes in hematopoietic progenitors (HSC/myeloblasts). PAFc and MLL recruitment promotes H2B mono-ubiquitination and H3K4 and H3K79 methylation resulting in transcriptional activation. (Middle) More differentiated myeloid cells down regulate PAFc expression resulting in decreased recruitment and transactivation by MLL. (Bottom) In leukemic cells harboring MLL fusion proteins the fusion protein synergizes with PAFc resulting in robust transcriptional activation of target genes. This is associated with increased H2B mono-ubiquitination and histone H3K79 methylation by DOT1L recruited through the MLL fusion partner. (See also Figure S6)
PAFc stimulates transcriptional activity by MLL and MLL fusion proteins
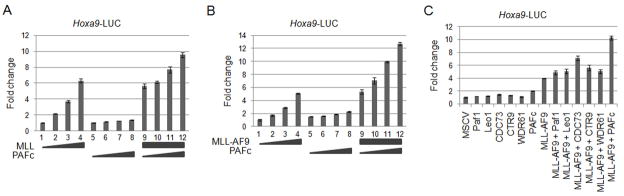
We then tested whether PAFc affects the transcriptional output mediated by MLL and the MLL-AF9 fusion protein. Dual luciferase assays were performed in 293 cells transfected with a luciferase reporter construct under the transcriptional control of the murine Hoxa9 promoter (Hoxa9-LUC). These experiments showed that transcriptional activation by wild type MLL is enhanced by co-expression of the five PAFc subunits (Figure 4A). Consistent with our earlier finding (Milne et al., 2002), we observed a dose dependent transcriptional activation of the Hoxa9 promoter by expression of increasing amounts of MLL-AF9 (Figure 4B). Furthermore, we observed a dose-dependent augmentation of MLL-AF9 dependent transcription of the Hoxa9 promoter when increasing amounts of PAFc were expressed. Notably, expression of PAFc alone had little effect on the Hoxa9 promoter in our assay (Figure 4A, 4B). A similar trend was observed when using an MLL-AF9 responsive luciferase construct containing a thymidine kinase promoter and multimerized Myc E-boxes (Figure S3A, S3B). Furthermore, we did not observe augmented transcription when single PAF components were introduced (Figure 4C). Deletion of the RD2 region to amino acid 1258 (which abrogates PAFc binding [Figure 3]) in an MLL-AF9 fusion protein also diminishes transcriptional activation (Figure S3C, compare lanes 5 and 6). Deletion of RD2 past aa1299 diminishes transactivation despite the more proximal PAFc interaction site and CxxC domain remaining intact, demonstrating the importance of the multivalent interaction for transcription. Together, these findings show MLL and MLL-AF9 synergize with PAFc to augment transcription of target genes.
Figure 4. The PAF complex synergizes with MLL and MLL-AF9 to augment transcriptional activity.
A) Luciferase assays were performed with the Hoxa9-LUC reporter construct and increasing doses of full length MLL (lanes 1–4) (0 – 0.6 μg) or PAFc (lanes 5–8) (0 – 0.6 μg). PAFc includes equal amounts of PAF1, LEO1, CDC73, CTR9 and WDR61. Lanes 9–12 show constant MLL (0.6 μg) with increasing doses of PAFc (0 – 0.6 μg). All changes are shown relative to lane 1 which includes Hoxa9-LUC and an empty expression vector. Error bars indicate +/− SD. Results of one of more than three representative experiments performed are shown. B) Experiment was performed as described in A except increasing doses of MLL-AF9 (0 – 0.6 μg) were used in lanes 1–4. Lanes 5–8 show increasing doses of PAFc (0 – 0.6 μg). Lanes 9–12 show increasing doses of PAFc (0 – 0.6 μg) in the presence of constant MLL-AF9 (0.6 μg). Error bars indicate +/− SD. One of three representative experiments is shown. C) Luciferase assay performed in transfected 293 cells using the Hoxa9-LUC reporter construct and individual PAFc components in lanes 1–6, the PAF complex (lane 7), MLL-AF9 alone (lane 8), MLL-AF9 plus individual PAF components (lanes 9–13) and MLL-AF9 with the PAF complex (lane 14). Error bars indicate +/− SD. Results of one of more than three representative experiments are shown. (See also Figure S3)
MLL fusion protein mediated transformation is dependent upon interaction with PAFc
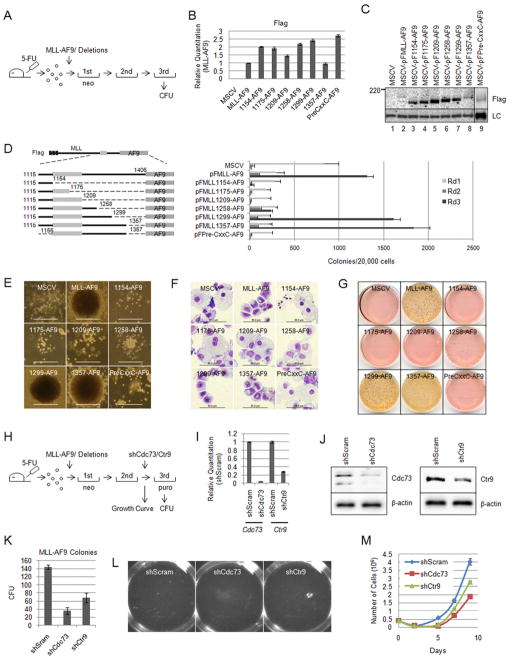
We then tested whether the transforming potential of MLL fusion proteins is dependent on the MLL-PAFc interaction. The deletions in the MLL RD2 region tested in immunoprecipitations described above (Figure 3A) or the pre-CxxC region were cloned into full length MLL-AF9 in MSCV-based retroviral vectors (Figure 5D). These were packaged in Plat-E cells and transduced into 5-FU primed bone marrow and analyzed in methylcellulose replating assays as previously described (Morita et al., 2000). Briefly, transduced cells were cultured under G418 selection in the presence of IL3, IL6, GM-CSF and SCF and colonies quantitated after the first, second and third rounds of replating (Figure 5A). Western blotting confirmed proteins of the predicted molecular weights were expressed following transient transfection of Plat-E cells (Figure 5C). In addition Real Time PCR confirmed expression of fusion gene mRNA in retrovirally transduced bone marrow (Figure 5B).
Figure 5. The PAFc interaction surface on RD2 is necessary for bone marrow transformation by MLL-AF9 fusion proteins.
A) Schematic diagram for MLL-AF9 and MLL-AF9 deletion colony forming assay. B) Real time qPCR using primers for the Flag tag on the MLL-AF9 fusion proteins. qPCR was performed on cDNA bone marrow cells after retroviral transduction. Expression levels are shown relative to MLL-AF9 transduced cells. Error bars indicate +/− SD. C) Western blot for the MSCV-based Flag tagged MLL-AF9 deletion constructs shown in D. Protein was extracted from transfected Plat-E cells and immunoprecipitated with M2 agarose beads. Proteins were detected with Flag antibodies. The loading control (LC) shows equal loading of protein. D) The constructs used for the retroviral infection and bone marrow colony assay are shown on the left. Final amino acids of the MLL deletions are shown. Primary, secondary and tertiary colony counts are shown for methylcellulose colony assays performed with the indicated MLL-AF9 fusion proteins. Error bars indicate standard deviation from duplicate experiments. One of more than three representative experiments is shown. E) Representative colony morphology is shown for each transduced MLL-AF9 fusion protein. Dense colonies are indicative of transformation while diffuse colonies indicate differentiation. Scale bar = 500μm F) Wright-Giemsa stained cytospins on cells isolated after the third round of methylcellulose plating. Scale bars indicate 50 μm. G) p-iodonitro tetrazolium violet (INT) stained colonies after three rounds of colony assay replating. Dense red colonies are visible from MLL-AF9, MLL-1299-AF9 and MLL-1357-AF9 transduced bone marrow. H) Schematic diagram for MLL-AF9 colony assay with shRNA mediated knock down of Cdc73 and Ctr9. Colony assays were performed as described in A except for an additional transduction after the second replating with shScram, shCdc73 and shCtr9 retrovises followed by plating in methylcellulose with puromycin selection. Colonies were scored following the third plating. I) Real Time qPCR was used to confirm knock down of Cdc73 and Ctr9 mRNA compared to shScram following transduction of 3T3 cells. Error bars indicate +/− SD. J) Protein lysate was collected from 3T3 cells transduced with shScram, shCdc73 and shCtr9 and separated by SDS-PAGE. Immunoblotting with Cdc73 and Ctr9 confirms knock down of the respective proteins. β-actin was probed as a loading control. K) Third round colony counts following transduction with shScram, shCdc73 or shCtr9. Error bars indicate +/− SD. L) Dense colonies are visible from colony assay plates containing shScram transduced cells. Significantly reduced colony numbers were visible from the shCdc73 and shCtr9 transduced cells. M) Cells collected after the second transduction with shRNA were grown in liquid culture in the presence of IL3, SCF and puromycin. A significant proliferative advantage was observed from MLL-AF9 cells transduced with shScram compared with shCdc73 or shCtr9. (See also Figure S4)
As shown in Figure 5D and 5G, similar numbers of tertiary colonies were observed when cells were transduced with MLL-AF9, MLL-1357-AF9 or MLL-1299-AF9. Further deletions of RD2 that extended more proximally than amino acid 1299, which markedly reduced PAFc interaction in our immunoprecipitation experiments (Figure 3), resulted in marked decreases in colony numbers (Figure 5D and 5G). Importantly, the morphology of these colonies was also dramatically different. Tertiary colonies from MLL-AF9, MLL-1357-AF9 and MLL-1299-AF9 all displayed a dense, compact morphology indicative of transformation (Lavau et al., 1997) (Figure 5E). Wright Giemsa-stained cytospins showed these compact colonies were composed of myeloblasts (Figure 5F). In contrast, transductions of constructs with more extensive deletions resulted in diffuse colonies composed of differentiating myeloid cells including monocytes and macrophages (Figure 5E, 5F and 5G). Of note, MLL-1258-AF9 retained a limited capacity to produce dense colonies after tertiary replating, but colony numbers were significantly reduced compared to MLL-AF9, MLL-1357-AF9 and MLL-1299-AF9 (Figure 5D, 5E and 5G). In keeping with this, minimal binding of PAFc was observed with the Cx-1258-AF9 construct (Figure 3). As expected, MLL-AF9, as well as, 1357 and 1299 deletions markedly up regulated Hoxa9 expression, while forms incapable of PAFc interaction did not (Figure S4B). The loss of MLL fusion protein mediated transformation upon deletion of the pre-CxxC and RD2 domains is comparable to the result of deleting the DNA binding CxxC domain (MLL-1154-AF9), which has been previously shown to be required for transformation (Ayton et al., 2004; Bach et al., 2009). To eliminate the possibility that the deletions introduced prevent interaction with the enzymatic activities necessary for leukemogenesis, we performed immunoprecipitations with the RD2 deletions constructs described in Figure 3. These experiments showed that association of DOT1L and the pTEFb component, Cyclin T1, with AF9 is maintained in all the deletion constructs used (Figure S4A). This suggests improper transformation activity is due solely to functions dependent on the RD2 region and not misfolding of proteins or deficient EAP complex recruitment (Figure S4A). To confirm PAFc is necessary for MLL-AF9 mediated transformation we performed colony assays by transducing bone marrow cells with MLL-AF9 followed by a second round of transduction with shRNA retroviruses directed against Cdc73 or Ctr9 (Figure 5H). Both shCdc73 and shCtr9 were confirmed to knock down Cdc73 and Ctr9, respectively, at the mRNA and protein level (Figure 5I and 5J). Knock down of either Cdc73 or Ctr9 resulted in significantly reduced colony formation compared to a scrambled control shRNA (Figure 5K and 5L). This data was confirmed using an established MLL-AF9 cell line (Figure S4C, S4D). We also observed a significantly reduced proliferation rate when either Cdc73 or Ctr9 was knocked down in primary cells grown in liquid culture in the presence of IL3 and SCF (Figure 5M). Reduced proliferation was also observed when Cdc73 or Ctr9 was knocked down in the MLL-AF9 cell line (Figure S4E). Together, these data suggest the PAFc interaction with MLL is crucially important for both MLL fusion protein mediated Hox deregulation and transformation.
PAFc promotes MLL and MLL fusion protein recruitment to target loci
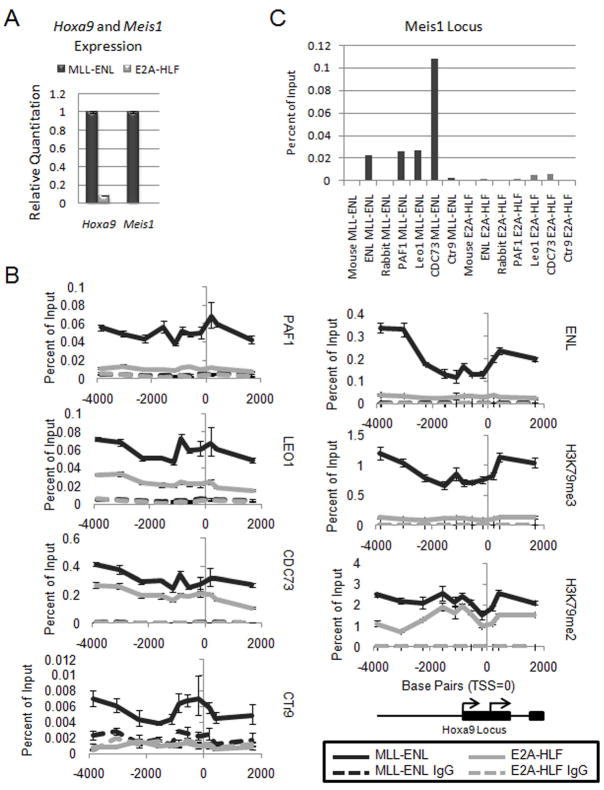
We then examined the localization of PAFc to a leukemogenic target gene of MLL such as Hoxa9. For these experiments we generated cell lines by transducing mouse bone marrow with either MLL-ENL or E2A-HLF. MLL-ENL cells express much higher levels of both Hoxa9 and Meis1 compared to E2A-HLF cells, which are not dependent on Hoxa9 expression for transformation (Ayton and Cleary, 2003) (Figure 6A). We performed chromatin immunoprecipitation (ChIP) experiments for PAF components, PAF1, LEO1, CDC73 and CTR9 at the Hoxa9 locus in both cell lines (Figure 6B). These experiments showed robust binding of MLL-ENL as detected by the ENL antibody in MLL-ENL cells compared to E2A-HLF cells (Figure 6B). Furthermore, levels of histone H3K79 di- and tri-methylation are markedly elevated in MLL-ENL cells consistent with ENL mediated recruitment of DOT1L (Mueller et al., 2007). Importantly, the pattern of binding of PAFc components and MLL-ENL across the locus is similar, consistent with an MLL-PAFc interaction (Figure 6B). Sequences downstream of the Hoxa9 locus also show a dramatic decrease in binding of both MLL-ENL and PAFc (Figure S5). MLL-ENL binding is somewhat more abundant upstream and downstream of the transcriptional start site which may reflect a PAFc independent function for MLL fusion proteins during transcriptional elongation (Figure 6B). Similarly, we also observed co-localization of MLL-ENL and PAFc at the Meis1 locus in MLL-ENL cells, with minimal binding seen in E2A-HLF cells (Figure 6C). Together, these data suggest that PAFc localizes with MLL fusion proteins to augment Hox and Meis1 transcription.
Figure 6. The PAF complex co-localizes with MLL-ENL across the Hoxa9 locus.
A) qPCR for Hoxa9 and Meis1 expression in MLL-ENL and E2A-HLF cell lines. Error bars indicate +/− SD. B) ChIP experiment performed in mouse bone marrow cell lines established with the MLL-ENL or E2A-HLF fusion protein. MLL-ENL IPs are shown in black and H2A-HLF IPs are shown in gray. Solid lines indicate the binding pattern of the component or histone modification listed to the right. The dotted lines indicate control IgG IPs for each cell line. The Hoxa9 locus is shown schematically at the bottom. C) Same experiment as described in B, but binding was determined on the Meis1 locus. The MLL-ENL IPs are shown in black (left) and the E2A-HLF IPs are shown in gray (right). (See also Figure S5)
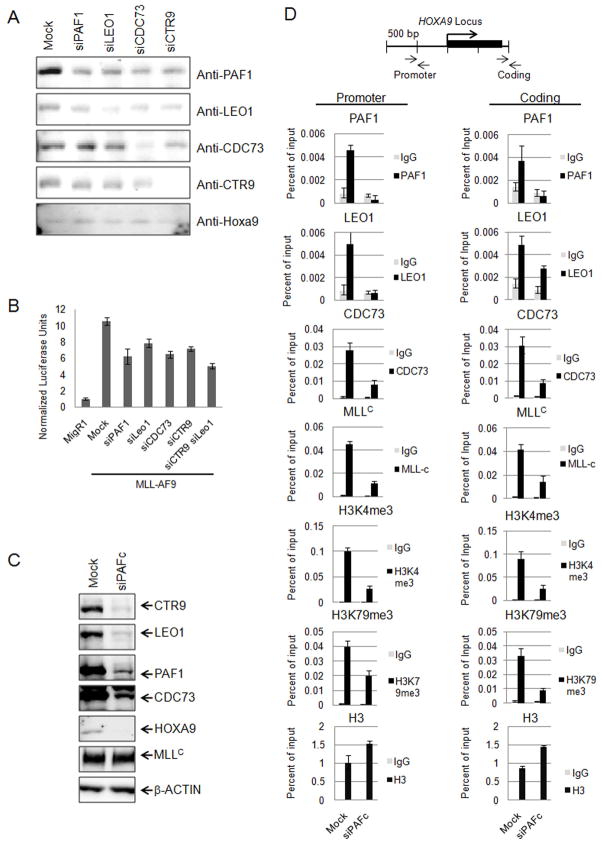
To further evaluate the role of PAFc in transcriptional activation we measured the effect of PAFc subunit knock down on MLL-AF9 mediated transcriptional activation. Knock down of PAF1, LEO1, CDC73 and CTR9 was successfully achieved in HeLa cells using siRNA transfection (Figure 7A). As previously reported, PAFc knock down decreases HOXA9 expression in HeLa cells (Figure 7A) (Zhu et al., 2005b). Importantly, MLL-AF9 transcriptional activation of Hoxa9-LUC is also impaired by PAF1, LEO1, CDC73 or CTR9 knock down (Figure 7B). Furthermore, we saw an additive effect by knocking down both CTR9 and LEO1 suggesting MLL fusion proteins require PAFc for efficient transcription of target genes (Figure 7B).
Figure 7. Knock down of PAFc reduces MLL-AF9 mediated transactivation and MLL binding to the HOXA9 locus.
A) siRNA mediated knock down of individual PAF components was verified by western blotting. Antibodies used for western blotting are shown on the right and siRNA is indicated on top. B) Luciferase assays were performed with the Hoxa9-LUC reporter construct and MLL-AF9 in HeLa cells after transfection with the indicated siRNA. Luciferase units are shown relative to the MigR1 control transfected cells. C) Simultaneous siRNA mediated knock down of PAFc (siCTR9, siLEO1, siPAF1, siCDC73) is shown by western blotting for PAF components after siRNA transfection of HeLa cells. β-ACTIN was detected as a loading control. D) ChIP experiments were performed in HeLa cells following simultaneous knock down of the PAF components shown in C. PAF1, LEO1, CDC73, MLLC, H3K4me3, H3K79me3 and H3 were immunoprecipitated from HeLa cells treated with siPAFc or non-targeting siRNA (Mock). Binding was assessed in both the promoter and coding region as indicated. A schematic of the HOXA9 locus and location of the primer-probe sets used for qPCR are shown. Error bars indicate +/− SD. One of more than three representative experiments is shown.
We then determined the effect of knock down of the PAF complex on MLL recruitment to the HOXA9 locus by performing ChIP assays on HeLa cells after simultaneous knock down of CTR9, PAF1, CDC73 and LEO1 (Figure 7C). We observed a significant decrease in binding of CDC73, PAF1 and LEO1 in both the promoter and coding region of the HOXA9 locus following knock down, as expected, while histone H3 levels remain unchanged or elevated in siPAFc-treated cells (Figure 7D). PAFc knock down resulted in a marked decrease in wild type MLL binding compared to mock treated cells (Figure 7D), without affecting MLL protein levels (Figure 7C), suggesting PAFc is necessary for MLL recruitment to HOXA9. Consistent with reduced binding of MLL, knock down of the PAF complex also resulted in a decrease in H3K4 tri-methylation at the HOXA9 locus (Figure 7D). We also observed a decrease in histone H3K79 tri-methylation (Figure 7D).
PAFc expression is coordinately down regulated during myeloid differentiation
The above results suggest that the PAF complex recruits MLL to the HOXA9 locus and that modulating PAFc levels may be an important mechanism for modulating MLL activity. It is noteworthy in this regard that a recent unbiased genome-wide siRNA screen identified PAFc subunits Ctr9, Wdr61 and Rtf1 amongst the 30 top genes regulating Oct4 expression and stem cell renewal (Ding et al., 2009). In this study PAFc was found to bind to key pluripotency genes, which is remarkable because MLL fusion protein transformed cells show, in addition to HOX gene over expression, a distinctive embryonic stem cell (ESC)-like pluripotency signature (Somervaille et al., 2009). Furthermore, expression of PAFc subunits is strongly regulated upon differentiation of ESCs into embryoid bodies (Ding et al., 2009). Collectively these findings suggested that PAFc maintains pluripotency in hematopoietic progenitors.
We established two differentiation models to explore the potential role of PAFc in regulation during hematopoietic differentiation. First, we created a conditional AML cell line by immortalizing murine bone marrow by transduction with Hoxa9-ER in the presence of tamoxifen (4-OHT). Upon 4-OHT withdrawal, these cells undergo differentiation and cell cycle arrest, which is largely complete by 120 hours (Figure S6A, S6B). 4-OHT withdrawal is accompanied by marked up regulation of c-Fos and c-Jun transcripts indicative of myeloid differentiation (Figure S6C). Microarray expression profiling was performed in triplicate at 24, 48, 72, 96 and 120 hours following 4-OHT withdrawal (only results for 72, 96 and 120 hours are shown in Figure 8A). These experiments showed a marked down regulation of all PAFc subunits and MLL target genes after induction of differentiation accompanied by up regulation of genes associated with myeloid differentiation (Figure 8A). The down regulation of PAFc expression was confirmed by qPCR in independently differentiated Hoxa9-ER cells (Figure 8B). We also analyzed the human HL-60 cell line, which rapidly differentiate into macrophages after exposure to phorbol 12-myristate 13-acetate (PMA) (Figure S6D). PMA treatment also led to a dramatic down regulation of PAFc in HL-60 cells (Figure S6E). To test the role of PAFc in hematopoietic differentiation, we enforced expression of PAFc in THP-1 cells with or without PMA induced differentiation. THP-1 cells were more resistant to differentiation, as determined by CD11b surface expression, when PAFc was over expressed (Figure 8C). Importantly, GFP negative cells showed no significant difference in CD11b expression (Figure S6F). Together, these data show PAFc expression is specifically down regulated during myeloid differentiation and that high level PAFc expression inhibits differentiation. Thus, PAFc may play an important role in regulating MLL binding to target genes during differentiation.
Discussion
The work presented here establishes PAFc as an important cofactor for both transcriptional regulation by MLL as well as for leukemogenesis mediated by MLL fusion proteins. Interestingly several PAFc components have been previously implicated in carcinogenesis. Most notably mutations in CDC73 (Parafibromin), encoded by HRPT-2 (hereditary hyperparathyroidism type 2), are responsible for the familial hyperparathyroidism-jaw tumor (HPT-JT) syndrome (Szabo et al., 1995). HPT-JT is an autosomal dominant disorder associated with hyperparathyroidism (HPT) and a high incidence of parathyroid adenomas, hyperplasias and carcinomas as well as renal abnormalities and uterine tumors (Newey et al., 2009). The mutations in HRPT-2 are predicted to lead to loss of function due to premature termination. The chromosome 1q25-q31 region spanning HRPT-2 frequently undergoes loss of heterozygosity in tumors arising in HPT-JT patients, suggesting that CDC73 functions as a tumor suppressor (Newey et al., 2009). Consistent with this role, over expression of wild type CDC73, but not a mutant form found in HPT, blocks cell proliferation and inhibits the cell cycle regulator cyclin D1 (Woodard et al., 2005).
However, other studies have implicated over expression of PAFc subunits in tumorigenesis. For example, CDC73 over expression in 293FT and COS7 cells increases S-phase entry and promotes cellular proliferation. This induction of CDC73-mediated cell dependent proliferation requires the SV40 large T-antigen, which is directly bound by CDC73 (Iwata et al., 2007). In a study to identify genes involved in pancreatic tumor progression, PAF1 was found to be over expressed as a result of a double minute amplification involving chromosome 19q13 (Batra et al., 1991). Furthermore, over expression of PAF1 results in transformation of NIH3T3 cells (Moniaux et al., 2006).
The dual roles of CDC73 as a tumor suppressor and an “aider and abettor” of an oncoprotein (see above) are shared with another MLL-interacting protein, Menin. Menin interacts with the N-terminus of MLL and is required for transformation by MLL fusion proteins (Caslini et al., 2007; Yokoyama et al., 2005). However, MEN1 mutations, which lead to loss of function of the Menin protein, are found in a variety of endocrine tumors including parathyroid hyperplasias and adenomas, as well as pancreatic islet tumor cells establishing Menin as a tumor suppressor (Chandrasekharappa et al., 1997; Lemmens et al., 1997). The similarities in the diseases associated with Menin and PAF subunit mutations (parathyroid and pancreatic islet tumors) raise the possibility that the mechanisms of oncogenicity may also be similar, perhaps mediated through cyclin dependent kinase deregulation as we have previously defined for Menin (Milne et al., 2005b). These results also suggest that MLL may play a broader role in tumorigenesis than previously expected. Additional experiments will be necessary to assess what role MLL plays in cancers associated with disruption or amplification of PAF components.
Recently, genome-wide siRNA screens identified components of the PAF complex as important in maintaining an embryonic stem cell identity. PAFc was found to bind and regulate several key pluripotency genes including Oct4 (Ding et al., 2009). Down regulation of PAFc caused embryonic stem cell differentiation suggesting its expression is required to maintain an ESC identity. Our experiments show a similar coordinate regulation of PAFc in hematopoietic cells, where PAF component expression is strongly down regulated during myeloid differentiation (Figure 8). Recent gene expression profiling experiments have shown that MLL fusion proteins enforce expression of an ESC gene expression signature including the expression of Myb, Hmgb3 and Cbx5, which is necessary for the maintenance of the leukemic stem cell phenotype (Somervaille et al., 2009). Our data suggests PAFc promotes MLL or MLL fusion protein recruitment to target loci in addition to its known role as a platform for recruitment of RAD6/BRE1 required for histone H2B ubiquitination and downstream histone H3K4 and H3K79 methylation (Figure 4 and 7). In this way MLL may be properly targeted to gene promoters in primitive hematopoietic cells, such as myeloblasts, through interaction with PAFc. During differentiation, down regulation of PAFc may be a mechanism for attenuating MLL binding to target genes as well as decreasing the histone H2B ubiquitination that is required for histone H3K4 and H3K79 methylation (Figure 8E). PAFc expression is high in leukemic cells allowing for synergistic activation of key ECS pluripotency genes that appear to underlie MLL mediated oncogenesis. It remains to be seen if MLL plays a role in enforcing the ECS signature that has been described for other particularly aggressive solid tumors (Ben-Porath et al., 2008; Gentles et al., 2009).
Our study identified two interaction sites between MLL and PAFc. The first involves an interaction with the MLL pre-CxxC domain between residues 1115–1154 and the PAFc subunit PAF1. In addition under our immunoprecipitation conditions we observed a second interaction with CTR9 that occurs through the RD2 region (Figure 2, S2 and 3). These findings are interesting in light of structural studies showing the CxxC domain of MLL forms a loop that interacts with DNA (Allen et al., 2006) thereby bringing the pre-CxxC sequence and the RD2 region into close juxtaposition in three dimensional space and creating a single binding surface for PAF1 and CTR9 (Figure 8D). It is intriguing in this regard that the region of RD2 necessary for a stable PAFc interaction is invariably conserved in MLL translocations, which involve breakpoints no more proximal than exon 8.
Our transformation assays indicate that both the pre CxxC and RD2 interaction domains of MLL are required for transformation. We hypothesize that although the interaction between MLL and PAFc can occur through single contact points, it is only fully stable when both sites are intact. Given the importance of each of these MLL-PAFc domains for transformation (Figure 5), small molecule inhibitors blocking either interaction would be expected to block oncogenesis by MLL fusion proteins. Given the general dependence of HOX expression on MLL, it is likely that inhibitors of this interaction will be effective in other leukemias showing high level HOX and MEIS1 expression, either as a result of MLL amplification or conceivably through other pathways. The experiments presented here lay the groundwork for determining the structure of the MLL-PAFc interacting domain and the ultimate development of effective therapy based on targeting this interaction.
Experimental Procedures
Cell Culture
293 and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%FBS and 1X non-essential amino acids. MLL-ENL and E2A-HLF cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 15% fetal calf serum (FBS) (Stem Cell Technologies). Hoxa9ER cells were cultured in IMDM supplemented with 15% FBS and 0.1% IL3. Plat-E cells were cultured in DMEM supplemented with 10% FBS. HL-60, THP-1, KOPN8 and K562 cells were cultured in RPMI-1640 medium supplemented with 10% FBS. Differentiation of HL-60 and THP-1 cells was induced by 10nM PMA treatment.
Luciferase Assay
293 cells were transiently transfected with MSCV MLL-AF9 (and derivatives), CMV-Renilla, and Hoxa9-LUC (or Myc-E box-LUC) constructs using FuGene 6 (Roche) according to manufacturer’s instructions. Cells were then serum starved in 0.5% FBS in OPTI-MEM media for 48 hours. Luciferase assays were performed using the Dual Luciferase assay kit (Promega) according to manufacturer’s instructions. Emission was detected using a Monolight 3010 (BD Biosciences).
Chromatin Immunoprecipitation
ChIP was performed as described previously (Milne et al., 2005a) using primary antibodies specific for MLLC (gift from Dr. Yali Dou), ENL (gift from Dr. Robert Slany), histone H3, H3K4 dimethylation, H3K4 trimethylation and H3K79 tri-methylation (Abcam) and Paf1, Leo1, Parafibromin and Ctr9 (Bethyl Laboratories Inc,. as described above). Quantitative real-time PCR was performed on the precipitated DNAs with TaqMan fluorescent labeling using primers and qPCR probes described in Supplemental Experimental Procedure. Binding was quantitated as follows: ΔCT = CT(input) − CT(Chromatin IP), % total = 2ΔCT.
siRNA Knockdown of PAFc
siRNA smart pools were obtained from Dharmacon for CTR9, CDC73, PAF1 and LEO1. siRNA transfection of HeLa cells was achieved using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions for analysis in luciferase assays. Oligofectamine (Invitrogen) was used according to manufacturer’s instructions for siRNA transfection of HeLa cells and PAFc knock down for ChIP assays.
Accession Numbers
Microarray data has been deposited in the Gene Expression Omnibus (GEO) repository from the National Center for Biotechnology Information (NCBI) with accession code GSE21299.
Additional information on Experimental Procedures can be found in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
A.G.M. is supported by a postdoctoral training grant from the National Institutes of Health (T32 HL07622). Y.H. is supported by a University of Michigan Computational Medicine and Biology Pilot Research Grant. J.L.H. is supported by the National Institutes of Health (CA92251) and by a SCOR grant from the Leukemia and Lymphoma Society. We thank Dr. David Allis for sharing data prior to publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. The EMBO journal. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplan PD. Chromosomal translocations involving the MLL gene: molecular mechanisms. DNA repair. 2006;5:1265–1272. doi: 10.1016/j.dnarep.2006.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nature genetics. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Molecular and cellular biology. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes & development. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK. Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene. 2009;28:815–823. doi: 10.1038/onc.2008.443. [DOI] [PubMed] [Google Scholar]
- Batra SK, Metzgar RS, Hollingsworth MA. Isolation and characterization of a complementary DNA (PD-1) differentially expressed by human pancreatic ductal cell tumors. Cell Growth Differ. 1991;2:385–390. [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer research. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science (New York, NY) 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia ZB, Stuart T, Diaz MO, Bushweller JH, Zeleznik-Le NJ. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7517–7522. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Alizadeh AA, Lee SI, Myklebust JH, Shachaf CM, Shahbaba B, Levy R, Koller D, Plevritis SK. A pluripotency signature predicts histologic transformation and influences survival in follicular lymphoma patients. Blood. 2009;114:3158–3166. doi: 10.1182/blood-2009-02-202465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends in molecular medicine. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Iwata T, Mizusawa N, Taketani Y, Itakura M, Yoshimoto K. Parafibromin tumor suppressor enhances cell growth in the cells expressing SV40 large T antigen. Oncogene. 2007;26:6176–6183. doi: 10.1038/sj.onc.1210445. [DOI] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Molecular cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. The EMBO journal. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Lemmens I, Van de Ven WJ, Kas K, Zhang CX, Giraud S, Wautot V, Buisson N, De Witte K, Salandre J, Lenoir G, et al. Identification of the multiple endocrine neoplasia type 1 (MEN1) gene. The European Consortium on MEN1. Hum Mol Genet. 1997;6:1177–1183. doi: 10.1093/hmg/6.7.1177. [DOI] [PubMed] [Google Scholar]
- Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Molecular and cellular biology. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, Schnepp RW, Krankel C, Livolsi VA, Gibbs D, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniaux N, Nemos C, Schmied BM, Chauhan SC, Deb S, Morikane K, Choudhury A, Vanlith M, Sutherlin M, Sikela JM, et al. The human homologue of the RNA polymerase II-associated factor 1 (hPaf1), localized on the 19q13 amplicon, is associated with tumorigenesis. Oncogene. 2006;25:3247–3257. doi: 10.1038/sj.onc.1209353. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene therapy. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Molecular cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Newey PJ, Bowl MR, Thakker RV. Parafibromin--functional insights. J Intern Med. 2009;266:84–98. doi: 10.1111/j.1365-2796.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Molecular cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Molecular and cellular biology. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Szabo J, Heath B, Hill VM, Jackson CE, Zarbo RJ, Mallette LE, Chew SL, Besser GM, Thakker RV, Huff V, et al. Hereditary hyperparathyroidism-jaw tumor syndrome: the endocrine tumor gene HRPT2 maps to chromosome 1q21-q31. Am J Hum Genet. 1995;56:944–950. [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Werel W, Fentzke RC, Thompson NE, Leykam JF, Burgess RR, Jaehning JA, Burton ZF. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Molecular and cellular biology. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer cell. 17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA repair. 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes & development. 2005a;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Molecular cell. 2005b;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.