Abstract
Supv3L1 is an evolutionarily conserved helicase that plays a critical role in the mitochondrial RNA surveillance and degradation machinery. Conditional ablation of Supv3L1 in adult mice leads to premature aging phenotypes including loss of muscle mass and adipose tissue and severe skin abnormalities. To get insights into the spatial and temporal expression of Supv3L1 in the mouse, we generated knock-in and transgenic strains in which an EGFP reporter was placed under control of the Supv3L1 native promoter. During development, expression of Supv3L1 begins at the blastocyst stage, becomes widespread and strong in all fetal tissues and cell types, and continues during postnatal growth. In mature animals reporter expression is only slightly diminished in most tissues and continues to be highly expressed in the brain, peripheral sensory organs, and testis. Together, these data confirm that Supv3L1 is an important developmentally regulated gene, which continues to be expressed in all mature tissues, particularly the rapidly proliferating cells of testes, but also in the brain and sensory organs. The transgenic mice and cell lines derived from them constitute a valuable tool for the examination of the spatial and temporal aspects of Supv3L1 promoter activity, and should facilitate future screens for small molecules that regulate Supv3L1 expression.
Keywords: Supv3L1, Suv3, mouse, expression pattern, retina, EGFP reporter
Introduction
The SUV3 gene was first described in yeast (Butow et al. 1989; Zhu et al. 1989; Conrad-Webb et al. 1990), where the gene product was found to be localized in the mitochondrial matrix and shown to display RNA helicase activity (Stepien et al. 1992). It is believed that together with the ribonuclease DSS1, yeast SUV3 forms a RNA degradosome complex (Dziembowski et al. 2003) that is engaged in several key processes including mitochondrial RNA decay and surveillance. The SUV3 gene was found to be conserved throughout evolution with orthologs documented in bacteria, plants, nematodes, fruit fly and mammals (Dmochowska et al. 1999). The mammalian ortholog of the yeast SUV3 gene, Supv3L1, is a member of the Ski2 family of DExH-box RNA helicases capable to unwind both RNA/RNA and DNA/DNA duplexes (Minczuk et al. 2002; Shu et al. 2004). It is believed to operate primarily in mitochondria although some evidence suggests it may also function in the nuclear compartment (Shu et al. 2004; Minczuk et al. 2005; Szczesny et al. 2007). The primary molecular function of Supv3L1 in mammalian cells appears to involve mtRNA processing (Wang et al. 2009). In the absence of Supv3L1, aberrant mtRNA turnover has been shown to impair mitochondrial protein synthesis, decrease ATP production, lower the mtDNA copy number and membrane potential, and increase the generation of ROS, collectively leading to cellular senescence and cell death (Khidr et al. 2008). We reported that mouse Supv3L1 is a developmentally essential gene whose mutation leads to early embryonic lethality (Pereira et al. 2007). Subsequently, using a conditional knockout strategy, we have shown that Supv3L1 plays critical role in adult tissues (Paul et al. 2009) and its disruption leads to premature aging phenotypes such as sarcopenia, loss of adipose tissue, kyphosis, skin defects and premature death.
The expression of Supv3L1 was assessed by Northern blotting in a small sampling of human tissues (Dmochowska et al. 1999). However, a comprehensive examination of Supv3L1 expression patterns during development and in adulthood has not been undertaken in any mammalian organism. To examine the spatio-temporal expression patterns of Supv3L1 in developing embryos as well as adult mice, and to facilitate future non-invasive monitoring of gene expression in a whole-animal model and isolated cell lines, we generated transgenic and knock-in strains of mice in which the native Supv3L1 promoter drives the enhanced green fluorescent protein (EGFP) reporter.
Materials and Methods
Targeting vector construction
Genomic DNA for vector construction was derived from the MICER MHPP321i21 vector, obtained from The Welcome Trust Sanger Institute (Adams et al., 2004). The mouse genomic segment of MHPP321i21 spans 7.6kb and includes exon 1 of the Supv3L1 gene. The final targeting vector, pME1, was obtained using recombineering methods (Liu et al. 2003; Warming et al. 2005) and contains an enhanced green fluorescent protein (EGFP) coding sequence under control of the Supv3L1 promoter (Fig. 1A) followed by a SV40 polyadenylation site and a Neo/Kan resistance cassette. In this construct, the translation initiation codon of EGFP was placed precisely in the position of the translation initiation codon of Supv3L1.
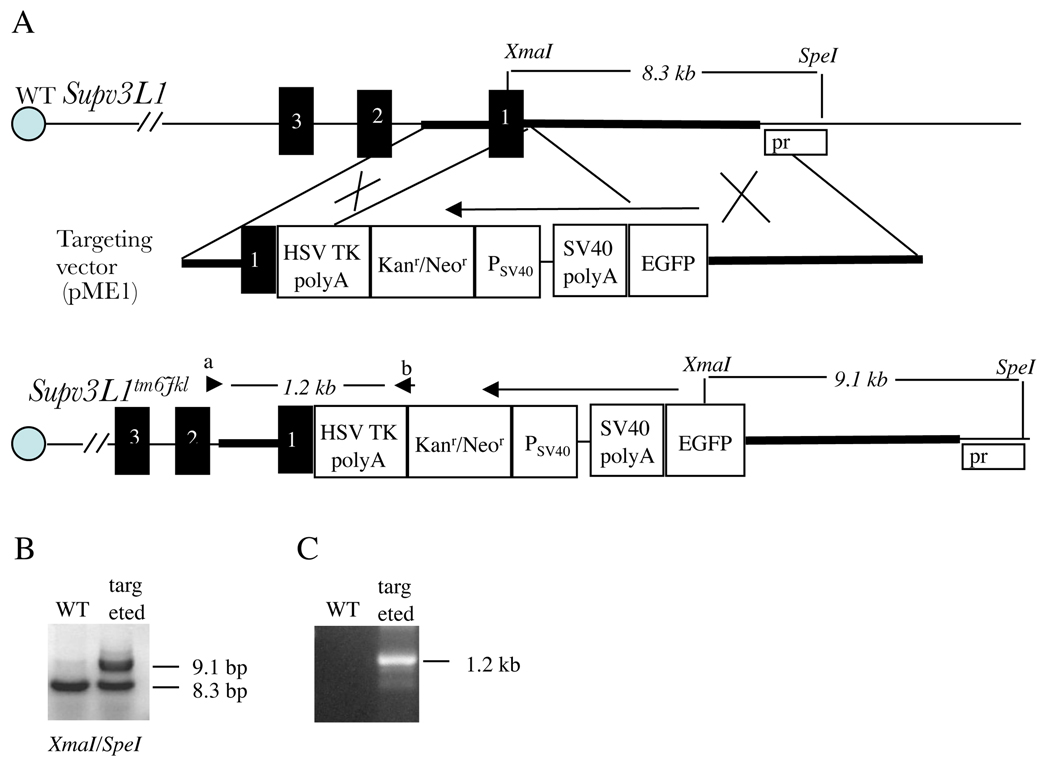
Figure 1.
Generating the knock-in EGFP allele (Supv3L1tm6Jkl). (A) Genomic structure of the wild-type Supv3L1 locus, targeting vector, and allelic modification generated in this study. Regions of homology between the genomic sequence and the vector are shown in bold. XmaI, XmaI restriction site; SpeI, SpeI restriction site; Pr, probe used in Southern blotting; EGFP, enhanced green fluorescent protein coding sequence; SV40 polyA, SV40 polyadenylation signals; PSV40, SV40 promoter; Kanr/Neor, neomycin/kanamycin resistance gene of Tn5; HSV TK polyA, polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene. Black boxes represent the first three exons of Supv3L1 (composed of total 16 exons). Arrows indicate the direction of transcription. Arrowheads designate the positions of PCR primers. (B) Southern hybridization analysis of targeted ES cells showing a correct integration event at the 5’ arm. Probe was generated by PCR using genomic DNA and primer pair c + d (ATGTCCGCACGCTGCGACTGTCGTCAGCC and GGCTGACGACAGTCGCAGCGTGCGGACAT). (C) PCR analysis of a targeted ES cell clone using primers a+b (CACGGCACTGCGTGCTTGGCAAGGCATAT and GACAGAATAAAACGCACGGTGTTGGGTCGT) confirming correct integration event at the 3’ arm.
Generation of transgenic mice
Pronuclear injections were performed according to standard procedures (Palmiter and Brinster 1986; Hanahan et al. 1989; Beddington et al. 1992; Kollias and Grosveld 1992). pME1 plasmid was digested with AscI to remove the bacterial part of the sequence of the vector. The 10.7 kb fragment containing the EGFP cassette (Fig. 1A) was used for injections of fertilized eggs isolated from FVB/N mice. The strain carrying the transgene was designated as Tg(EGFP/Supv3L1)-1.
Gene targeting of ES cells
129Ola ES cells derived from a male embryo were grown on mitotically inactive SNL76/7 feeder cells. 107 ES cells were electroporated with 20µg of pME1 linearized with AscI, and G418 selection was initiated after 24 hours. 200-G418 resistant clones were selected for further analysis. Correctly targeted ES cell clones (Fig. 1B and C) were identified as described (Ramirez-Solis et al. 1995; Klysik and Singer 2005).
Generation of targeted mice
All breeding and procedures were carried out at the Brown University Animal Facility according to institutional regulations and the NIH Guide for the Use and Care of Laboratory Animals. Targeted ES cells were grown to 80% confluency and used for injection. E3.5 blastocysts were isolated from C57BL/C2J female mice and injected with 20–25 ES cells. The injected blastocysts were implanted into the uteri of day 2.5 pseudo-pregnant females for the generation of chimeras. About 10 injected embryos were implanted per uterine horn. The male chimeras were mated with C57BL/C2J females to obtain F1 progeny. Germ line transmission of the targeted allele, designated Supv3L1tm6Jkl (Fig, 1A), was obtained with several targeted cell lines. Heterozygous Supv3L1tm6Jkl mice were maintained by mating with C57BL/C2J animals.
Genotyping
Genomic DNA was prepared from ES cells, tail biopsies or tissues and PCR was performed using primers a+b (CACGGCACTGCGTGCTTGGCAAGGCATAT and GACAGAATAAAACGCACGGTGTTGGGTCGT). The expected size of the PCR product detecting the targeted allele is 1268 bp. Amplification conditions consisted of an initial incubation at 94°C for 2 min, followed by 35 cycles at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 90 sec. Genotyping by Southern hybridization (Fig. 1B) was performed using a 288 bp PCR-generated probe (primers: ATGTCCGCACGCTGCGACTGTCGTCAGCC and GGCTGACGACAGTCGCAGCGTGCGGACAT).
Timed matings
Matings were set up and the day on which the copulation plug was found was designated as E0.5. To obtain one-cell, two-cell or blastocyst stage embryos, Supv3L1tm6Jkl males and wild-type females were mated and embryos were released from the oviducts or uteri of pregnant females. At later stages, embryos were dissected away from fetal membranes, washed in PBS and used for DNA extraction, PCR analysis, and fluorescence detection. Tissues from experimental and control littermate animals used in these studies were matched according to sex and age.
Blastocyst outgrowth
Heterozygous males and wild-type females were mated and blastocysts were isolated from pregnant females at E3.5. Blastocysts were cultured individually in ES cell medium without LIF in a humidified incubator at 37°C and 5% CO2. Media were changed every other day. On the fourth day, the cultures were washed with 5 ml of sterile PBS and used for the detection of fluorescence.
Green fluorescence detection
Animals, embryos, dissected tissues, and tissue sections were photographed using a dissection microscope equipped with GFP optics and a UV source. Visual genotyping was performed using an external UV-light source and EGFP-goggles (BLS).
Western blots
Protein extracts were prepared using RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EGTA, 1% NP40) supplemented with protein inhibitors (1 mM PMSF, 1mM Na3Vo4, 1mM NaF). The lysates were then centrifuged for 40 minutes at 16,000 × g and 4°C, and the supematants were collected and centrifuged a second time for 15 minutes at 16,000 × g and 4°C. Total protein concentrations were determined using the Biorad D/C protein assay kit. The extracts were mixed with Laemli sample buffer (containing SDS and β-mercaptoethanol) and thirty micrograms of total protein per lane were electrophoresed on a 7.5% SDS-polyacrylamide gel, transferred to Immobilon-P, and probed with rabbit anti-EGFP (Abeam) or rabbit anti-mouse Supv3L1 antibodies. The Supv3L1 antibody was raised against a synthetic peptide corresponding to the 15 C-terminal amino acids of the mouse protein (custom made, Invitrogen). After probing with a secondary antibody, HRP-conjugated goat-anti rabbit IgG (Jackson Immunoresearch, Inc.), the filters were incubated with a chemiluminescent substrate and exposed to KODAK Biomax film.
Tissue processing and staining
Tissues from experimental and control littermate animals used in these studies were matched according to sex and age. Frozen skin sections were imaged for EGFP fluorescence and subsequently processed for hematoxylin and eosin (H&E) staining. Eyes were enucleated, fixed with 4% parafromaldehyde (PFA) and processed for sectioning and microscopy as described (Khanna et al. 2009).
Results and Discussion
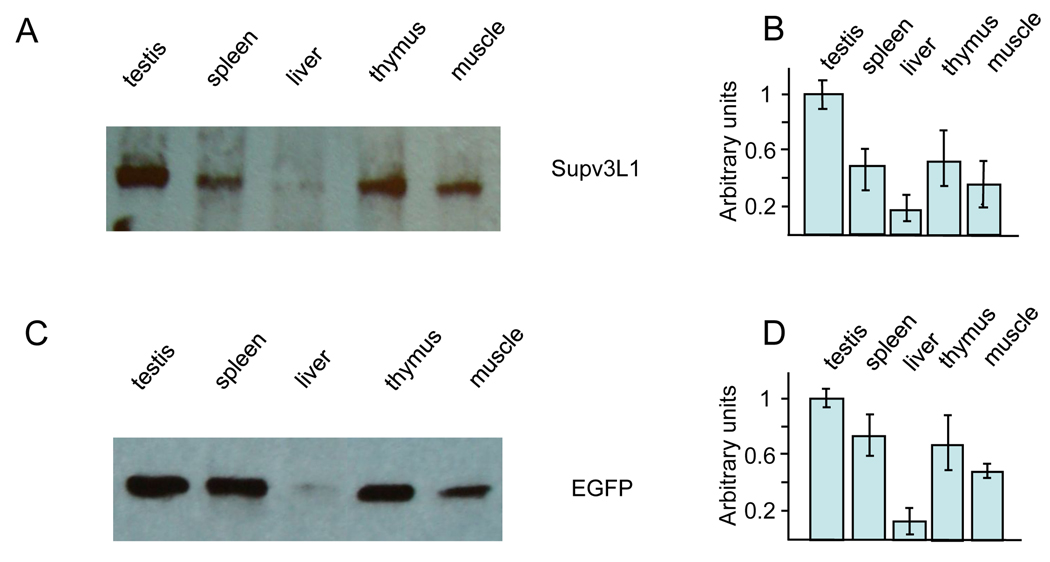
To assess whether the EGFP-reporter faithfully reflects the expression of the native Supv3L1 gene, Western blotting was performed on protein extracts from a variety of tissues of wild-type and Supv3L1tm6Jkl/+ animals using both anti-mSupv3L1 and anti-EGFP antibodies (Fig. 2). Expression was evident in all tissues tested with both antibodies, and the highest levels of Supv3L1 and EGFP proteins were detected in the testes, spleen, thymus and muscle. Somewhat lower expression was observed in the liver. Since the relative levels of the Supv3L1 and EGFP proteins were very similar, the EGFP-reporter reflects closely the expression of a native gene. Consistent with our prior studies (Paul et al. 2009) the presence of the Neo cassette in the targeted locus appears to have no affect on Supv3L1 promoter activity.
Figure 2.
Expression levels of Supv3L1 and EGFP reporter as assayed by Western blotting. (A) Tissue extracts were prepared from 10 weeks old wild-type mouse and analyzed using rabbit antibody against mouse Supv3L1. (B) Quantification of the data shown in (A) after correcting for loading variations using GAPDH as an internal standard. The results represent an average of three gels and are plotted in arbitrary units/mg of the protein extract. (C) Western blotting performed using tissue extracts obtained from 10 weeks old targeted (Supv3L1tm6Jkl/+) animal and rabbit antibodies against EGFP. (D) Quantification of the data shown in (C) representing an average of three gels.
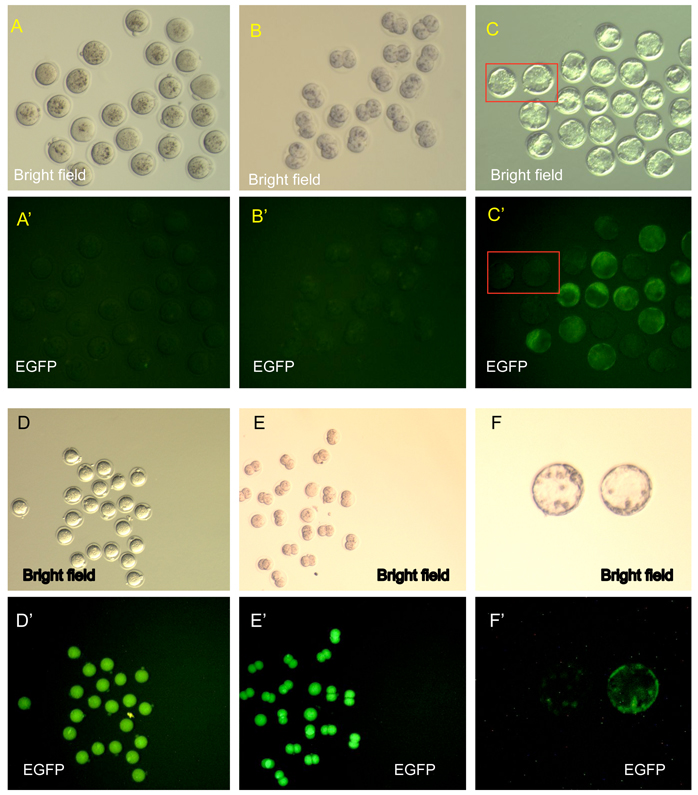
Taking advantage of this EGFP reporter, paternally inherited Supv3L1 gene expression was assessed at different stages of development. In timed mating experiments, heterozygous males (Supv3L1tm6Jkl/+) were crossed with wild-type females and fertilized eggs were isolated at E0.5 (Fig. 1A). Among 54 one-cell zygotes examined, none were found to express EGFP (Fig. 3A and 3A’). Two-cell embryos isolated from oviducts at E1.5 (or obtained by overnight culture of fertilized eggs) showed no EGFP fluorescence signal (Fig. 3B and 3B’). Thus, the reporter gene is silent at these stages of development, or its expression is below the sensitivity of this detection method. First signs of expression were seen at the blastocyst stage (E3.5). Although the signal was weak, approximately 50% of blastocysts were found to display EGFP fluorescence above background (Fig. 3C and 3C’). The signal was detectable both within the inner cell mass and the trophectoderm. Blastocyst outgrowths in culture showed expression primarily in inner cell mass derived cells, but weak fluorescence was also detectable in the surrounding cells of the trophoblast lineage (not shown). Consistently, targeted ES cell clones used to derive the Supv3L1tm6Jkl/+ F1 mice were found positive for EGFP-reporter expression (not shown).
Figure 3.
The EGFP reporter expression at preimplantation stages. (A, A’) One-cell embryos derived from matings of Supv3L1tm6Jkl/+ males and wild type females (paternal transmission of the reporter allele). Note no detectable expression of the EGFP transgene. (B, B’) Two-cell embryos derived from matings of Supv3L1tm6Jkl/+ males and wild type females. Note no EGFP expression. (C, C’) Blastocysts derived from matings of Supv3L1tm6Jkl/+ males and wild type females. Rectangles indicate examples of wild-type blastocysts negative for EGFP expression. (D, D’) One-cell embryos obtained from Supv3L1tm6Jkl/+ females mated with wild-type males (maternally transmitted reporter). Note that all embryos are EGFP positive while only half of them can be EGFP-transgene positive. (E, E’) Two-cell embryos derived from Supv3L1tm6Jkl/+ females mated with wild-type males. Note that all E1.5 embryos are EGFP positive while only half of them are positive for the transgene. (F, F’) Blastocysts isolated from Supv3L1tm6Jkl/+ females after mating with wild-type males (maternally transmitted reporter). Roughly 50% of these balstocysts are EGFP positive and 50% are negative. One of each kind is shown.
One-cell embryos (E0.5) obtained from matings of Supv3L1tm6Jkl/+ females and wild-type males, in which the EGFP reporter is maternally transmitted, were uniformly EGFP positive (Fig. 3D and D’), and high level expression continued at the two-cell stage (E1.5; Fig. 3E and E’). Since half of these embryos do not carry the EGFP knock-in allele, the EGFP reporter protein must have accumulated during pre-meiotic or meiotic divisions of oogenesis at which cells of this lineage existed at 2N stage. High levels of EGFP persisted in the zygotic cytoplasm until E1.5. At the blastocyst stage (E3.5), approximately half of these embryos were positive for EGFP, showing low levels of expression similar to the paternally inherited allele (Fig. 3F and F’). In this context, it might be important to note that meiotic reduction of 2N chromosomes to N chromosomes in Supv3L1tm6Jkl/+ mice does not affect the viability of gametes. Matings of Supv3L1tm6Jkl/+ females with wild-type males as wells as matings of Supv3L1tm6Jkl/+ males with wild-type females both produced Supv3L1tm6Jkl/+ and wild-type pups at the expected 1:1 Mendelian ratios.
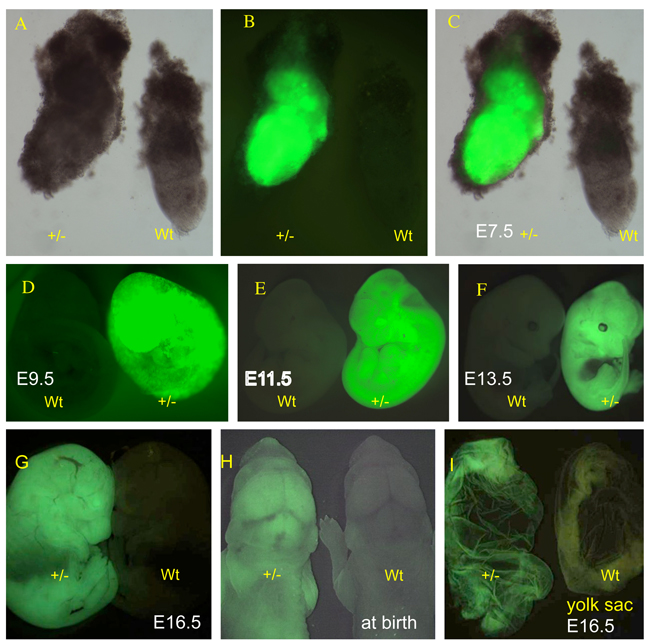
At later developmental stages, the expression patterns of the paternally and maternally transmitted reporter alleles were indistinguishable (not shown). Paternally inherited expression at the late primitive streak stage (E7.5) is shown in Fig. 4A, B and C. The expression is strong in all cell layers, including extra-embryonic tissues such as the visceral yolk sac. As expected, extra-embryonic tissues of maternal origin that do not carry the transgene, including spongy layers of endometrial tissue, appeared negative. Widespread expression of the reporter continued through the stage of 21–29 somites (E9.5; Fig. 4D and D’), 43–48 pairs of somites (E11.5; Fig. 4E and E’), and 60 pairs of somites (E13.5; Fig. 4F and F’). The liver at E13.5 displayed detectable but lower fluorescence (Fig. 4F’). At E16.5 and at birth, all tissues examined expressed the reporter (Fig. 4G and 4G’ and Fig. 4H and H’), including extraembryonic membranes such as yolk sac (Fig. 4I and I’) and amnion (not shown).
Figure 4.
The EGFP reporter expression in postimplantation embryos. (A) Supv3L1tm6Jkl/+ embryo at E7.5 (+/−) and wild-type embryo (wt) photographed in the bright field. (B) EGFP emission of the same embryos shown in (A). (C) Merged images of (A) and (B). (D) EGFP expression in wild-type (wt) and Supv3L1tm6Jkl/+ (+/−) embryos at E9.5. (E) Fluorescent light emission in wild-type (wt) and Supv3L1tm6Jkl/+ (+/−) embryos at E11.5. (F) Wild-type (wt) and Supv3L1tm6Jkl/+ (+/−) embryos showing EGFP expression at E13.5. Note rather weak signal in the area of the developing liver. (G) The EGFP reporter expression at E16.5. (H) The EGFP reporter expression at birth. (I) Yolk sac from Supv3L1tm6Jkl/+ (+/−) and wild-type embryos at E16.5.
Supv3L1 expression after birth was assessed by dissecting out a variety of organs from 5 days old pups and examined for EGFP fluorescence emission. The EGFP reporter continued to be expressed widely and was detectable in all organs examined. Relatively low fluorescence was observed in the liver. The strongest expression of the reporter at 5 days of age was found in the skin, skeletal muscle, heart, brain, and eyes (not shown).
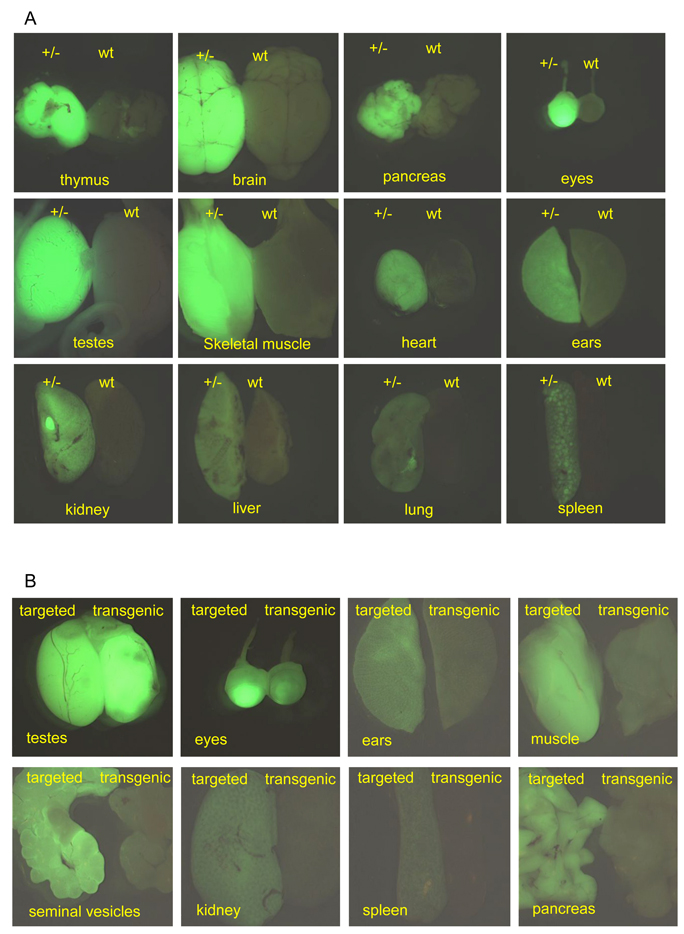
This widespread expression continued into adult life. At 10 weeks of age EGFP emission was detectable in all tissues tested (Fig. 5A), although the levels of fluorescence appeared somewhat diminished in most of the organs relative to the expression seen at 5 days. The lowest signals were seen in the liver, lung and skin, while the highest signals were found in the testes, brain, eyes and thymus. The testes displayed the highest levels of EGFP fluorescence light emission, suggesting an important role for Supv3L1 in the spermatogenetic proliferation of cells taking place within the seminiferous tubules. The bright fluorescent eyes could actually be used for visual genotyping of both the mature transgenic and knock-in mice using an external 488 nm excitation source and EGFP-goggles. Expression of the EGFP reporter in coronal cross-sections of the brain was widespread. However, the hippocampal formations, thalamus, hypothalamus and amygdala were noticeably more fluorescent relative to the neocortices (not shown).
Figure 5.
(A) Expression of EGFP reporter in tissue dissected out from 10 weeks old wild-type (wt) and Supv3L1tm6Jkl/+ (+/−) male. Images were taken at the same conditions of magnification, brightness, gain and exposure time, to reflect relative intensities of the fluorescence light emission in different tissues. Note remarkably higher reporter expression in the brain, testes, eyes, muscle and the thymus. (B) Relative expression levels of EGFP reporter in tissue dissected out from 10 weeks old targeted (Supv3L1tm6Jkl/+) and heterozygous transgenic (Tg(EGFP/Supv3L1)-1) animals.
While the EGFP reporter in the knock-in animals was integrated precisely into the Supv3L1 locus and is expressed from the native promoter, in the transgenic lines the genomic location and copy number of the transgene are not known. In these animals the Supv3L1 promoter sequence driving EGFP reporter expression is limited to 6.5 kb. Thus, it was of interest to compare the EGFP reporter expression patterns in these two lines. Similar to the knock-in mice, we found that the expression of the paternally transmitted reporter in the transgenic strain begins early during development. One-cell fertilized eggs and two-cell embryos did not express the reporter. The EGFP emission was detectable in blastocysts and became widespread at later stages (not shown). With the exception of the testes, however, the intensity of reporter expression in many tissues isolated from mature transgenic mice was found to be lower (Fig. 5B). In fact, lung, spleen, kidney and liver from 10 weeks old transgenic animals showed EGFP emission close to the limit of detection. The reasons for the lower levels of EGFP reporter expression in adult transgenic mice are not known at this time. Although the copy number of the transgene has not been determined, pronuclear injection of DNA typically results in the tandem integration of multiple copies. Thus, the copy number of the reporter is likely to be higher in the transgenic mice relative to the single copy in knock-in animals. The factors affecting the expression EGFP levels in the transgenic mice may include position effects at the genomic locus of transgene integration (which is currently unknown), or promoter functions that may be missing in the 6.5 kb sequence used to drive reporter expression.
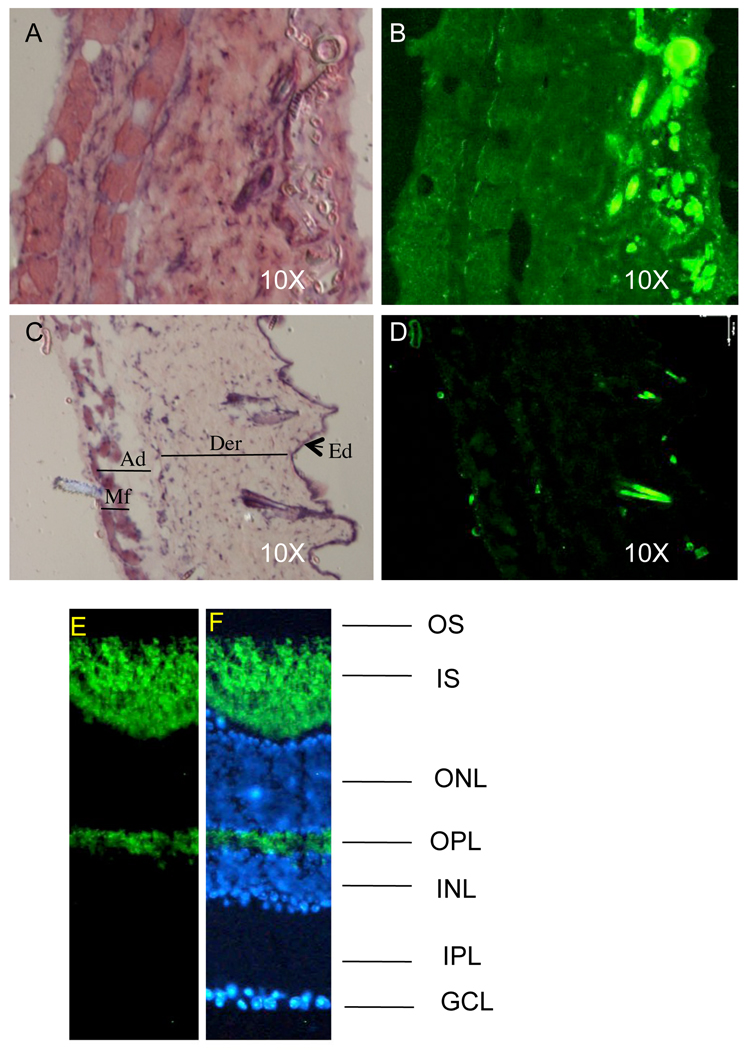
Finally, we tested whether the levels of reporter expression in the knock-in animals were sufficient to detect clear fluorescence signals in thin (5 microns) tissue cross-sections. The eye was chosen as an example of a highly fluorescent tissue and skin as an example of a tissue with lover levels of the reporter expression. Fig. 6A and B demonstrate that EGFP fluorescence is detectable in all layers of the skin relative to the wild-type skin (Fig. 6C and D). In the retina, expression of EGFP was detected predominantly in the photoreceptor layer. As shown in Fig 6E and F, the EGFP was observed in the photoreceptor inner segment and the synaptic (outer plexiform) layer. Photoreceptors are highly metabolically active neurons as they undergo periodic outer segment disc shedding and renewal (Young 1976). Hence, photoreceptors consume large amounts of ATP generated by oxidative phosphorylation in the mitochondrial electron transport chain (Yu et al. 2001; Yu et al. 2005). The fact that mitochondria are predominantly located in the photoreceptor inner segment (Hoang et al 2002; Perkins et al. 2003) is consistent with an important role of Supv3L1 in enabling high mitochondrial activity during light-dependent signal transduction cascades.
Fig. 6.
Detecting EGFP emission in tissue sections. (A) H&E stained section of the skin derived from Supv3L1tm6Jkl/+ animal. (B) Fluorescent light emission from the same section before H&E staining. Note that all cell layers including subcutaneous muscle fibers are positive. Bright fluorescent spots on the epidermal side of the skin are the hair shafts. (C) H&E stained section of the wild-type skin. Ad, adipose layer; Der, dermis; Ed, epidermis; Mf, subcutaneous muscle fibers. (D) Same section before H&E staining that shows no EGFP emission. Bright spots on the epidermal side of the crossection are the hair shafts that fluorize in the EGFP-independent manner. (E) Fluorescent light emission in mouse retina cryosections and (F) co-staining with hoechst (blue; nuclear stain). The signal is predominantly detected in photoreceptor inner segment (IS) and outer plexiform layer (OPL). OS: outer segment; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer.
In summary, we have generated both transgenic and a knock-in mouse strains that express an EGFP reporter under control of the Supv3L1 promoter. We found that paternally inherited reporter expression begins early in development, being first detectable at the blastocyst stage. Abundant and widespread expression persists during all developmental stages as well as during postnatal growth. Expression diminishes somewhat in most adult tissues, but it remains high in testes, brain, and certain types of sensory organs and cells, such as the retina. The spatial and temporal patterns of expression were indistinguishable in the transgenic and knock-in animals. Expression levels were more faithfully maintained during adulthood in the knock-in mice and this line is likely to produce more reliable results in future experiments. However, both the transgenic and knock-in strains constitute valuable research tools that will facilitate future non-invasive studies of the intriguing biology of Supv3L1 in whole animals, as well as in ex vivo cell culture models derived from them.
Acknowledgements
This work is supported by grants 5P20RR015578-07 and EY007961 from the National Institute of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, Jonkers J, Smith J, Plumb B, Taylor R, Nishijima I, Yu Y, Rogers J, Bradley A. Mutagenic insertion and chromosome engineering resource (MICER) Nat Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- Beddington RSP. Transgenic strategies in mouse embryology and development. In: Grosveld FG, Kollias G, editors. Transgenic Animals. San Diego: Academic Press; 1992. pp. 79–98. [Google Scholar]
- Butow RA, Zhu H, Perlman P, Conrad-Webb H. The role of a conserved dodecamer sequence in yeast mitochondrial gene expression. Genome. 1989;31:757–760. doi: 10.1139/g89-134. [DOI] [PubMed] [Google Scholar]
- Conrad-Webb H, Perlman PS, Zhu H, Butow RA. The nuclear SUV3-1 mutation affects a variety of post-transcriptional processes in yeast mitochondria. Nucleic Acids Res. 1990;18:1369–1376. doi: 10.1093/nar/18.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmochowska A, Kalita K, Krawczyk M, Golik P, Mroczek K, Lazowska J, Stepien PP, Bartnik E. A human putative Suv3-like RNA helicase is conserved between Rhodobacter and all eukaryotes. Acta Biochim Pol. 1999;46:155–162. [PubMed] [Google Scholar]
- Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Transgenic mice as probes into complex systems. Science. 1989;246:1265–1275. doi: 10.1126/science.2686032. [DOI] [PubMed] [Google Scholar]
- Hoang QV, Linsenmeier RA, Chung CK, Curcio CA. Photoreceptor inner segments in monkey and human retina: mitochondrial density, optics, and regional variation. Vis Neurosci. 2002;19:395–407. doi: 10.1017/s0952523802194028. [DOI] [PubMed] [Google Scholar]
- Khanna H, Davis EE, Murga-Zamalloa CA, Estrada-Cuzcano A, Lopez I, den Hollander AI, Zonneveld MN, Othman MI, Waseem N, Chakarova CF, Maubaret C, Diaz-Font A, Macdonald I, Muzny DM, Wheeler DA, Morgan M, Lewis LR, Logan CV, Tan PL, Beer MA, Inglehearn CF, Lewis RA, Jacobson SG, Bergmann C, Beales PL, Attié-Bitach T, Johnson CA, Otto EA, Bhattacharya SS, Hildebrandt F, Gibbs RA, Koenekoop RK, Swaroop A, Katsanis N. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidr L, Wu G, Davila A, Procaccio V, Wallace D, Lee WH. Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J Biol Chem. 2008;283:27064–27073. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J, Singer JD. Mice with the enhanced green fluorescent protein gene knocked in to chromosome 11 exhibit normal transmission ratios. Biochem Genet. 2005;43:321–333. doi: 10.1007/s10528-005-5223-6. [DOI] [PubMed] [Google Scholar]
- Kollias F, Grosveld FG. Transgenic strategies in mouse embryology and development. In: Grosveld FG, Kollias G, editors. Transgenic Animals. San Diego: Academic Press; 1992. pp. 47–77. [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, Mroczek S, Pawlak SD, Stepien PP. Human ATP-dependent RNA/DNA helicase hSuv3p interacts with the cofactor of survivin HBXIP. Febs J. 2005;272:5008–5019. doi: 10.1111/j.1742-4658.2005.04910.x. [DOI] [PubMed] [Google Scholar]
- Minczuk M, Piwowarski J, Papworth MA, Awiszus K, Schalinski S, Dziembowski A, Dmochowska A, Bartnik E, Tokatlidis K, Stepien PP, Borowski P. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–5086. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Brinster RL. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul E, Cronan R, Weston PJ, Boekelheide K, Sedivy JM, Lee S-Y, Wiest DL, Resnick MB, Klysik JE. Disruption of Supv3L1 damages the skin and causes sarcopenia, loss of fat, and death. Mamm Genome. 2009;20:92–108. doi: 10.1007/s00335-008-9168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M, Mason P, Szczesny RJ, Maddukuri L, Dziwura S, Jedrzejczak R, Paul E, Wojcik A, Dybczynska L, Tudek B, Bartnik E, Klysik J, Bohr VA, Stepien PP. Interaction of human SUV3 RNA/DNA helicase with BLM helicase: loss of the SUV3 gene results in mouse embryonic lethality. Mech Aging Dev. 2007;128:609–617. doi: 10.1016/j.mad.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Ellisman MH, Fox DA. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: bioenergetic and functional implications. Mol Vis. 2003;9:60–73. [PubMed] [Google Scholar]
- Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- Shu Z, Vijayakumar S, Chen CF, Chen PL, Lee WH. Purified human SUV3p exhibits multiple-substrate unwinding activity upon conformational change. Biochemistry. 2004;43:4781–4790. doi: 10.1021/bi0356449. [DOI] [PubMed] [Google Scholar]
- Stepien PP, Margossian SP, Landsman D, Butow RA. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc Natl Acad Sci USA. 1992;89:6813–6817. doi: 10.1073/pnas.89.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny RJ, Obriot H, Paczkowska A, Jedrzejczak R, Dmochowska A, Bartnik E, Formstecher P, Polakowska R, Stepien PP. Down-regulation of human RNA/DNA helicase SUV3 induces apoptosis by a caspase- and AIF-dependent pathway. Biol Cell. 2007;99:323–332. doi: 10.1042/BC20060108. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DD-H, Shu Z, Lieser SA, Chen P-L, Lee W-H. Human mitochondrial SUV3 and PNPase form a 330kDa heteropentamer to cooperatively degrade dsRNA with 3'-to-5' directionality. J Biol Chem. 2009 doi: 10.1074/jbc.M109.009605. doi/10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Visual cells and the concept of renewal. Invest Ophthalmol Vis Sci. 1976;15:700–725. [PubMed] [Google Scholar]
- Yu DY, Cringle SJ. Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Prog Retin Eye Res. 2001;20:175–208. doi: 10.1016/s1350-9462(00)00027-6. [DOI] [PubMed] [Google Scholar]
- Yu DY, Cringle SJ. Retinal degeneration and local oxygen metabolism. Exp Eye Res. 2005;80:745–751. doi: 10.1016/j.exer.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Zhu H, Conrad-Webb H, Liao XS, Perlman PS, Butow RA. Functional expression of a yeast mitochondrial intron-encoded protein requires RNA processing at a conserved dodecamer sequence at the 3' end of the gene. Mol Cell Biol. 1989;9:1507–1512. doi: 10.1128/mcb.9.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]