Abstract
In this review we discuss the potential expectations, validity, predictive ability, and reality of pharmacogenetics in (i) titration of medication dose; (ii) prediction of intended (efficacy) drug response; and (iii) dose prediction of unintended (adverse) drug response. We expound on what these potential genetic predictors tell us and more importantly what they cannot tell us.
Although pharmacogenetic markers have been hailed as promising tools; these proclamations are based mainly on associations rather than their evaluation as predictors. To put the expectations of the promise of pharmacogenetics in a realistic perspective we review three examples. First warfarin pharmacogenetics, wherein although the validity of the genetic variant-dose is established and there is a validity of genetic variant-hemorrhage association, the clinical utility of testing is not clear. Second, the strong and clinically relevant HLA-Stevens Johnson syndrome/Toxic epidermal necrolysis association highlights the role of ethnicity. Third, the influence of CYP2D6 on tamoxifen efficacy, a model candidate with potential clinical utility, but unclear validity.
These examples highlight both the challenges and opportunities of pharmacogenomics. First, establishing a valid association between a genetic variation and drug response; second, doing so for a clinically meaningful outcome and third, providing solid evidence or rationale for improvement in patient outcomes compared to current standard of care
Keywords: Pharmacogenetics, Carbamazepine, Warfarin, Tamoxifen, CYP2C9, CYP2D6, HLA-B*1502
Introduction
It has long been recognized that patients have varied responses to drugs, both beneficial and adverse. Serious adverse drug reactions represent an important clinical issue and are an important cause of hospital admissions,(1–3) whereas lack of response to drug therapy, while not uncommon, leads to inefficient use of healthcare resources and delay in patients receiving appropriate alternative therapies.
Our increasing understanding of influences such as environmental exposures, nutritional status, co-morbidities, severity of disease, and concomitant medications has helped explain heterogeneity in drug response. In addition, the profound contribution of genetics has been appreciated for some time and is receiving greater emphasis. The technological advances spearheaded by the Human Genome Project now offer the opportunity for using genetic information to predict disease risk and drug response. Pharmacogenetics is the study of how genetic differences affect variation in response to medication. The promise (expectation) of pharmacogenetics is to be able to deliver “personalized medicine” by making decisions that optimize patient health outcomes based on a patient’s genetic makeup.(4)
Despite this promise, as with disease genetics, various widely cited pharmacogenomic association studies have not be reproduced and confirmed. For example, one study indicated a significant relationship between an alpha-adducin gene variant and diuretic antihypertensive response,(5) but several recent, larger studies failed to confirm such an association,(6–8) and the association between the CETP polymorphisms and statin therapy outcomes has been widely studied, but a recent meta-analysis failed to validate the association.(9) Furthermore, several pharmacogenomic associations that have not been consistently replicated to date including: ACE gene polymorphisms and antihypertensives,(10) beta-receptor polymorphisms and both asthma (11, 12) and heart failure medications,(13) and serotonin transporters and antidepressants.(14, 15)
The importance of sound epidemiologic approaches to assessing genetic associations has been verified by these experiences, including appropriately powered studies, assessment of potential selection bias and confounding, adjustment for multiple comparisons, careful assessment of phenotypes, and caution regarding publication bias.(16–18) More importantly, the recognition of the promise of genotype-guided therapy has fostered the development of multi-center, multinational consortiums such as the International Warfarin Pharmacogenomics Consortium (IWPC).(19) Such large efforts will continue to serve as a critical mechanism for providing the necessary sample sizes to identify and validate pharmacogenomic associations and evaluate their predictive ability.
Herein we discuss the potential expectations, validity, predictive ability, and reality of pharmacogenetics in (i) titration of medication dose; (ii) prediction of intended (efficacy) drug response; and (iii) dose prediction of unintended (adverse) drug response. We expound on what these potential genetic predictors can tell us and more importantly what they cannot tell us based on the current evidence and how this knowledge can set the research direction in informing the development of novel therapeutics. To this end we review several examples to highlight pharmacogenetic associations from an epidemiologic perspective. First warfarin pharmacogenetics, wherein although the validity of the gene-dose (surrogate endpoint) is established and there is a validity of gene-outcome (hemorrhage) association, the clinical utility of testing is not clear. Second, the strong and clinically relevant HLA-Stevens Johnson syndrome/Toxic epidermal necrolysis association highlights the role of ethnicity. Third, the association of CYP2D6 with the efficacy of tamoxifen highlights a model candidate with unclear validity but potential for clinical utility.
The long road from Association to Prediction
The extensive research efforts undertaken over the past decade have identified several genetic markers that are strongly associated with outcomes of interest. Although these pharmacogenetic markers have been hailed as promising tools; these proclamations are based mainly on associations rather than their evaluation as predictors. Therefore the expectations of their performance and ultimately the ability to improve drug therapy, patient outcomes, and healthcare spending need to be put in a realistic perspective.
At the crux of this debate are three questions:
Can a genetic risk factor (genetic marker) associated with an adverse (or beneficial) outcome be a clinically useful predictor of that outcome? (clinical validity)
Can incorporation of the genetic factor predict risk of the outcome more accurately than existing clinical models? (clinical utility)
Will the risks predicted for individuals be sufficiently different to warrant a change in treatment decisions? (degree of clinical utility)
Evaluating the relationship between variation in genetic factors and outcomes can be particularly challenging due to the varying study designs, differences in outcomes evaluated, variation in outcome definitions. Therefore the readers should familiarize themselves with evaluation of epidemiological studies with regard to potential sources of error: chance, bias and confounding. The readers should also understand the characteristics of predictive tests (sensitivity, specificity, positive predictive value and negative predictive value) and summary statistical measures that enable assessment of improvement in the predictive ability.(20)
Pharmacogenetics as a tool for predicting drug dosage
A majority of the early research in pharmacogenetics focused on drug metabolizing enzymes and identified common polymorphisms in patients exhibiting unusual adverse drug response to conventional doses. Many of these gene-dose associations have been replicated in independent populations and provide perhaps the greatest potential for realization of the “Personalized Medicine” promise.
There are relatively few examples of genetic variation influencing drug dosage that are well-validated across different racial/ethnic/geographic groups as with the case of warfarin. The effect of Cytochrome P450 2C9 (CYP2C9, the principal enzyme in warfarin metabolism) and Vitamin K epoxide reductase complex 1 (VKORC1, the target protein inhibited by warfarin to produce therapeutic anticoagulation) variants on warfarin dose requirements is probably the most well studied.(21)
Current warfarin dosing practice involves administration of a standard “one size fits all” starting dose (e.g. 5mg/day) or estimation of initial dose based on clinical characteristics (age, gender, medications, liver function, etc.). Dose adjustment is then based on anticoagulation (as measured by the international normalized ratio; INR) response with the goal of maintaining INR in the target range. However these dosing strategies result in over-anticoagulation or under-anticoagulation in a significant proportion of patients. Therefore the ability to improve the accuracy of dose prediction could potentially improve anticoagulation control and decrease the risk of thrombotic or hemorrhagic events associated with under-anticoagulation or over-anticoagulation.
CYP2C9 genotype alone accounts for 2% to 10% of the variance in warfarin dose,(22, 23) and VKORC1 genotype alone accounts for 10% to 25%, and non-genetic factors (including age, body size, and concomitant medications) account for 20% to 25%. Integration of these factors further improves the explanatory power, accounting for up to 60% of the variability in dose. Although the explanatory power is higher for Caucasians and Asians, compared to African Americans, the direction of the associations and the predictive ability has been validated. The randomized clinical trail by Anderson and colleagues showed that incorporation of CYP2C9 and VKORC1 genotype significantly improved the variation in warfarin dose explained (47% versus 32% for non-genetic factors, p<0.0001).(24)
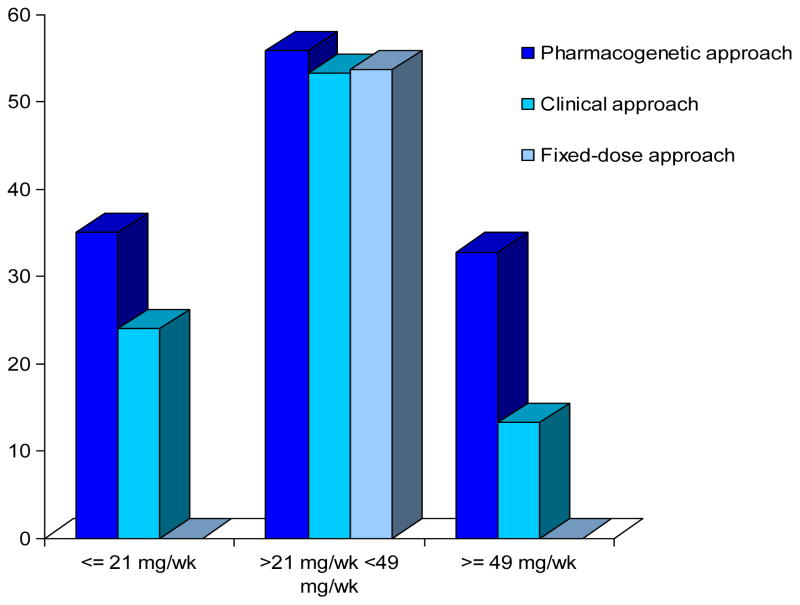
The validity and predictive ability of these associations is further evidenced by the seminal work of the IWPC,(19) providing evidence of the usefulness of genotype information across racial/ethnic/geographic populations. Dose prediction by the pharmacogenetic algorithm (incorporating CYP2C9 and VKORC1) was significantly superior to clinical algorithm as indicated by the mean absolute error (MAE is a quantity used to measure how close forecasts or predictions are to the eventual outcomes). Specifically, the pharmacogenetic algorithm improved ability to accurately predict patients requiring ≤ 3 mg/day (54.3% versus 33.4%) and those requiring ≥ 7 mg/day (26.4% versus 9.1%). Moreover this study showed that genotype-guided therapy exhibited the greatest benefits in patients ultimately requiring ≤ 3 mg/day (33.9% of cohort) or ≥ 7 mg/day (12.4% of cohort). This highlights that genotype-guided warfarin dosing can more accurately predict dose for the individual patient and this benefit can be realized by a significant proportion (46%) of the patients (Figure 1)
Figure 1.
Percent of patients with predicted dose that was within 20% of the actual stable therapeutic dose of warfarin. (19)
The dose estimates are shown according to three actual-dose groups: low dose (≤21 mg per week), intermediate-dose (>21 to <49 mg per week), and high-dose (≥49 mg per week). The fixed dose was 35 mg per week. With the fixed-dose approach, none of the estimates for the patients in the low dose and high-dose groups were within 20% of the actual dose.
This example highlights the important characteristics of pharmacogenetic predictors that address the crux of the debate:
CYP2C9 and VKORC1 genotypes are clinically useful predictors of dose.
-
Incorporation of CYP2C9 and VKORC1 genotype provided superior warfarin dose prediction compared to the clinical algorithm (or the fixed- 5mg dose algorithm). These findings are consistent to those of a recent study of 200 patients by Anderson et al.(24)The variance in warfarin dose explained by CYP2C9 and VKORC1 genotypes was 2-fold higher than that explained by clinical factors alone. Most of the benefits were derived by appropriately dosing the patient groups possessing no variant alleles and those possessing multiple variant alleles. However, the study failed to demonstrate an increase in the percentage of INRs within the therapeutic range by institution of genotype-guided therapy.
Despite the negative primary end point, exploratory analyses identified two genotypic subgroups that benefited from genotype-guided therapy: wild-type patients (whose dose requirements are greater than average) and carriers of multiple variant alleles (whose dose requirements are lower than average). In these patient subgroups, pharmacogenetic guidance yielded a 10% increase in within-range INRs (p=0.03).(24) The upcoming Clarification of Optimal Anticoagulation with Genetics (COAG) trial will randomize patients to clinical warfarin dosing versus pharmacogenetically guided dosing to test whether the latter improves anticoagulation control.
The reduction in mean absolute error is sufficiently different to warrant a change in dosing strategies/decisions if testing has been done. However, given the lack of evidence on improvement of outcomes, the clinical relevance falls short. Therefore testing is recommended, not mandatory.
Pharmacogenetics as a tool for predicting drug response with regards to toxicity and efficacy
Although the bulk of efforts have focused on understanding gene-dose associations, the ultimate goal and promise of personalized medicine is to improve clinical outcomes (drug safety and efficacy). Although studies have reported gene-outcome (beneficial and adverse) associations, they have been not been extensively replicated in independent populations. However, it is important for the reader to realize that “hard endpoints” such as events (e.g. myocardial infarction, cancer recurrence, death) are infrequently encountered in prospective studies with a short follow-up duration.
This has tilted the balance in favor of ascertaining surrogate endpoints, allowing researchers to conduct shorter and smaller trials. Although this may lead to the rapid and appropriate dissemination of new treatments, the use of surrogates is based on crucial assumptions. We highlight this issue with warfarin pharmacogenetics synthesizing the evidence from surrogate outcomes such as INR and dose and hard endpoints of major bleeding events. In this still emerging field, multiple ongoing efforts that result in adequate accrual of such “hard endpoints” will be needed to shed light on the utility of gene-outcome associations.
The association of CYP2C9 with risk of hemorrhage among warfarin users
First, we caution the reader to attend to the definition of the hemorrhagic event. The definition varies from ‘over-anticoagulation’(25–31) to a clinically defined ‘hemorrhagic event.’(32–35) Moreover hemorrhagic events may be have varying definitions; minor events (e.g. nose bleeds) are those that do not require intensive medical/surgical interventions; major events (e.g. retroperitoneal hematoma) are those that do require intensive medical/surgical interventions.
Four studies have reported the risk of hemorrhage associated with CYP2C9,(32–35) with two assessing risk of major hemorrhage by genotype (Table 1).(34, 35) Higashi et al demonstrated the risk of major hemorrhage associated with variant CYP2C9 genotype was higher during initiation of therapy [RR 3.9, 95%CI: 1.3, 12.1] and during the entire follow-up period (Table 1).(34) This CYP2C9-hemorrhage association was confirmed in both African and European American patients after accounting for the variation in vitamin K epoxide reductase (VKORC1) and clinical covariates.(35) Perhaps more importantly, this study found the risk associated with the variant CYP2C9 genotype persists even after stabilization of anticoagulation therapy. The persistence of risk after stabilization indicates that CYP2C9 genotype information may be clinically useful throughout the duration of therapy.
Table 1.
Selected studies evaluating the influence of CYP2C9 and/orVKORC1 on risk of hemorrhage among warfarin users
| Reference | Study Design | Polymorphisms assessed | Risk of Hemorrhage RR [95% CI] | |
|---|---|---|---|---|
| Minor | Major | |||
| Aithal 1999 | Case Control (36 cases, 52 controls) | CYP2C9 *2, *3 | 2.7 [0.9, 8.1] | 3.7 [1.4, 9.5] |
| Margaglione 20001 | Retrospective cohort (n=180) | CYP2C9 *2, *3 | Minor and Major Combined 2.6 [1.2, 5.7] | |
| Higashi 2002 | Retrospective Cohort (n=186) | CYP2C9 *2, *3 | NE | 2.4 [1.2, 4.9] |
| Limdi 2008 | Prospective cohort (n=446, 227 AA) |
CYP2C9 *2, *3 VKORC1–1173 |
CYP2C9 1.3 [0.8, 1.9] VKORC1 0.8 [0.5, 1.3] |
CYP2C9 3.0 [1.2, 7.5] VKORC1 1.7 [0.7, 4.4] |
All comparisons are presented for variant vs. wild-type genotypes at a non-directional statistical significance of 0.05.
NE – not evaluated or not reported.
AA: African American.
Minor hemorrhage included mild nosebleeds, microscopic hematuria, mild bruising, and mild hemorrhoidal bleeding.
Major hemorrhage combined serious, life-threatening and fatal bleeding episodes as defined by Fihn et al.
rarity of serious/life-threatening bleeding (n=10) necessitated evaluation of major and minor hemorrhage as a composite outcome.
Therefore in the case of warfarin, VKORC1 and variants explain approximately 25% of warfarin dose requirement variability, compared to approximately 10% for CYP2C9 variants.(19) Yet CYP2C9 variants have been associated with a 2–4 times higher risk of major hemorrhage,(34, 35) while VKORC1 appears to confer a higher risk of over-anticoagulation (INR >4)(31, 36) but does not appear to confer as significant a hemorrhagic risk.(35) Although over-anticoagulation (INR>4) increases the risk of hemorrhage, its use as a surrogate assumes that a high INR usually (or always) precedes a hemorrhagic event, failing to recognize the multiple complex pathways involved in its occurrence.
These findings illustrate the challenge of relying on intermediate outcomes (such as dose requirements, or anticoagulation level) to model (or implicitly infer) the influence of genetic variants on clinical events, life expectancy, and quality of life.
Warfarin pharmacogenetics: expectation, current status, reality, and potential promises (Table 2):
Table 2.
Risk of first hemorrhagic complication among warfarin users by CYP2C9 genotype
| Higashi (N=185) | Limdi (N=446) | |||||
|---|---|---|---|---|---|---|
| Risk associated with CYP2C9 variant; HR [95%CI] | 2.4 [1.2, 4.9] | 3.0 [1.2, 7.5] | ||||
| CYP2C9 genotype | CYP2C9 genotype | |||||
| Number of patients | Variant | Wild-type | Variant | Wild-type | ||
| With hemorrhage | 16 | 16 | 32 | 15 | 22 | 37 |
| Without hemorrhage | 42 | 111 | 153 | 73 | 336 | 409 |
| 58 | 127 | 185 | 88 | 358 | 446 | |
| Sensitivity | 50.0% | 40.5% | ||||
| Specificity | 72.5% | 82.1% | ||||
| Positive Predictive Value | 27.6% | 17.0% | ||||
| Negative Predictive Value | 72.5% | 82.1% | ||||
The sensitivity of a test is defined as the proportion of people with disease who will have a positive result.
The specificity of a test is the proportion of people without the disease who will have a negative result.
The positive predictive value (PPV) of a test is defined as the proportion of people with a positive test result who actually have the disease.
The negative predictive value (NPV) of a test is the proportion of people with a negative test result who do not have disease.
CYP2C9 Variant genotype includes one or two copies of *2, *3 alleles among European Americans and *2, *3, *5, *6 and *11 alleles among African Americans.
HR [95%CI] denotes the Hazard ratio and 95% Confidence Interval
Although CYP2C9 has been associated with risk of hemorrhage its ability to predict hemorrhage is modest given the low sensitivity and positive predictive value. Moreover these findings from observational studies of relatively small sample size, and although interacting medications were accounted for, the study design is not completely free of the confounding due to unmeasured variables.
Although the CYP2C9-hemorrhage association has spurred analyses of clinical utility and cost-effectiveness of pharmacogenetically guided warfarin therapy, (37–39)they have not adequately evaluated if incorporation of CYP2C9 improves risk prediction.
Currently CYP2C9 has limited influence on treatment decisions, as it has not been shown to improve risk prediction. Even if we identity a genetic marker (CYP2C9 or another gene) that improves the prediction of hemorrhage, the application of this information to drug therapy is limited as there are currently no alternatives to coumarins.
In summary, variation in the CYP2C9 and VKORC1 genes clearly impact warfarin dosing requirements, but given that anticoagulation status is (or should be) already closely monitored and individualized in warfarin patients, the incremental benefits of pharmacogenomics knowledge are less clear. (31, 37) If genetic factors were to prove to be useful in significantly improving our ability to predict disastrous hemorrhagic events, perhaps the decision to institute antiplatelet therapy versus warfarin therapy could be influenced. However such clinical decisions, to be balanced, would require the relative risk reduction in thromboembolism afforded by these therapies in patients with different genotypes. Thus, the results of the upcoming COAG trial will be crucial to assess the clinical utility of warfarin pharmacogenetics, and to impact current clinical care.
The association of HLA-B*1502 with risk of serious hypersensitivity reaction among patients on carbamazepine
Drug hypersensitivity is an important clinical problem, affecting more than 7% of the general population. It commonly involves the skin and mucosal surfaces, and in rare, severe cases can lead to hepatitis, renal failure, gastrointestinal bleeding, pneumonitis, bone marrow suppression, blindness and death. (40)
Carbamazepine (CBZ) is a widely used drug for the treatment of epilepsy, bipolar disorder, trigeminal neuralgia and chronic pain. In rare cases CBZ causes life-threatening hypersensitivity reactions, namely Stevens-Johnson syndrome (SJS) and Toxic Epidermal Necrolysis (TEN). The risk of these events is estimated to be about 1 to 6 per 10,000 new users in countries with mainly Caucasian populations. However, the risk in some Asian countries is estimated to be about 10 times higher (http://www.pharma.us.novartis.com/product/pi/pdf/tegretol.pdf). Although rare, these SJS/TEN are associated with 30% mortality. (41, 42)A case-control study conducted in Han Chinese residing in Taiwan first identified HLA-B*1502 as the genetic marker for CBZ induced SJS/TEN (Table 3).(43)
Table 3.
Association of HLA-B*1502 with Stevens-Johnson syndrome (SJS) and Toxic Epidermal Necrolysis (TEN)
| Chung et al Han Chinese | |||
|---|---|---|---|
| HLA-B*1502 OR [95%CI] | 2504 [126, 49,522] | ||
| HLA-B*1502 | |||
| Number of patients | Present | Absent | |
| SJS/TEN | 44 | 0 | 44 |
| Without SJS/TEN | 3 | 98 | 101 |
| 47 | 98 | 145 | |
| Sensitivity | 100% | ||
| Specificity | 97.0% | ||
| PPV | * | ||
| NPV | * | ||
The sensitivity of a test is defined as the proportion of people with disease who will have a positive result.
The specificity of a test is the proportion of people without the disease who will have a negative result.
The positive predictive value (PPV) of a test is defined as the proportion of people with a positive test result who actually have the disease.
The negative predictive value (NPV) of a test is the proportion of people with a negative test result who do not have disease.
These values cannot be calculated based on OR from a case-control study as Sensitivity and specificity are estimated values, not actual values
OR [95%CI] denotes the odds ratio and 95% Confidence Interval
Although this was a case control study, the exquisitely detailed definition of the phenotype (44) enabled identification of this genetic marker. This importance of outcome definitions was further illustrated by Hung et al who extended the genetic study to different types of CBZ induced reactions including 60 patients with SJS/TEN, 13 patients with hypersensitivity syndrome and 18 with maculopapular exanthema and 144 CBZ- tolerant controls. The association of HLA-B*1502 with SJS/TEN was confirmed (OR=1357; 95% CI: 193.4–8838.3). In contrast HLA-B*1502 association was not observed in patients with hypersensitivity syndrome or maculopapular exanthema.(45)
A European study of 12 carbamazepine-induced SJS/TEN cases did not confirm the prior association. (46) Among these, only four had a HLA-B*1502 allele. Remarkably, all four patients reported Asian ancestry, whereas the other eight did not. This shows that although the HLA region may contain important genes for SJS, the HLA-B*1502 allele is not a universal marker for this disease and that its ability to predict SJS/TEN is ethnicity dependent.(46) The two European case-control studies of 15 patients with CBZ-induced SJS/TEN(46, 47) revealed the presence of HLA-B*1502 in five patients originally from China, Cambodia, Vietnam, Reunion Island and Thailand all of whom had a parent of Asian origin. The remaining 10 patients, who were white, did not possess the HLA-B*1502 variant.
These data suggest that Chinese patients who carry the HLA-B*1502 allele are at a substantially high risk of SJS/TEN when exposed to CBZ. Although no published data have confirmed the HLA-B*1502-SJS/TEN association, the higher prevalence of this variant in Asian populations, including individuals of Han Chinese, Filipino, Malaysian, South Asian Indian, or Thai decent,(48) resulted in the Food and Drug Administration decision to recommend testing in patients of Asian origin.
Across Asian populations, notable variation exists in the prevalence of HLA-B*1502. Greater than 15% of the population is reported positive in Hong Kong, Thailand, Malaysia, and parts of the Philippines, compared to about 10% in Taiwan and 4% in North China. South Asians, including Indians, appear to have intermediate prevalence of HLA-B*1502, averaging 2 to 4%, but higher in some groups. HLA-B*1502 is present in <1% of the population in Japan and Korea. HLA-B*1502 is largely absent in individuals not of Asian origin (e.g., Caucasians, African-Americans, Hispanics, and Native Americans).
Carbamazepine pharmacogenetics: expectation, current status, reality, and potential promises: Although HLA-B*1502 is a useful predictor for SJS/TEN associated with carbamazepine, its usefulness may be restricted to Han Chinese patients (or those of Asian descent):
-
Among Han Chinese patients SJS/TEN HLA-B*1502 is very strongly associated with risk of SJS/TEN. The strength of the association is directly related to the performance of the test. Although this characteristic contributes to the high sensitivity and specificity (Table 3) of the test, the positive and negative predictive value cannot be estimated from this case-control study.
As 3% carbamazepine tolerant patients and 8.6% of Chinese controls possess the HLA-B*1502 allele, it can be argued that screening could potentially eliminate a useful drug from the treatment choices. However given the high odds of SJS/TEN among carriers, the availability of other antiepileptic drugs of similar efficacy (and fewer drug interactions), perhaps a false positive rate of 3 to 9% for patients of Asian descent is palatable.
Among these patients incorporation of HLA-B*1502 information would allow prediction of SJS/TEN. This would be an improvement in clinicians ability to predict SJS/TEN over the current used method which involves a detailed history of past hypersensitivity reactions to other medications, particularly anti-epileptic drugs.
As there are many alternative equally efficacious anti-epileptic drugs available with more favorable adverse-effect profiles, this information would likely change treatment decisions.
Patients who test positive for HLA-B*1502 may be at increased risk of SJS/TEN from carbamazepine and other AEDs (phenytoin, lamotrigine) that have been associated with SJS/TEN. Therefore, in HLA-B*1502-positive patients, doctors should consider avoiding use of other AEDs associated with SJS/TEN when alternative therapies are equally acceptable. Tested patients who are found to be negative for HLA-B*1502 have a low risk of SJS/TEN from CBZ, but SJS/TEN can still rarely occur, so healthcare professionals should still watch for symptoms in these patients. For more details on pharmacogenetics of antiepileptic drugs we refer the reader to two recent reviews.(48, 49)
The association of CYP2D6 with lack of response to tamoxifen
Tamoxifen is a commonly used adjuvant therapy for the treatment and prevention estrogen-receptor positive (ER+) breast cancer, with an approximate 30% reduction in annual breast cancer death compared to placebo.(50) It appears, however, that a subpopulation of ER+ premenopausal women with breast cancer may have a lower response to tamoxifen.
Tamoxifen is a prodrug that undergoes extensive first pass oxidative metabolism by various cytochrome P450 enzymes to active metabolites. Briefly, tamoxifen is hydrolyzed or demethylated to various intermediates by CYP2D6, 2B6, 2C9, 2C19, and 3A. It has recently been shown that the main anti-estrogenic active metabolite is endoxifen, and the key liver enzyme catalyzing the conversion from intermediates to endoxifen is CYP2D6.(51) CYP2D6 is a highly polymorphic enzyme, with eighty different single nucleotide polymorphisms identified to date that influence enzymatic activity or levels, resulting in a wide spectrum of phenotypic patterns. However, to simplify this otherwise complex CYP2D6 phenotypic pattern, subjects are often categorized as extensive (‘normal’) metabolizers (EM), poor metabolizers (PM), or ultra rapid metabolizers (UM). Thus, patients carrying low-activity alleles (PMs) have lower levels of the active metabolite endoxifen.
There is growing clinical evidence of the pharmacogenetic effect of CYP2D6 variants on tamoxifen treatment outcomes.(52, 53) Goetz et al evaluated post-menopausal women following resection of estrogen-receptor positive (ER+) tumors receiving tamoxifen as part of the tamoxifen-only arm of a North Central Cancer Treatment Group (NCCTG).(52, 54) The women were retrospectively genotyped for the most frequent CYP2D6 inactivating allele (CYP2D6 *4). Extensive metabolizers were defined as patients without a *4 allele (i.e., wt/wt) who were not co-prescribed a CYP2D6 inhibitor. Patients with decreased CYP2D6 metabolism were classified as intermediate or poor metabolizers (PM) based on the presence of one or two CYP2D6*4 alleles or the co-administration of a moderate or potent CYP2D6 inhibitor. The authors found that compared to EM women, PM women had significantly shorter time to recurrence (p = 0.034; HR = 1.91; 95% CI 1.05–3.45) and worse relapse-free survival (p = 0.017; HR = 1.74; 1.10–2.74), although there was not statistically significant difference in overall survival.
The implications of these findings may have high clinical relevance because there are alternative therapies available – namely aromatase inhibitors. In the Arimidex, Tamoxifen, Alone or in Combination (ATAC) trial, the aromatase inhibitor anastrozole was compared with tamoxifen for 5 years in 9366 postmenopausal women with localized breast cancer. After a median follow-up of 68 months, anastrozole significantly prolonged disease-free survival (hazard ratio 0.87, 95% CI 0.78–0.97, p=0.01).(55) In the BIG 1–98 trial, 4003 women treated with letrozole were compared to 4007 in the tamoxifen group. After a median follow-up of 25.8 months, as compared with tamoxifen, letrozole significantly reduced the risk of an event ending a period of disease-free survival (hazard ratio, 0.81; 95 percent confidence interval, 0.70 to 0.93; P=0.003). There were also differences in the side effect profiles, with thromboembolism, endometrial cancer, and vaginal bleeding more common with tamoxifen, and skeletal and cardiac events and hypercholesterolemia more common with aromotase inhibitors. Thus, there are clearly two viable treatment options to tamoxifen for postmenopausal women.
Punglia and colleagues recently conducted a population-based decision analysis to quantify the potential clinical implications of, and uncertainty surrounding, the use of CYP2D6 testing, utilizing the results of Goetz et al and the BIG 1–98 trial.(56) The authors estimated 5-year disease-free survival of tamoxifen-treated patients with no mutations (wt/wt) was 83.9%, similar to that (84.0%) for genotypically unselected patients who were treated with aromatase inhibitors. With stronger genetic association estimates, disease-free survival with tamoxifen exceeded that with aromatase inhibitors in wt/wt patients. Thus, not only might CYP2D6 variant patients benefit from switching from tamoxifen to an aromotase inhibitor, but wild type patients may benefit equally, or better, with tamoxifen compared to aromotase inhibitors. These results help clarify the need for further information on this association and may be useful for guideline development. Patient-level decision support tools could also be developed using a generally similar approach, although greater clarity of the validity of the pharmacogenomic association, as discussed below, is probably needed.
However, there are some differing results in regard to the association between CYP2D6 variants and clinical outcomes with tamoxifen therapy (Table 4). In a study that confirms the findings of Goetz et al, Schroth and colleagues in Germany conducted a similar study in 206 women.(53) They found that PM women had a higher recurrence rate (HR=2.24; P=0.02) and shorter event-free survival (HR=1.89; P=0.02) compared to EM women, and (similarly), there was not a significant effect on overall survival, potentially a result of low statistical power given the sample size. In contrast, studies by Nowell et al (57) and by Wegman et al(58) indicated CYP2D6 poor metabolizers had an increase in disease free survival. There are several potential explanations for these discrepant findings. While the studies of Goetz (52)and Schroth(53) were based on data from clinical trials, the populations in the studies by Nowell and Wegman (57, 58) were not necessarily from controlled studies. Thus, adherence may have been more variable, and EM women that were able to activate tamoxifen to endoxifen could have experienced a higher incidence of side effects that led them to discontinue treatment or decrease dose intensity. Indeed, one recent study has suggested that women who are poor metabolizers of tamoxifen may have higher adherence.(59) However, the conflicting findings cannot be readily dismissed, and highlight the need for retrospective genotyping analyses of large tamoxifen RCTs.(60)
Table 4.
Association of CYP2D6 and survival among patients with breast cancer on tamoxifen therapy.
| Study | Size (n) | Poor metabolizers | DFS HRadj (95% CI) | OS HRadj (95% CI) |
|---|---|---|---|---|
| Goetz 2007 | 180 | 36%1 | 1.60 (1.06–2.43) | 1.34 (0.83–2.16) |
| Schroth 2007 | 206 | 40%2 | 1.89 (1.10–3.25) | 1.73 (0.88–3.41) |
| Nowell 2005 | 162 | 30%3 | 0.77 (0.32–1.81) | n/a |
| Wegman 2007 | 119 | ~27%3 | 0.33 (0.08–1.43) | n/a |
DFS, disease free survival; OS, overall survival; HRadj, adjusted hazard ratio
One or more *4 alleles or CYP2D6 inhibitor
One or more *4, *5, *10, or *41 alleles
One or more *4 alleles
Tamoxifen pharmacogenetics: expectation, current status, reality, and potential promises: This example highlights that CYP2D6 testing for postmenopausal women taking tamoxifen as adjuvant therapy for breast cancer may address the clinically important questions:
Women with CYP2D6 variants with lower activity likely have a decreased response to tamoxifen, but results are not consistent across studies to date.
As there is no current, alternative approach to predicting response to tamoxifen, a test based on a valid association would be a clear improvement
The availability of aromatase inhibitors significantly increases the probability that testing would lead to an alteration in treatment.
In summary, the use of CYP2D6 genotyping to help guide adjuvant chemotherapy selection for early-stage premenopausal women offers significant promise, and may have dramatic clinical importance, but additional studies are needed to validate direction and strength of the association with disease recurrence and overall survival before testing can be recommended in routine practice.
Genome Wide Association studies (GWAS)
The goal of GWAS is to test the link between changes in the DNA sequence (genotype) of individuals in a population and a trait (phenotype) by assaying the majority of common polymorphisms across the genome. GWAS, typically ‘hypothesis-free’ experiments, have lead to discovery of new risk alleles associated with disease susceptibility.(61) However, most of the risk alleles have small effects with unclear clinical value. Although there are very few published GWAS related to drug response, the potential for gaining insight into pharmacogenomics of drug efficacy and toxicity is promising.(62) Two recent examples are: CYP2C8 polymorphisms associated with bisphosphonate-related osteonecrosis in patients with multiple myeloma(63) and the association of genes in the gamma amino-butyric acid signaling pathway and neuroleptic induced tardive dyskinesia.(64)
Conclusion
These examples highlight both the challenges and opportunities of pharmacogenomics. First, establishing a valid association between genetic variation and drug response; second, doing so for a clinically meaningful outcome and third, providing solid evidence or rationale for improvement in patient outcomes compared to current standard of care. Over the past decade, the field has moved from high expectations, with many initial ‘false positive’ association findings, to more careful epidemiological work. The current focus is on establishing clinically relevant associations instead of surrogate outcomes.
Although pharmacogenetics may not impact most drugs in the near future, valid and clinically meaningful associations have been established for some. Efforts are now shifting to collaborative studies involving researchers, regulators, clinicians, and payers. Although these stakeholders sometimes have very different perspectives on the utility of pharmacogenomic tests,(38, 65) there has been recognition that both research and development are needed to successfully evaluate and bring appropriate tests to patient care. The Clarification of Optimal Anticoagulation with Genetics (COAG trial) which will randomize patients to clinical warfarin dosing versus pharmacogenetically guided dosing, the TailorRx trial being conducted by the NCI with the OncotypeDx test, and the recently completed abacavir trial (PREDICT-1) are evidence of these efforts. The next decade promises to be fruitful for pharmacogenomics, as multi-center collaborative consortiums are established to validate association findings, and RCTs conducted as necessary for pharmacogenomic tests with promising clinical utility. Although randomized clinical trials are underway to for some drugs candidates such as warfarin, they may be unrealistic for other drug candidates such as carbamazepine given the rarity of SJS/TEN outcomes. Therefore consortia of investigators pooling, data and expertise will play a crucial role in the years to come.
Acknowledgments
Supported in part by grants from the National Heart Lung and Blood Institute (RO1HL092173), the National Institute of Neurological Disorders and Stroke (K23NS45598), and Centers for Disease Control (CDC) National Office of Public Health Genomics (U18GD000005-01)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med. 2007 Dec 4;147(11):755–65. doi: 10.7326/0003-4819-147-11-200712040-00006. [DOI] [PubMed] [Google Scholar]
- 2.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. Jama. 1998 Apr 15;279(15):1200–5. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 3.Moore TJ, Cohen MR, Furberg CD. Serious adverse drug events reported to the Food and Drug Administration, 1998–2005. Arch Intern Med. 2007 Sep 10;167(16):1752–9. doi: 10.1001/archinte.167.16.1752. [DOI] [PubMed] [Google Scholar]
- 4.Ingelman-Sundberg M. Pharmacogenomic biomarkers for prediction of severe adverse drug reactions. N Engl J Med. 2008 Feb 7;358(6):637–9. doi: 10.1056/NEJMe0708842. [DOI] [PubMed] [Google Scholar]
- 5.Manunta P, Bianchi G. Pharmacogenomics and pharmacogenetics of hypertension: update and perspectives--the adducin paradigm. J Am Soc Nephrol. 2006 Apr;17(4 Suppl 2):S30–5. doi: 10.1681/ASN.2005121346. [DOI] [PubMed] [Google Scholar]
- 6.Davis BR, Arnett DK, Boerwinkle E, Ford CE, Leiendecker-Foster C, Miller MB, et al. Antihypertensive therapy, the alpha-adducin polymorphism, and cardiovascular disease in high-risk hypertensive persons: the Genetics of Hypertension-Associated Treatment Study. Pharmacogenomics J. 2007 Apr;7(2):112–22. doi: 10.1038/sj.tpj.6500395. [DOI] [PubMed] [Google Scholar]
- 7.Gerhard T, Gong Y, Beitelshees AL, Mao X, Lobmeyer MT, Cooper-DeHoff RM, et al. Alpha-adducin polymorphism associated with increased risk of adverse cardiovascular outcomes: results from GENEtic Substudy of the INternational VErapamil SR-trandolapril STudy (INVEST-GENES) Am Heart J. 2008 Aug;156(2):397–404. doi: 10.1016/j.ahj.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schelleman H, Klungel OH, Witteman JC, Hofman A, van Duijn CM, de Boer A, et al. The influence of the alpha-adducin G460W polymorphism and angiotensinogen M235T polymorphism on antihypertensive medication and blood pressure. Eur J Hum Genet. 2006 Jul;14(7):860–6. doi: 10.1038/sj.ejhg.5201632. [DOI] [PubMed] [Google Scholar]
- 9.Boekholdt SM, Sacks FM, Jukema JW, Shepherd J, Freeman DJ, McMahon AD, et al. Cholesteryl ester transfer protein TaqIB variant, high-density lipoprotein cholesterol levels, cardiovascular risk, and efficacy of pravastatin treatment: individual patient meta-analysis of 13,677 subjects. Circulation. 2005 Jan 25;111(3):278–87. doi: 10.1161/01.CIR.0000153341.46271.40. [DOI] [PubMed] [Google Scholar]
- 10.Mellen PB, Herrington DM. Pharmacogenomics of blood pressure response to antihypertensive treatment. J Hypertens. 2005 Jul;23(7):1311–25. doi: 10.1097/01.hjh.0000173510.52987.68. [DOI] [PubMed] [Google Scholar]
- 11.Ferdinands JM, Mannino DM, Gwinn ML, Bray MS. ADRB2 Arg16Gly polymorphism, lung function, and mortality: results from the Atherosclerosis Risk in Communities study. PLoS ONE. 2007;2(3):e289. doi: 10.1371/journal.pone.0000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tantisira KG, Weiss ST. The pharmacogenetics of asthma: an update. Curr Opin Mol Ther. 2005 Jun;7(3):209–17. [PubMed] [Google Scholar]
- 13.Feldman DS, Carnes CA, Abraham WT, Bristow MR. Mechanisms of disease: beta-adrenergic receptors--alterations in signal transduction and pharmacogenomics in heart failure. Nat Clin Pract Cardiovasc Med. 2005 Sep;2(9):475–83. doi: 10.1038/ncpcardio0309. [DOI] [PubMed] [Google Scholar]
- 14.Binder EB, Holsboer F. Pharmacogenomics and antidepressant drugs. Ann Med. 2006;38(2):82–94. doi: 10.1080/07853890600551045. [DOI] [PubMed] [Google Scholar]
- 15.Matchar DB, Thakur ME, Grossman I, McCrory DC, Orlando LA, Steffens DC, et al. Testing for cytochrome P450 polymorphisms in adults with non-psychotic depression treated with selective serotonin reuptake inhibitors (SSRIs) Evid Rep Technol Assess (Full Rep) 2007 Jan;(146):1–77. [PMC free article] [PubMed] [Google Scholar]
- 16.Ioannidis JP, Bernstein J, Boffetta P, Danesh J, Dolan S, Hartge P, et al. A network of investigator networks in human genome epidemiology. Am J Epidemiol. 2005 Aug 15;162(4):302–4. doi: 10.1093/aje/kwi201. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis JP, Gwinn M, Little J, Higgins JP, Bernstein JL, Boffetta P, et al. A road map for efficient and reliable human genome epidemiology. Nat Genet. 2006 Jan;38(1):3–5. doi: 10.1038/ng0106-3. [DOI] [PubMed] [Google Scholar]
- 18.Khoury MJ, Little J, Higgins J, Ioannidis JP, Gwinn M. Reporting of systematic reviews: the challenge of genetic association studies. PLoS Med. 2007 Jun;4(6):e211. doi: 10.1371/journal.pmed.0040211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009 Feb 19;360(8):753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeechan K, Macaskill P, Irwig L, Liew G, Wong TY. Assessing new biomarkers and predictive models for use in clinical practice: a clinician’s guide. Arch Intern Med. 2008 Nov 24;168(21):2304–10. doi: 10.1001/archinte.168.21.2304. [DOI] [PubMed] [Google Scholar]
- 21.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008 Sep;28(9):1084–97. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008 Jun 5; doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limdi NA, Beasley TM, Crowley MR, Goldstein JA, Rieder MJ, Flockhart DA, et al. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008 Oct;9(10):1445–58. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007 Nov 27;116(22):2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 25.Schalekamp T, Brasse BP, Roijers JF, Chahid Y, van Geest-Daalderop JH, de Vries-Goldschmeding H, et al. VKORC1 and CYP2C9 genotypes and acenocoumarol anticoagulation status: interaction between both genotypes affects overanticoagulation. Clin Pharmacol Ther. 2006 Jul;80(1):13–22. doi: 10.1016/j.clpt.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Schalekamp T, Brasse BP, Roijers JF, van Meegen E, van der Meer FJ, van Wijk EM, et al. VKORC1 and CYP2C9 genotypes and phenprocoumon anticoagulation status: interaction between both genotypes affects dose requirement. Clin Pharmacol Ther. 2007 Feb;81(2):185–93. doi: 10.1038/sj.clpt.6100036. [DOI] [PubMed] [Google Scholar]
- 27.Schalekamp T, Oosterhof M, van Meegen E, van Der Meer FJ, Conemans J, Hermans M, et al. Effects of cytochrome P450 2C9 polymorphisms on phenprocoumon anticoagulation status. Clin Pharmacol Ther. 2004 Nov;76(5):409–17. doi: 10.1016/j.clpt.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Schalekamp T, van Geest-Daalderop JH, de Vries-Goldschmeding H, Conemans J, Bernsen Mj M, de Boer A. Acenocoumarol stabilization is delayed in CYP2C93 carriers. Clin Pharmacol Ther. 2004 May;75(5):394–402. doi: 10.1016/j.clpt.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 29.Schelleman H, Chen Z, Kealey C, Whitehead AS, Christie J, Price M, et al. Warfarin Response and Vitamin K Epoxide Reductase Complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007 Feb 28;81(5):742–7. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 30.Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood. 2000 Sep 1;96(5):1816–9. [PubMed] [Google Scholar]
- 31.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008 Mar 6;358(10):999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999 Feb 27;353(9154):717–9. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 33.Margaglione M, Colaizzo D, D’Andrea G, Brancaccio V, Ciampa A, Grandone E, et al. Genetic modulation of oral anticoagulation with warfarin. Thromb Haemost. 2000 Nov;84(5):775–8. [PubMed] [Google Scholar]
- 34.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. Jama. 2002 Apr 3;287(13):1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 35.Limdi NA, McGwin G, Goldstein JA, Beasley TM, Arnett DK, Adler BK, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008 Feb;83(2):312–21. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meckley LM, Wittkowsky AK, Rieder MJ, Rettie AE, Veenstra DL. An analysis of the relative effects of VKORC1 and CYP2C9 variants on anticoagulation related outcomes in warfarin-treated patients. Thromb Haemost. 2008 Aug;100(2):229–39. [PubMed] [Google Scholar]
- 37.Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009 Jan 20;150(2):73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 38.Lesko LJ. The critical path of warfarin dosing: finding an optimal dosing strategy using pharmacogenetics. Clin Pharmacol Ther. 2008 Sep;84(3):301–3. doi: 10.1038/clpt.2008.133. [DOI] [PubMed] [Google Scholar]
- 39.McWilliam A, Lutter R, Nardinelli; C. Healthcare impact of personalized medicine using genetic testing: an exploratory analysis for warfarin. Personalized Medicine. 2008;5(3):279–84. doi: 10.2217/17410541.5.3.279. [DOI] [PubMed] [Google Scholar]
- 40.Roujeau JC, Stern RS. Severe adverse cutaneous reactions to drugs. N Engl J Med. 1994 Nov 10;331(19):1272–85. doi: 10.1056/NEJM199411103311906. [DOI] [PubMed] [Google Scholar]
- 41.Gerdts B, Vloemans AF, Kreis RW. Toxic epidermal necrolysis: 15 years’ experience in a Dutch burns centre. J Eur Acad Dermatol Venereol. 2007 Jul;21(6):781–8. doi: 10.1111/j.1468-3083.2006.02082.x. [DOI] [PubMed] [Google Scholar]
- 42.Gravante G, Delogu D, Marianetti M, Trombetta M, Esposito G, Montone A. Toxic epidermal necrolysis and Steven Johnson syndrome: 11-years experience and outcome. Eur Rev Med Pharmacol Sci. 2007 Mar–Apr;11(2):119–27. [PubMed] [Google Scholar]
- 43.Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004 Apr 1;428(6982):486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 44.Roujeau JC. The spectrum of Stevens-Johnson syndrome and toxic epidermal necrolysis: a clinical classification. J Invest Dermatol. 1994 Jun;102(6):28S–30S. doi: 10.1111/1523-1747.ep12388434. [DOI] [PubMed] [Google Scholar]
- 45.Hung SI, Chung WH, Jee SH, Chen WC, Chang YT, Lee WR, et al. Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions. Pharmacogenet Genomics. 2006 Apr;16(4):297–306. doi: 10.1097/01.fpc.0000199500.46842.4a. [DOI] [PubMed] [Google Scholar]
- 46.Lonjou C, Thomas L, Borot N, Ledger N, de Toma C, LeLouet H, et al. A marker for Stevens-Johnson syndrome …: ethnicity matters. Pharmacogenomics J. 2006 Jul–Aug;6(4):265–8. doi: 10.1038/sj.tpj.6500356. [DOI] [PubMed] [Google Scholar]
- 47.Alfirevic A, Jorgensen AL, Williamson PR, Chadwick DW, Park BK, Pirmohamed M. HLA-B locus in Caucasian patients with carbamazepine hypersensitivity. Pharmacogenomics. 2006 Sep;7(6):813–8. doi: 10.2217/14622416.7.6.813. [DOI] [PubMed] [Google Scholar]
- 48.Ferrell PB, Jr, McLeod HL. Carbamazepine, HLA-B*1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics. 2008 Oct;9(10):1543–6. doi: 10.2217/14622416.9.10.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loscher W, Klotz U, Zimprich F, Schmidt D. The clinical impact of pharmacogenetics on the treatment of epilepsy. Epilepsia. 2009 Jan;50(1):1–23. doi: 10.1111/j.1528-1167.2008.01716.x. [DOI] [PubMed] [Google Scholar]
- 50.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005 May 14–20;365(9472):1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 51.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006 Jul;80(1):61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 52.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007 Jan;101(1):113–21. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 53.Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007 Nov 20;25(33):5187–93. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 54.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005 Dec 20;23(36):9312–8. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 55.Howell A, Cuzick J, Baum M, Buzdar A, Dowsett M, Forbes JF, et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005 Jan 1–7;365(9453):60–2. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 56.Punglia RS, Burstein HJ, Winer EP, Weeks JC. Pharmacogenomic variation of CYP2D6 and the choice of optimal adjuvant endocrine therapy for postmenopausal breast cancer: a modeling analysis. J Natl Cancer Inst. 2008 May 7;100(9):642–8. doi: 10.1093/jnci/djn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005 Jun;91(3):249–58. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 58.Wegman P, Vainikka L, Stal O, Nordenskjold B, Skoog L, Rutqvist LE, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7(3):R284–90. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rae J, Rae JMS. Cytochrome P450 2D6 activity predicts adherence to tamoxifen therapy. Breast Cancer Res Treat. 2007;106(suppl 1):521. [Google Scholar]
- 60.Hayes DF, Stearns V, Rae J, Flockhart D. A model citizen? Is tamoxifen more effective than aromatase inhibitors if we pick the right patients? J Natl Cancer Inst. 2008 May 7;100(9):610–3. doi: 10.1093/jnci/djn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008 Oct 15;17(R2):R156–65. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldstein DB, Tate SK, Sisodiya SM. Pharmacogenetics goes genomic. Nat Rev Genet. 2003 Dec;4(12):937–47. doi: 10.1038/nrg1229. [DOI] [PubMed] [Google Scholar]
- 63.Sarasquete Me Fau - Garcia-Sanz R, Garcia-Sanz R Fau - Marin L, Marin L Fau - Alcoceba M, Alcoceba M Fau - Chillon MC, Chillon Mc Fau - Balanzategui A, Balanzategui A Fau - Santamaria C, et al. Bisphosphonate-related osteonecrosis of the jaw is associated with polymorphisms of the cytochrome P450 CYP2C8 in multiple myeloma: a genome-wide single nucleotide polymorphism analysis. (1528-0020 (Electronic)).
- 64.Inada T Fau - Koga M, Koga M Fau - Ishiguro H, Ishiguro H Fau - Horiuchi Y, Horiuchi Y Fau - Syu A, Syu A Fau - Yoshio T, Yoshio T Fau - Takahashi N, et al. Pathway-based association analysis of genome-wide screening data suggest that genes associated with the gamma-aminobutyric acid receptor signaling pathway are involved in neurolepticinduced, treatment-resistant tardive dyskinesia. (1744-6872 (Print)).
- 65.Garcia DA. Warfarin and pharmacogenomic testing: the case for restraint. Clin Pharmacol Ther. 2008 Sep;84(3):303–5. doi: 10.1038/clpt.2008.131. [DOI] [PubMed] [Google Scholar]