Summary
This article proposes saddlepoint approximations to the expectation and variance-covariance function of multitype age-dependent branching processes. The proposed approximations are found accurate, easy to implement, and much faster to compute than by simulating of the process. Multiple applications are presented, including the analyses of clonal data on the generation of oligodendrocytes from their immediate progenitor cells, and on the proliferation of Hela cells. New estimators are also constructed to analyze clonal data. The proposed methods are finally used to approximate the distribution of the generation, which has recently found several applications in cell biology.
Keywords: Cell kinetics, Discretely observed stochastic process, Oligodendrocytes, Pseudo-likelihood, Quasi-likelihood, Saddlepoint approximation
1. Introduction
1.1 The general problem
Age-dependent branching processes have become well-established means to study the dynamics of complex biological systems. Recently, they have found successful applications in such fields as stem cell biology to study the processes of division, differentiation and death of progenitor cells (Yakovlev et al, 1998; Boucher et al, 1999, 2001; Zorin et al, 2000; Hyrien et al, 2005a, 2005b), or to investigate the propagation of prions in yeast (Morgan, Ridout, and Ruddock, 2003). Additional examples can be found in relevant monographs (Jagers, 1975; Yakovlev and Yanev, 1989; Kimmel and Axelrod, 2002; Haccou, Vatutin and Jagers, 2005).
The practical implementation of age-dependent branching processes, however, remains hindered by the fact that most characteristics of the process, including the moments of the population size at any given time, do not have closed-form expressions. Three approaches are available to mitigate this limitation. A first approach, which is rarely satisfactory, consists in using a simplified version of the model (such as discrete- or continuous-time Markov branching processes) that allows computations to be carried out explicitly. A second approach is to use asymptotic approximations of the characteristics of interest as time goes to infinity. For instance, Hoel and Crump (1974) used the asymptotic form of the expected population size to estimate parameters of the distribution of the cell cycle time. More recently, Ridout et al (2006) and Cole et al (2007) used a similar approach to approximate the malthusian parameter and the expected generation number of age-dependent branching processes. However, the asymptotic behavior of the process may be diffcult to study for multitype processes with complex branching structures, and the accuracy of asymptotic approximations may not always prove satisfactory for transient properties. A third approach is to resort to computer simulations of the process to generate the desired approximations. Zorin et al (2000), Boucher et al (2001), Hyrien et al (2005a, 2005b), Hanin et al (2006), Golinelli et al. (2006), and Hyrien (2007) resorted to such a technique to estimate cell kinetics parameters when the process is observed at discrete points in time (a situation sometimes referred to as clonal experiments in cell biology). The simulation-based approach does not have the limitations of the above two methods, but it may break down for several reasons.
Firstly, it is time-consuming when a large number of cells have to be simulated, either because the number of initial ancestors is large, or because simulations have to be conducted on an extended period of time, or because the process is supercritical (that is, the expected number of offsprings per cell is greater than one). Supercritical processes provide reasonable models of the earliest stages of cell proliferation where the population size may grow exponentially (Jagers, 1975). Consider for instance the branching process for which every cell divides into two cells upon completion of its mitotic cycle, and assume that the duration of the cycle follows a gamma distribution with mean 36 hours and standard deviation 45 hours. To approximate moments of the population size at day 8, starting with a single cell at time 0, we could consider simulating 10, 000 realizations of the process (in our past work we have often used 100,000 simulations; Hyrien et al, 2005a, 2005b), and compute relevant empirical moments of cell counts. These simulations would take us about 1/2 hour on average. When decreasing the mean mitotic cycle duration to 34 or 32 hours, we would need 1.6 and 6.1 hours instead to perform the same task. The computing time is thus very sensitive to how fast the population grows. If such approximations were used to estimate model parameters (as is the case with the simulated pseudo-likelihood estimator (Hyrien, 2007)), the above 10,000 simulations would have to be repeated thousands of times to optimize the estimating function, which would make the approach very time consuming in that particular case.
The second limitation has to do with the discrete nature of branching processes which makes simulation-based approximations non-differentiable (Hyrien, 2007). This precludes the use of a variety of estimation techniques, including quasi-likelihood estimators (Hyrien, 2007) which have been proven to enjoy optimality properties (Heyde, 1997).
The objective of the present paper is to propose a novel approach to approximate moments of multitype age-dependent branching processes, which does not suffer from the above-mentioned limitations. The proposed approach rests on the technique of saddlepoint approximations, which is applied to suitable decompositions of the moments. Our investigations demonstrated that this approach can be thousand times faster than the method using direct computer simulations of the process, while still providing very accurate approximations (see Supplementary Materials). In addition, the use of saddlepoint approximations makes it available a wider range of estimation techniques for discretely observed branching processes, such as the methods of quasi-likelihood and generalized estimating equation.
1.2 A motivating example from stem cell biology
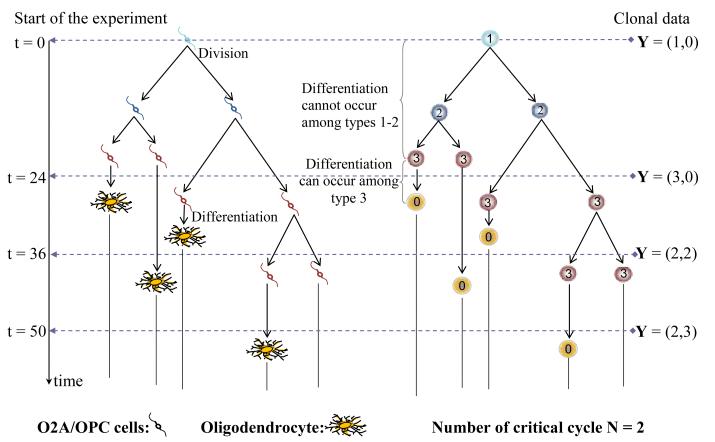
One cellular system that has been extensively studied using age-dependent branching processes is that of oligodendrocytes (the cells of the central nervous system (CNS) that enwrap axons with myelin sheaths) generated from their immediate ancestral progenitor cells (Raff et al, 1983). The oligodendrocyte-type 2 astrocyte progenitor cell (also referred to as an oligodendrocyte precursor cell and here abbreviated as O2A/OPC) can be isolated from multiple regions of the CNS and studied in vitro. The O2A/OPC lineage has proven to be exceptionally suitable for studying differentiation at the clonal level. Dividing O2A/OPCs can be readily distinguished from oligodendrocytes on the basis of cell morphology (the former being bipolar with small elliptical cell bodies and the latter being multipolar with large round cell bodies), enabling unambiguous identification of every cell in a clone (e.g, Noble et al, 1988). Moreover, O2A/OPCs readily establish clones from single founder cells, and the subsequent development of these clones can be followed over time. (see Figure 1 for an illustration of the development of a clone of O2A/OPCs). Clonal analyses have been of enormous value in investigating the control of differentiation at the single cell level. Despite the power of clonal analyses in the study of differentiation, they also have an important limitation. The complete histories of the cellular trees are not fully observed, but instead the composition of the clones at the times of observation are used to provide information on the proliferative patterns of the cells. The quantitative analysis of such experiments calls for specific estimation methods. One objective of this article is to propose new methods for such data built on the proposed saddlepoint approximations.
Figure 1.
The remainder of this article is organized as follows. Section 2 describes the process under consideration along with the proposed saddlepoint approximations. New estimation methods for clonal data using branching processes are presented in section 3. These methods are used in section 4.1 to analyze the generation of oligodendrocytes from cultured O2A/OPCs. The subsequent sections present two additional applications. In section 4.2 we analyze a second set of clonal data on the proliferation of cultured Hela cells, where we investigate the assumption that the distribution of the time to division does not depend on the generation, an assumption that is not often assessed in cell kinetic studies. Finally, our last illustration (section 5) considers the problem of approximating the distribution of the generation, which has found recent applications in the analysis of cell proliferation kinetics (Cole et al, 2006; Hyrien and Zand, 2008).
2. A multitype age-dependent branching process
2.1 The process
We model cell kinetics using a multitype age-dependent branching process presented by Hyrien et al (2005a), which generalizes the classical multitype Bellman-Harris process. The process under consideration is akin to a two-type process earlier proposed by Jagers (1975), and it can also be viewed as a multitype extension of the Sevastyanov’s process.
The process under consideration describes a population consisting of K cell types (1 ≤ K ≤ ∞). Without loss of generality, this process begins at time 0 with a single initiator cell of type 1 and age zero. Upon completing its lifespan (that is, the time to transformation of the cell, such as division, differentiation or death, computed from its birth), every type-k cell, k = 1, …, K, generates ξk,1 type-1 cells, …, ξk,K type-K cells. The vector ξk = (ξk,1, …, ξk,K) is assumed to be a random vector with probability distribution function . Each realization of this vector corresponds to a particular transformation of the cell. Let Sk denote the support of ξk. To illustrate and motivate the construction of this process, consider a simple two-type process, with type-1 and type-2 cells corresponding to progenitor cells and to terminally differentiated cells, respectively. Upon completion of its lifespan, every type-1 cell either divides into two type-1 cells with probability p, or it differentiates into a single type-2 cell with probability q = 1 − p, while every type-2 cell dies with probability one. In this example, ξ1 and ξ2 are 1 × 2-vectors with respective supports S1 = {(2, 0); (0, 1)} (for whether type-1 cells divide or differentiate) and S2 = {(0, 0)}.
Let τk denote the lifespan of any type-k cell. In the Bellman-Harris process, ξk and τk are mutually independent. Put in the context of our two-type process, this means that the times to division and the times to differentiation of type-1 cells are identically distributed. However, the processes of division and differentiation involve different molecular mechanisms, so that the occurence of these two events would likely take place at distinct points in time, as suggested by our past studies on O2A/OPCs (Hyrien et al, 2005a; 2006) and an analysis presented in section 4.1 of this paper, where it was found that the lifespans of cells that divide and those of cells that differentiate do not follow the same distributions. The assumption behind the Bellman-Harris process seems therefore biologically unrealistic, and our process relaxes it by introducing a different distribution for every possible transformation of the cell, and this for every cell type. Thus, for every x = (x1, …, xK) ∈ Sk, let denote the conditional distribution of the lifespan of any type-k cell, given the cell undergoes the transformation defined by x. For instance, in our example, we would specify distributions F1,(2,0)(t) and F1,(0,1)(t) for the time to division and for the time to differentiation of type-1 cells (under a Bellman-Harris process, these distributions would be identical). Under the above specifications, the marginal distribution of the lifespan is clearly a mixture Fk(t)=Σx∈Skpk(x)Fk,x(t). For regularity of the process, assume further that Fk,x(0) = 0 (so the lifespan cannot be zero), and that the τk’s are non-lattice random variables (r.v.’s). Last, assume every cell of the population evolves independently of every other cell.
2.2 Expectation and variance-covariance of the process
For every a, j, k = 1, …, K, let , where denotes the number of type-k cells at time t ≥ 0 arising from a single ancestor cell of type-a (in subsequent sections, the superscript a shall be dropped whenever unecessary). Let and denote the expectation and covariance function of the process. Write and .
The expectations satisfy the system of functional equations
where 1{a=k} is the indicator function. The variance-covariance function can be written as
where
and where
These equations can be solved using renewal theory arguments (Athreya and Ney, 1972). For instance, for the binary splitting process, where K = 1 and where ξ1 = 2 with probability one, the expressions for the expectation and variance reduce to
and
where F★l(·) denotes the l-fold convolution of F (·), and where, by convention, F★0(·) = 1. In the most general case, these moments take more complicated expressions. However, just as in the above example, the expected values can be decomposed in terms of linear combinations of functions taking the form
| (1) |
for some xkl ∈ Skl, kl ∈ {1, …, K}, l = 1, …, M, and where denotes the convolution of Fk1,xk1 and Fk2,xk2. The variance-covariance functions may sometimes involve extra terms of the form
| (2) |
for some given function g(t), where c(t) = dC(t)/dt.
The coeffcients associated with the functions C(t) and I(t) in the expressions for and are solely defined through the offspring distributions, and are computed explicitly. The integral in (2) is easily handled by numerical integration, which does not present any real diffculty here since integration is over a one-dimensional space. The functions c(t) and C(t), however, have generally no closed-form expressions, and they remain the sole obstacle to the explicit computation of the expectation and variance-covariance function of the process. Hyrien et al. (2005a, 2005b) and Hyrien (2007) considered simulating the branching process to compute the needed approximations. We present an alternative method that takes advantage of the above decompositions of the moments.
2.3 Saddlepoint approximations
The functions c(·) and C(·) are the p.d.f. and c.d.f. of a r.v. S = τk1 + ⋯ + τkM, where τkl, l = 1, …, M, are independent r.v.’s with respective c.d.f.’s Fkl,xkl. Specifically, S represents the sum of the lifespan of a cell of type kM and of those of its kM − 1 immediate ancestors. A reasonable approach to approximate c(s) and C(s) is through saddlepoint approximation, a technique that has become well understood to approximate tail probabilities and distribution functions since Daniels (1954)’s seminal paper. Other notable contributions are the work by Barndorff-Nielsen and Cox (1979), Lugannani and Rice (1980), and Renshaw (2000), to name a few, where several saddlepoint approximations have been developed for p.d.f.’s and for c.d.f.’s of non-lattice r.v.’s. For reason of space, we limit our consideration to a single one for C(s) and for c(s).
Denoting by the cumulant generating function of any r.v. X, and by its r-th derivative (providing that these quantities exist), we have and Following Daniels (1987) we approximate c(s) by
| (3) |
where , and where denotes the root to the equation . The root always exists. Using the method proposed by Lugannani and Rice (1980), we approximate C(s) by
| (4) |
where φ(·) and Φ(·) denote the p.d.f. and c.d.f. of the standard normal distribution, and where and . When , (4) is replaced by
| (5) |
In our first approximation to the moments of Z(a)(s), the functions c(s) and C(s) are replaced by their respective saddlepoint approximations. When does not take an explicit expression, a root finder algorithm needs to be implemented.
3. Statistical inference from clonal data
Clonal experiment is an experimental setup that is commonly used to study the processes of cell division, cell differentiation and cell death under in vitro conditions. In such experiments, isolated founding initiator cells are placed in a culture medium with predefined growing conditions. Each founder cell initiates its own population (or clone), the composition of which will change over time in accordance with the processes of division, differentiation and death that govern the cellular system under consideration. The resultant clone is examined at a given point in time, and the number of cells for each observable cell type are reported. This is done for multiple cell clones and at different time points so one can observe what happens over time. For instance, in the context of cultured oligodendrocytes, two cell types can be visually distinguished based on their morphology (O2A/OPCs and oligodendrocytes); clonal experiments would provide the numbers of cells at the sampling time for each of these two cell types in individual clones. Thus in the clone depicted in Figure 1, the experimentalist would report 2 O2A/OPCs and 2 oligodendrocytes at t = 36 hrs. In subsequent experiments, each clone is only to be examined at a single time point.
Let Yil denote the number of cells of the lth observable type scored at time ti in the ith clone, where l = 1, …, L, and i = 1, …, n; write Yi = (Yi1, …, YiL)’. The notation L and n stand for the numbers of observable cell types and for the number of clones examined during the experiment, respectively. It is important to note that L need not be equal to K, the latter denoting the number of (modeling) types used to construct the model (see section 4.2 for an example where L = 2 and K = ∞). The clones grow separately during the experiment, and it is therefore reasonable to assume that the r.v.’s Yi, i = 1, …, n, are independent. In order to gain a quantitative insight into the dynamics of a given cell population, one formulates a branching process, the probability measure of which will be fully specified up to an unknown finite dimensional parameter vector θ. This vector will typically contain parameters of the distributions of the cell lifespans and of the offsprings. In what follows, we propose methods to estimate θ using our saddlepoint approximations.
In order to construct realistic models of the temporal development of any given cell population, observable cell types sometimes need to be described by multiple types in the branching process (so that K > L). For instance, the proliferative characteristics of O2A/OPCs are suspected to change with the generation, and these cells will be represented by 3 types in the branching process defined in section 4.1. In contrast, the pool of oligodendrocytes is more homogeneous, and it will be described by a single type. Thus, define a stochastic process , where Il, l = 1, …, L, are L subsets forming a partition of {1, …, K}; that is, and Il1 ⋂ Il2 = ∅ for all 1 ≤ l1 ≤ l2 ≤ L. Yl(t) denotes the number of cells of the lth distinguishable cell type (for simplicity, we shall assume that the process starts from a type-1 cell). Let and denote the expectation of and Yl(t) for the parameter vector θ. We have that . Write mY (t, θ) = {mY,1(t, θ), …, mY,L(t, θ)}’, and put for the saddlepoint approximation of mY (t, θ). Similarly, let denote the saddlepoint approximation to the variance-covariance matrix of Y(t) = {Y1(t), …, YL(t)}’ (the latter being denoted by ΩY(t, θ)).
Several methods have been proposed to analyze clonal data (Nedelman et al., 1985; Yakovlev et al., 1998a, 1998b, 2000; Boucher et al., 1999, 2001; von Collani et al., 1999 Zorin et al., 2000; Hyrien et al., 2005a, 2005b; Hyrien, 2007). All these publications resorted to branching processes to model clonal expansion. Diffculties in the statistical analysis of such data arise mainly because generally neither the distribution nor the moments of the process have tractable explicit expressions in the non-Markovian case. Alternatives to the Monte Carlo estimators proposed by Zorin et al. (2000), Hyrien et al. (2005a, 2005b) and Hyrien (2007) can be constructed using the proposed saddlepoint approximations.
A pseudo likelihood (PL) estimator will maximize
Altought the PL function takes the form of the log-likelihood of a Gaussian sample, the normality assumption is not required for this approach to be justified (Hyrien, 2007). In the absence of normality, PL estimators are not generally effcient however. More effcient estimators, also defined from the expectation and variance-covariance matrix of the process, can be constructed using the quasi-likelihood (QL) approach. We thus propose a QL estimator that solves the quasi-score equation Q(θ) = 0, where
QL estimators require to differentiate expected values of the process with respect to θ. When the moments are approximated through simulations, the resultant approximations are not differentiable. This is due to the discrete nature of Y(t) (Hyrien, 2007). Saddlepoint approximations to the moments do not suffer from this limitation.
When the branching process contains infinitely many types, the model-based variance-covariance matrix ΩY(t, θ) may have a cumbersome expression, making it diffcult to compute the above PL and QL estimators. The method of generalized estimating equation (GEE) provides an alternative approach in such cases (Liang and Zeger, 1986). The GEE estimator solves the equation QGEE(μ) = 0, where
and where denotes a working correlation matrix that does not depend on θ. When the experimental design provides independent replicated observations in suffcient number at every time point, can be taken as the empirical estimator of the variance-covariance matrix of Yi. In other situations the working “independence” correlation matrix (the L × L identity matrix) would also be acceptable, althought it is generally suspected to decrease effciency of estimators when observations are moderately or strongly dependent (Liang and Zeger, 1986).
When computed from exact moments, PL, QL and GEE estimators will generally be consistent and asymptotically Gaussian under mild regularity conditions. We refer to Hyrien (2007) for asymptotic properties of the PL estimator in the context of discretely observed age-dependent branching processes. When moments are approximated using saddlepoint approximations, these asymptotic properties will not generally hold true unless the order of the approximations increases with the sample size (which would happen if the sampling times increased with the sample size or if higher order terms were included in the saddlepoint approximations of section 2.3). If the approximations to the moments are in close agreement with their exact values, however, the resultant estimators are expected to be close to the “exact” estimators. The performance of the proposed approximate estimators can be assessed in simulation studies. One such studies is reported in Supplementary Materials, which suggested that the proposed estimators are essentially unaffected by approximation of the moments (e.g., the bias of the estimators is negligible), and that they remain virtually identical to their exact counterparts. Analysis of experimental data often involve the construction of confidence intervals for model parameters and the test of hypotheses (e.g., for assessing the effects of different culture conditions on cell proliferation or for comparing nested models). These can be accomplished using resampling and Monte Carlo techniques. We refer to Hyrien et al (2005) for a Monte Carlo Wald test, which can be easily adapted to the above estimators, and to Diccicio and Efron (1996) for confidence intervals.
4. Analysis of experimental clonal data
4.1 The generation of oligodendrocytes from cultured O2A/OPCs
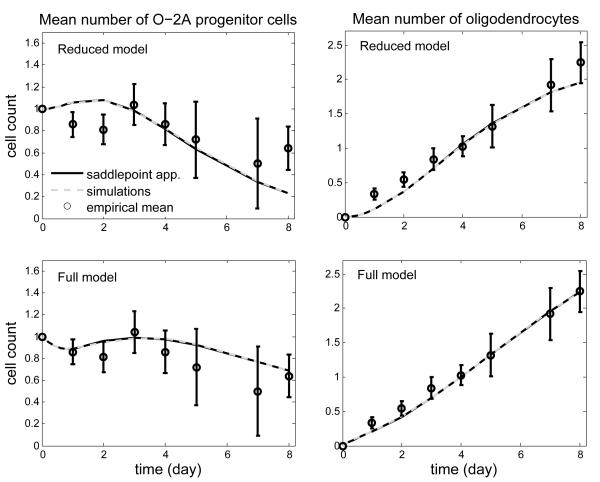
To illustrate the proposed methods we analyzed a set of clonal data on the generation of oligodendrocytes from O2A/OPCs cultured in vitro. In this experiment, O2A/OPCs isolated from the optical nerve of 7-day old rats were plated in cell culture in the presence of thyroid hormone. Cells were fed every other day with fresh PDGF. Clones were observed 1, 2, 3, 4, 5, 7 and 8 days after plating. At each time of observation, the composition of either 50, 100 or 150 clones (depending on the time point; total sample size n = 850) was individually examined to obtain the number of O2A/OPCs and the number of oligodendrocytes in each of them (as illustrated in Figure 1). The clones were discarded after examination. The mean number of O2A/OPCs and the mean number of oligodendrocytes versus time are showed in the left and right panels (respectively) of Figure 2. Each clone started with a single O2A/OPC and zero oligodendrocyte. The mean number of O2A/OPCs appears to decrease at days 1 and 2, increase at day 3, and decrease again after day 3. In contrast, the mean number of oligodendrocytes increases steadily in an almost linear fashion from the start of the experiment. On day 8, any clone would contain on average 0.7 O2A/OPC and 2.3 oligodendrocytes. The largest clone contained a total of 12 cells.
Figure 2.
The objective of our analyses was to gain a quantitative insight into how cell clones develop over time and to estimate cell kinetics parameters. The temporal development of clones of O2A/OPCs was modeled using a modified version of the branching process proposed by Boucher et al. (1999). This model assumes a random, clone-specific “critical” number, N, that identifies the generation up to which differentiation of O2A/OPCs into oligodendrocytes cannot occur. Differentiation may only happen among cells of generation greater than N. The number of modeling types is K = N + 2. The process was defined from the following assumptions (see Figure 1, right panel, for a schematic representation), where, again, lifespan is used as a generic term for the time to division or to differentiation computed from birth.
A1- Without loss of generality, the process begins with a single cell of generation one (a founder O2A/OPC). This cell is said to be of type 1.
A2- In clones such that N > 0, every type-k cell, k = 1, …, N, divides into two type-(k + 1) cells (O2A/OPCs) when it completes its lifespan.
A3- Upon completion of its lifespan, every type-(N + 1) cell either divides into 2 new type-(N + 1) cells with probability p, or it differentiates into one type-0 cell (an oligodendrocyte) with probability 1 − p.
A4- The time to division of any type-k cell, k = 1, …, N +1, is represented by a non-negative r.v. τ1 with c.d.f. .
A5- The time to differentiation of any type-(N + 1) cell is represented by a non-negative r.v. τ2 with c.d.f. .
A6- Type-0 cells (oligodendrocytes) have an infinite lifespan.
In the above model, the distributions G1(·) does not depend on k, the type of the cell. Boucher et al. (1999) further assumed that G1(·) = G2(·) (a Bellman-Harris process). Based on past investigations (Hyrien et al., 2005a; 2006), we relaxed this constraint to allow G1(·) and G2(·) to be dissimilar gamma distributions with respective means and variances ml and , l = 1, 2. Let and denote the corresponding shape and scale parameters. We shall refer to Boucher et al. (1999)’s model as the reduced model, and to the above one as the full model. Boucher et al. (1999) found that the support of N was the set {0, 1, 2}; we proceeded from this choice as well in our analyses, where we set , c = 0, 1, 2. Here we have θ = (m1, σ1, m2, σ2, p, π0, π1, π2)’.
Boucher et al. (1999) derived expressions for the expectation and variance of the number of O2A/OPCs and of the number of oligodendrocytes as a function of time in the case where G1(t) = G2(t). Using similar techniques, it is easy to obtain expressions for these moments in the full model. For instance the expected number of O2A/OPCs at any time t ≥ 0 is given by , where
The expression for the covariance between the numbers of O2A/OPCs and the number of oligodendrocytes is also derived in a similar fashion. These moments involve convolutions of the form , k = 0, 1 …, where, for k ≥ 1, is the c.d.f. of a gamma distribution with mean and variance km1 and . The associated cumulant generating function takes the form Kk(u) = −kα1 log(1 − β1u) − α2 log(1 − β2u). Its rth derivative is given by
and the solution to the equation is
| (6) |
Saddlepoint approximations to the moments can be easily constructed using the above quantities and equations (3-5).
While O2A/OPCs and oligodendrocytes are the only two distinguishible cell types, we enriched cell classification by defining L = 4 “observable” cell types: O2A/OPCs of first generation (type 1), O2A/OPCs of generation greater than one (type 2), oligodendrocytes that arise from an O2A/OPC of first generation (type 3), and all other oligodendrocytes (type 4). Based on the composition of the clones, the number of cells of each type can be easily determined. For instance, a clone containing a single O2A/OPCs will yield one cell of type 1 and 0 cells of types 2, 3, and 4. The expressions for the mean, variance and covariance of the number of cells of these four types can be determined as explained above. This enriched classifiation is expected to improve statistical inference since estimation is conducted conditional on whether the lifespan of the initiator cell has ended and on whether this cell ended up dividing or differentiating. In a simulation study, we compared the performance of the pseudo-likelihood estimator based on the original two cell types with the enriched classification. The study indicated that the variance of the estimator is substantially decreased in the latter case.
PL and QL estimates are reported in Table 1. The fitted model is shown in Figure 2. The full model provided a better fit than the reduced model. PL and QL estimates were relatively close. The full model suggested that the mean time to division of O2A/OPCs was substantially larger than their mean time to differentiation (23-30 hrs vs. 45-50 hrs). This is consistent with the results of a past analysis reported by Hyrien et al. (2005a) using a different model. Standard errors were computed for the parameter estimates of the full model fitted to the four cell types using bootstrap. The QL estimates of the mean and variance of the distribution of the time to events were smaller than their PL counterparts; the standard errors of the PL estimates for the parameters of the probability of division were smaller than those for the QL estimates. To assess the accuracy of saddlepoint approximations, we computed simulation-based approximations to the moments. The two approximations were almost perfectly superimposed (Figure 2).
Table 1.
Parameter estimates and their standard errors for the reduced and the full models
| m1 (hr) | σ1 (hr) | m2 (hr) | σ2 (hr) | p | π 0 | π 1 | π 2 | ||
|---|---|---|---|---|---|---|---|---|---|
| Reduced model (Similar distribution for time to transformation) |
|||||||||
| QL | estimate | 44.5 | 28.9 | = m1 | = σ1 | 0.21 | 0.47 | 0.45 | 0.08 |
| PL | estimate | 45.4 | 36.3 | = m1 | = σ1 | 0.32 | 0.49 | 0.51 | 0.00 |
| Full model (Dissimilar distribution for time to transformation) |
|||||||||
| QL | estimate s.e. |
51.9 1.8 |
28.2 1.4 |
29.3 4.8 |
28.9 7.5 |
0.38 0.06 |
0.54 0.07 |
0.33 0.08 |
0.13 0.09 |
| PL | estimate s.e. |
47.2 3.1 |
23.7 3.1 |
30.9 6.8 |
49.8 10.3 |
0.31 0.04 |
0.42 0.03 |
0.29 0.05 |
0.29 0.05 |
4.2 The proliferation of Hela cells
In the quantitative analysis of dividing cell populations it is commonly assumed that the distribution of the mitotic cycle (MC) duration remains identical across generations, with the occasional exception of that of initiator cells (e.g., Hanin et al, 2006). There exists a number of biological reasons for questioning this assumption, however, since cell proliferation may slow down due to more limited access to nutrients or to cell signaling. In this second application the proposed method is used to investigate this assumption through analyzing a set of clonal data on the proliferation of Hela cells cultured in vitro. In contrast to the previous example, clonal cultures of these cells contained a single morphologically distinguishible cell type (L = 1), and every cell can be assumed to divide into two new cells upon completion of its MC with probability one. In the experiment under consideration, the number of cells was counted in multiple clones at the following time points: 10, 16, 18, 20, 24, 30, 35, 40, 48, 55, and 60 hours. About one hundred clones were examined at each of these time points, totaling n = 1077 clones in the entire experiment (See Hanin et al (2006) for further experimental details).
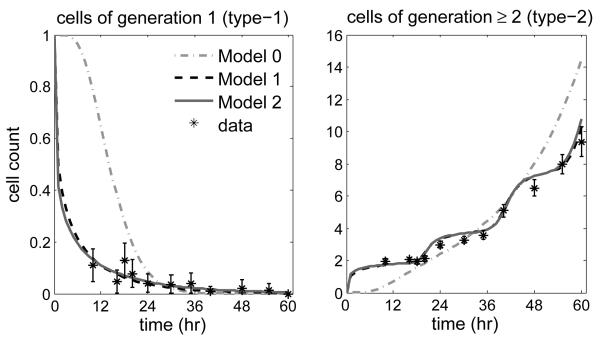
We modeled cell proliferation using a branching process with infinitely many types (K = 1), with each type representing a specific generation. This process begins with a single initiator cell of generation 1. The MC duration of any cell of generation g follows a gamma distribution with mean μg and variance , g ≥ 1. The corresponding c.d.f. shall be denoted by Gg. Hanin et al (2006) considered a model where the mean and variance for the MC duration were assumed identical across generations, except for cells of first generation, so that μ1 ≠ μ2 and μ2 = μ3 = ⋯, and similarly for the variances. We shall refer to this process as Model 1.
We first challenged Model 1 against a simpler model (Model 0) that assumes that the mean and the variance of the MC duration do not change with generation; that is, we tested the null hypothesis vs. . This was done using a Monte Carlo Wald test as described by Hyrien et al (2005), with simulated data generated using fitted Model 0. Our analysis strongly supported Model 1 (P < 0.001), and suggested that the mean MC duration was much shorter for cells of first generation (4.4 hrs vs. 20.1 hrs; see Table 2). This can be explained by the fact that founder cells are sampled at some point in their MC (not at the beginning), and μ1 would rather represent the expected residual MC duration.
Table 2.
Parameter estimates for Model 0, 1, and 2
| Model | μ 1 | σ 1 | μ 2 | σ 2 | γ | κ |
|---|---|---|---|---|---|---|
| 0 | 15.0 | 6.2 | - | - | - | - |
| 1 | 4.4 | 8.3 | 20.1 | 1.9 | - | 0.09 |
| 2 | 4.4 | 9.9 | 20.8 | 2.2 | -0.03 | 0.11 |
To further assess Model 1, we tested it against an extended model (Model 2), which allowed the means and variances for the MC duration of cells of all generations to be different. To perform this analysis we set μg = μ2eγ(g-2), ∀g = 2, 3⋯, so that the mean MC duration would increase or decrease with the generation according to whether γ > 0 or γ < 0. For g ≥ 2, the variance assumed the form , thereby yielding a coeffcient of variation constant across generations and equal to ρ (= σg/μg). The mean and variance for the first generation, μ1 and , were free parameters, just as in Model 1. Parameter estimates are reported in Table 2 (see below for computational details). In particular, we estimated γ as γ = −0.03, thereby suggesting that the mean MC duration would slightly decrease after the second generation. A Monte Carlo Wald test of the hypothesis vs. did not reject at the level of significance 5% (P = 0.56). Our analyses therefore suggested that the MC duration of cells of first generation was shorter than that of subsequent generations, but the MC duration of cells of generation equal to or larger than two were not significantly different.
To perform the above analyses, we proceeded as follows. Similarly to the analysis of O2A/OPCs, we distinguished cells of first and of subsequent generations, and refer to them as type-1 and type-2 cells, respectively. Let Yi = (Yi1, Yi2)’ denote the number of type-1 and the number of type-2 cells counted in the ith clone at time ti. Let Z1(t) and Z2(t) be stochastic processes denoting the number of type-1 and the number of type-2 cells at any time t, and let m1(t, θ) and m2(t, θ) denote their expectation. Write Z(t) = {Z1(t), Z2(t)} and m(t, θ) = {m1(t, θ), m2(t, θ)}’. We have that m1(t, θ) = 1 − G1(t), and . Because the model contains infinitely many types, the variance-covariance matrix of the numbers of cells takes a complicated form, and we did not use it to fit the models. We estimated θ = (μ1, σ1, μ2, γ, ρ)’ using the GEE approach, with each ΩY (ti) taken as the empirical variance-covariance matrix of Yi computed from clones observed at time ti (the experiment provided approximately 100 replicates for each time point). The expression for m1(t, θ) is explicitely known, and the value of m2(t, θ) only needs to be approximated. The cumulant generating function for G1 ★⋯★ Gg(t) is given by
The saddlepoint is the root to the equation . We deduced from equation (6) where we set k = 1; for g ≥ 3, the equation is solved numerically. The saddlepoint approximations to the expectation follows from equations (3-5). Figure 3 shows the experimental and fitted (Models 0-2) mean number of type-1 and type-2 cells versus time. Simulations were used to assess the accuracy of the saddlepoint approximations to the expected number of cells.
Figure 3.
5. Approximating the distribution of the generation
The distribution of the generation (that is, the number of divisions undergone since time 0) has found multiple uses in cell biology. For instance, Hyrien and Zand (2008) used it in the analysis of CFSE (carboxy-fluorescein succinimidyl ester)-labeling experiments, and Cole et al (2006) considered the expectation of this distribution to compare cell proliferation kinetics across a set of experimental conditions. This distribution does not generally have an explicit expression, which motivated Cole et al (2006) to approximate its expectation using asymptotic arguments. Asymptotic formulae such as the one used by Cole et al (2006) have not been derived for general models, and saddlepoint approximations offer a good alternative to approximate the distribution of the generation or its expectation.
Let κ(t) denote the generation of any cell randomly sampled in the population at time t, given the population has not become extinct by time t, and let Zl(t) denote the number of cells in generation l. Let , g = 1, 2 ⋯, denote the corresponding conditional probability distribution of κ(t). When the number of initiator cells tends to infinity, it can be shown that pg(t) → mg(t)/∑l≥1 ml(t), where mg(t) denotes the mean number of cells of generation g at time t arising from a single initiator cell. Of note these expectations are not conditioned on {∑l≥1 Zl(t) > 0}, and this approximation is valid for the general process defined in section 2. The expected generation number at any time t corresponding to the above distribution is therefore given by μgen(t) = ∑g≥1 gmg(t)/∑l≥1 ml(t). Thus, we propose to approximate pg(t) and μgen(t) by replacing the mg(t)’s in their respective expressions by saddlepoint approximations.
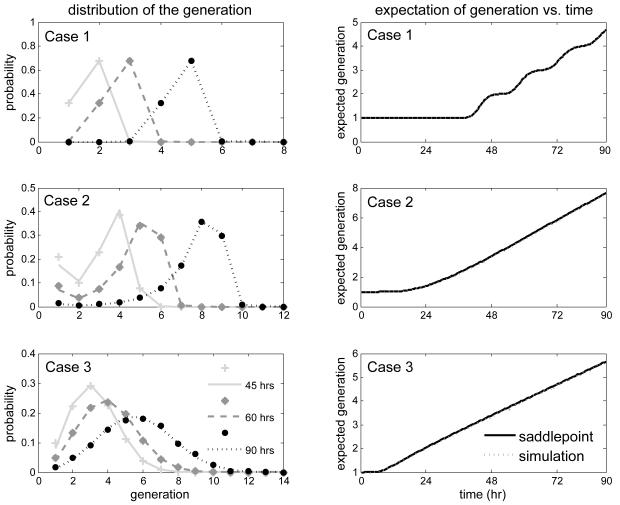
We tested this approach using a two-type process that begins at time 0 with a large number of type-1 cells. Upon completion of its lifespan, every cell of generation g (g = 1, 2 ⋯) either divides into two cells of generation (g + 1) with probability p, or it dies (that is, disappears from the population) with probability q = 1 − p. The duration of the lifespan of any cell of first generation is represented by the sum of two r.v.s τ0 + τ1, where τ0 and τ1 follow Inverse Gaussian distributions G0 and G with respective means μ0 and μ, variances and σ2, and c.d.f.s G0 and G, while, for every g = 2, 3 ⋯, the duration of the lifespan of any cell of generation g is described by a r.v. τg that follows the Inverse Gaussian distribution with c.d.f. G. This model extends a model considered by Hyrien and Zand (2008) to describe the activation and proliferation of lymphocytes. The r.v. τ0 models the time that is needed for lymphocytes to become activated, while τg, g = 1, 2 ⋯, models the time that is needed for any activated lymphocytes to die or divide. It can be shown that the mean number g of cells in any given generation takes the form mg(t) = {1 − G0(t)}1{g=1} + (2p)g {G0 ★ G★(g-1)(t) − G0 ★ G★g(t)}. The cumulant generating function for G0★ G★g is , the rth derivative of which is given by
where the double factorial r!! is defined as
The saddlepoint which solves the equation is approximated numerically. Plugging the above expressions into equations (3-5) yields the desired quantities. Depicted in Figure 4 are the saddlepoint approximations to pg(t), g = 1, 2 ⋯, for three time points (left panels), and its expectation as a function of time (right panels). Each row corresponds to a particular set of parameter values (see figure legend for the parameter values). The saddlepoint approximations were compared to simulation-based approximations (computed using direct simulations of the branching process). As can be seen, the saddlepoint approximations are in excellent agreement with their simulated counterparts, and capture remarkably well transient features of the expected generation number. The only exception is for p1(45) in Case 2, where we convoluted two distributions with very dissimilar variances.
Figure 4.
6. Discussion
Age-dependent branching processes, and their multitype extensions, have become important tools in the analysis of cell population dynamics. To get around the diffculties that arise in their practical use, we have proposed saddlepoint approximations to their expectation and variance-covariance function. Our work suggests that this approach provides quite accurate, easy to implement approximations to these moments. The only requirement is existence of the cumulant generating function for the lifespan distributions, as well as the differentiability of these functions. This method also proved to be much faster than the method based on simulations of the process, and it offers the possibility to resort to a wider range of estimation techniques (such as QL and GEE estimators). The proposed approach allows complex branching processes to be fitted quite easily to experimental data. However, when fitting such models one should keep in mind that the fit will never worsen as the structure of the model is enriched, and that any improvement in the fit will not necessarily reflect that a better description of the biological process has been achieved. Whenever possible, the findings should be verified in independent experiments. In this spirit, Hyrien et al (2006) used time-lapse experiments to validate the results of a previous analysis of clonal data that suggested that the distributions of the time to division and of the time to differentiation of O2A/OPCs could be dissimilar (Hyrien et al, 2005).
Acknowledgment
This research was supported by NIH grants 2R01 NS039511, R01 CA134839, 1R01 AI069351, 2P30 ES001247, and NIH/NIAID grant N01-AI-050020.
Footnotes
Supplementary Materials The Web Appendix is available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
References
- Athreya KB, Ney PE. Branching Processes. Springer-Verlag; Berlin: 1972. [Google Scholar]
- Barndorff-Nielsen O, Cox DR. Edgeworth and saddle-point approximations with statistical applications. J. R. Statist. Soc. B. 1979;41:279–312. [Google Scholar]
- Boucher K, Yakovlev AY, Mayer-Pröschel M, Noble M. A stochastic model of temporarily regulated generation of oligodendrocytes in vitro. Math. Biosci. 1999;159:47–78. doi: 10.1016/s0025-5564(99)00010-3. [DOI] [PubMed] [Google Scholar]
- Boucher K, Zorin A, Yakovlev AY, Mayer-Pröschel M, Noble M. An alternative stochastic model of generation of oligodendrocytes in cell culture. J. Math. Biol. 2001;43:22–36. doi: 10.1007/s002850100085. [DOI] [PubMed] [Google Scholar]
- Cole DJ, Morgan BJT, Ridout MS, Byrne LJ, Tuite MF. Approximations for Expected Generation Number. Biometrics. 2007;63:1023–1030. doi: 10.1111/j.1541-0420.2007.00780.x. [DOI] [PubMed] [Google Scholar]
- von Collani E, Tsodikov A, Yakovlev A, Mayer-Pröschel M, Noble M. A random walk model of oligodendrocyte generation in vitro and associated estimation problems. Math. Biosci. 1999;159:189. doi: 10.1016/s0025-5564(99)00017-6. [DOI] [PubMed] [Google Scholar]
- Daniels HE. Saddlepoint approximations in statistics. Ann. Math. Statist. 1954;25:631–650. [Google Scholar]
- DiCiccio TJ, Efron B. Bootstrap confidence intervals. Statistical Science. 1996;3:189–228. [Google Scholar]
- Golinelli D, Guttorp P, Abkowitz JA. Bayesian inference in a hidden stochastic two-compartment model for feline hematopoiesis. Mathematical Medicine and Biology. 2006;23:153–172. doi: 10.1093/imammb/dql008. [DOI] [PubMed] [Google Scholar]
- Haccou P, Jagers P, Vatutin VA. Branching Processes: Variation, Growth and Extinction of Populations. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- Hanin L, Hyrien O, Bedford J, Yakovlev A. A comprehensive stochastic model of irradiated cell population in cell culture. Journal of Theoretical Biology. 2006;239:401–416. doi: 10.1016/j.jtbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Heyde CC. Quasi-Likelihood and Its Applications. A General Approach to Optimal Parameter Estimation. Springer-Verlag; New-York: 1997. [Google Scholar]
- Hoel DG, Crump KS. Estimating the generation-time distribution of an age-dependent branching process. Biometrics. 1974;30:125–135. [PubMed] [Google Scholar]
- Hyrien O. A pseudo maximum likelihood estimator for discretely observed multi-type Bellman-Harris branching processes. J. Statist. Plann. Inference. 2007;137:1375–1388. doi: 10.1016/j.jspi.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Ambeskovic I, Mayer-Pröschel M, Noble M, Yakovlev A. Stochastic modeling of oligodendrocyte generation in cell culture: model validation with time-lapse data. Theor. Biol. Medical Model. 2006;3 doi: 10.1186/1742-4682-3-21. Art. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Mayer-Pröschel M, Noble M, Yakovlev A. A stochastic model to analyze clonal data on multi-type cell populations. Biometrics. 2005a;61:199–207. doi: 10.1111/j.0006-341X.2005.031210.x. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Mayer-Pröschel M, Noble M, Yakovlev A. Estimating the life-span of oligodendrocytes from clonal data on their development in cell culture. Math. Biosci. 2005b;193:255–274. doi: 10.1016/j.mbs.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Zand MS. A mixture model with dependent observations for the analysis of CFSE-labeling experiments. J. Am. Statist. Assoc. 2008;103:222–239. [Google Scholar]
- Jagers P. Branching Processes with Biological Applications. John Wiley and Sons; London: 1975. [Google Scholar]
- Kimmel M, Axelrod DE. Branching Processes in Biology. Springer; New York: 2002. [Google Scholar]
- Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Lugannani R, Rice S. Saddlepoint approximations for the distribution of the sum of independent random variables. Adv. Appl. Proba. 1980;12:475–490. [Google Scholar]
- Morgan BJT, Ridout MS, Ruddock L. Models for yeast prions. Biometrics. 2003;59:562–569. doi: 10.1111/1541-0420.00066. [DOI] [PubMed] [Google Scholar]
- Nedelman J, Downs H, Pharr P. Inference on an age-dependent, multitype branching-process model of mast cells. J. Math. Biol. 1985;25:203–226. doi: 10.1007/BF00276390. [DOI] [PubMed] [Google Scholar]
- Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature. 1988;333:560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on the culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- Renshaw E. Applying the saddlepoint approximation to bivariate stochastic processes. Math. Biosci. 2000;168:57–75. doi: 10.1016/s0025-5564(00)00037-7. [DOI] [PubMed] [Google Scholar]
- Ridout MS, Cole DJ, Morgan BJT, Byrne LJ, Tuite MF. New Approximations to the Malthusian parameter. Biometrics. 2006;62:1216–1223. doi: 10.1111/j.1541-0420.2006.00564.x. [DOI] [PubMed] [Google Scholar]
- Yakovlev AY, Boucher K, Mayer-Pröschel M, Noble M. Quantitative insight into proliferation and differentiation of oligodendrocyte-type 2 astrocyte progenitor cells in vitro. Proc. Natl. Acad. Sci. USA. 1998a;95(4):14164–14167. doi: 10.1073/pnas.95.24.14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev AY, Mayer-Pröschel M, Noble M. A stochastic model of brain cell differentiation in tissue culture. J. Math. Biol. 1998b;37:49–60. doi: 10.1007/s002850050119. [DOI] [PubMed] [Google Scholar]
- Yakovlev AY, Mayer-Pröschel M, Noble M. Stochastic formulations of a clock model for temporally regulated generation of oligodendrocytes in vitro. Math. Comput. Model. 2000;32:125–137. [Google Scholar]
- Yakovlev AY, Yanev N. Transient Processes in Cell Proliferation Kinetics. Springer; Heidelberg: 1989. [Google Scholar]
- Zorin AA, Yakovlev AY, Mayer-Pröschel M, Noble M. Estimation problems associated with stochastic modeling of proliferation and differentiation of O-2A progenitor cells in vitro. Math. Biosci. 2000;167:109–121. doi: 10.1016/s0025-5564(00)00040-7. [DOI] [PubMed] [Google Scholar]