Abstract
We carried out a QTL mapping experiment in two phenotypically similar inbred mouse strains, C57BL/6J and C58/J, using the open field assay, a well-established model of anxiety-related behavior in rodents. This intercross was initially carried out as a control cross for an ENU-mutagenesis mapping study. Surprisingly, although open field behavior is similar in the two strains, we identified significant QTL in their F2 progeny. Marker regression identified a locus on chromosome 8 having associations with multiple open field measures and a significant interaction between loci on chromosomes 13 and 17. Together, the chromosome 8 locus and the interaction effect form the core set of QTL controlling these behaviors with additional loci on chromosomes 1 and 6 present in a subset of the behaviors.
Keywords: quantitative trait locus, locomotor activation, anxiety, open-field assay
Introduction
Anxiety-related disorders are estimated to have a lifetime prevalence as high as 28%, making them the most common mental health problems affecting humans (Kessler et al., 2005). Epidemiological studies consistently identify a genetic component to these diseases, with heritability estimated at 30 to 40% for specific anxiety-related disorders (Hettema et al., 2001). A clear genetic basis for neuroticism, the personality dimension most highly correlated with anxiety-related disorders, exists as well, and the genetic factors associated with both exhibit substantial overlap (Hettema et al., 2004, Jardine et al., 1984, Kendler et al., 1993). For this reason, neuroticism is regarded as an endophenotype of anxiety-related disorders.
The neurobiological processes mediating fear and anxiety, and the genetic factors that influence them, are thought to be shared among species (Gray & Mcnaughton, 2000). While it is not possible to fully model anxiety disorders in animals, it is possible to study related behavioral responses and endophenotypes. The open-field assay was developed to measure emotionality in rodents, a trait considered analogous to neuroticism in humans (Hall, 1934, Hall, 1936). In this test, the animal is allowed to freely explore a novel arena. In this mildly stressful environment, the animal experiences the conflicting desires to explore the unfamiliar area and to avoid the brightly lit, anxiety-provoking open space. Animals exhibiting less emotionality will explore the space more freely, and show higher activity levels as well as an increase in the amount of time spent in the center of the arena. These behavioral parameters are appropriately responsive to anxiolytic agents, validating the use of the open-field assay for measuring anxiety-related behaviors (reviewed in (Prut & Belzung, 2003).
Genetically distinct inbred strains of mice differ markedly in their response to the open field (Bolivar et al., 2000, Bothe et al., 2005, Crabbe et al., 1999, Mcclearn, 1959, Thompson, 1953), and selection experiments have successfully produced rodent strains with heritable differences in open field behavior (reviewed in (Flint, 2004)). Heritability estimates range from 20-50% (Broadhurst, 1960, Defries et al., 1978), but clearly, genetic factors play an important role in modulating these traits. As such, this assay has been used extensively to identify genetic loci associated with anxiety. Quantitative trait loci (QTL) on chromosomes 1, 12 and 15 have been independently identified by several groups using several different mapping strategies (Flint et al., 1995, Gershenfeld et al., 1997, Kelly et al., 2003, Singer et al., 2005, Turri et al., 2004); additional chromosomal loci have been identified in these and other studies that await independent replication (De Ledesma et al., 2006, Gill & Boyle, 2005, Turri et al., 2001, Turri et al., 1999).
These previously reported mapping experiments utilized founder strains that are relatively distant genetically and show markedly different behavior in the open field. Here we report the identification of a set of QTL for open field behavior in the F2 intercross progeny of two phenotypically similar strains, C57BL/6J (B6) and C58/J (C58).
Methods
Subjects
All experiments followed the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee.
All mice were obtained from our in-house breeding colony. The parental strains were originally purchased from the Jackson Laboratory. Both strains are now maintained in the GNF breeding colony that is refreshed with breeders from the Jackson Laboratory every 6-7 generations to maintain the Jax (J) substrain. Female B6 and male C58 animals from our in-house colony were bred to generate F1 progeny, which were subsequently intercrossed to yield the F2 animals used for QTL mapping. Animals were maintained on a 12 h light:12 h dark cycle, housed in groups of 2-4 in standard HEPA-filtered polycarbonate mouse cages containing a layer of Bed-o-cob bedding and one cotton nestlet. Food (Pico rodent chow 20, Purina) and water were made available ad libitum. Animals were weaned at 3 weeks and all testing was performed on experimentally naïve animals. For assessment of open field behavior in parental strains, 24 B6 (12 each, males and females) and 23 C58 (11 female, 12 male) animals were tested at approximately 90 days of age. F1 animals (15 female, 11 male) were tested at approximately 60 days of age. For the mapping studies, 196 female and 166 male F2 intercross animals were tested at approximately 60 days of age.
Open Field Assay
The open field apparatus is a 17″×17″×13″ arena with a white Plexiglas floor and clear Plexiglas walls (ENV-515, Med Associates) which are surrounded by infrared detection beams on the X, Y and Z-axes that track the animals' position and activity over the course of the experiment. The apparatus is isolated within a 73.5 × 59 × 59 cm testing chamber fitted with overhead fluorescent lighting (lux level 14). All testing was conducted during the light cycle. Two hours before testing, cages were moved from the housing racks to a quiet anteroom adjacent to the testing room. Following this period of habituation, animals were removed from their home cage, immediately placed in the corner of the open field arena and allowed to freely explore the apparatus for a test interval of ten minutes. Animals were scored for a number of behaviors in the open field including total distance traveled (in cm), ambulatory episodes (number of times animal breaks user defined number of beams before coming to rest), percent time resting, average velocity (in cm per second), number of rearings and percent time spent in center of arena. These data are recorded during testing and scored in post-session analyses using commercially available software (Activity Monitor 5.1, Med Associates). The testing apparatus was cleaned between each animal with a 0.1% bleach solution.
Statistical Analyses
Correlational analysis on the six variables measured in the open field was conducted to determine if the measures were related. Since many of the variables were significantly correlated, a factor analysis using varimax rotation was conducted on both the parental strain and F2 data to examine the underlying structure. Factors with eigenvalues greater than one were extracted. All statistical analyses (excluding mapping analysis described below) were conducted using the JMP statistical software package for Mac (version 6.0.2). For parental strains, a two-way analysis of variance (ANOVA) was used to identify main and interaction effects of sex and strain on group means. For F1s, a one-way ANOVA identified main effects of sex on all behavioral phenotypes. Mean differences were considered significantly different at p < 0.05.
SNP Genotyping
SNP genotyping assays were performed using the Sequenom MassARRAY system as previously described (Wiltshire et al., 2003) except that following dephosphorylation of the PCR amplification product, the genotyping reaction was combined with 0.755 µl of water, 0.2 µl of Termination mix (Sequenom), 0.2 µl of 10x Buffer (Sequenom), 0.041 µl of 0.063 units/ µl Thermosequenase (Sequenom) and 0.804 µl of 7 µM −14µM extension primer. The MassEXTEND reaction was performed in a nested format: After holding at 94°C for 30 sec, 20 cycles were carried out, each consisting of a hold at 94°C for 5 sec, followed by 5 minicycles of 5 sec at 52°C, and 5 seconds at 80°C. The reaction mix was desalted by adding 6 µg of a cationic resin, SpectroCLEAN (Sequenom), and resuspended in 30 µl of water. Completed genotyping reactions were spotted in nanoliter volumes onto a matrix arrayed into 384 elements on a silicon chip (Sequenom SpectroCHIP), and the allele-specific mass of the extension product was determined by matrix-assisted laser desorption ionization, time-of-flight MS. Data were analyzed by automated allele calling using the SPECTROTYPER software.
Genetic Analysis and Mapping
The entire group of 196 female and 166 male F2 animals was genotyped using a panel of 211 SNP markers, yielding an average SNP density of 1 marker per 11 mega base pairs (Mb). Six chromosomal regions had gaps in SNP coverage greater than 40 Mb, the minimum density required for QTL mapping studies, resulting in regions for which analyses were not informative. These were Ch1 0-53 and 84-142 Mb, Ch3 63-116 Mb, Ch14 11-75 Mb, Ch15 0-57 Mb and X 0-45. Initial analysis of raw trait values indicated sensitivity to outliers, therefore all traits were transformed by converting the data ranks, rescaling these to zero-one range and converting these to normal scores. As a result, all traits are normally distributed. The transformation reduces the impact of outliers while only minimally perturbing the shape of the data.
The genetic map was estimated in R/qtl using the data from this cross and was converted back to Mb coordinates by linear interpolation between known physical positions of markers in the mouse genome (Build 33). All genomic positions listed in text are Mb positions.
A standard genome scan was conducted using R/qtl (Broman et al., 2003) to identify QTL with significant main effects. Because sex differences were observed for all of the open field traits studied, sex was included as an additive covariate in the genome scan. A model with sex as an interactive covariate (Solberg et al., 2006) was used, but indicated no sex-specific QTL and the results are not reported here. A logarithm of the odds ratio (LOD) score was computed at 4 Mb (2 cM) increments over the entire genome. Significance thresholds were estimated by analysis of 1000 permutations (Churchill & Doerge, 1994). The transformation of the data allows for permutation-based thresholds that are exactly the same for all traits. For main QTL scans with sex as an additive covariate, suggestive loci have a LOD score greater than 2.1 and significant loci have a LOD score greater than 3.3. These values correspond to genome-wide adjusted p-values of 0.63 and 0.05 respectively (Churchill & Doerge, 1994, Lander & Kruglyak, 1995).
To identify potential epistatic interactions, we also performed a search for significant pairs of loci using an exhaustive simultaneous search strategy. Each pair of loci in the genome was assessed for both additive and epistatic effects on the phenotype values. All significant QTL and interactions detected were used to construct a multiple QTL model to describe the simultaneous effects of all QTL on the traits. The method employed here is analogous to multiple interval mapping (Kao et al., 1999), but it was implemented using the algorithm described by Sen and Churchill (Sen & Churchill, 2001) and Sugiyama, et al (Sugiyama et al., 2001) (Sugiyama et al., 2002). Significance thresholds were calculated by permutation analysis using 600 permutations. (Nettleton & Doerge, 2000)
Finally, a backward elimination approach was used to refine the multiple-QTL model. QTL may be removed if they fail to achieve significance (at p < 0.01) after the effects of other QTL have been taken into account. The use of p < 0.01 as a cutoff is arbitrary but we have found that this effectively removes QTL that explain less than 1-2% of the total variance. This is a conservative approach, as only those QTL that were detected in the genome scans and retained in the final model are reported. Main scans are reported for all measures obtained in the open field, however specific QTL models were chosen based on consistent detection of the QTL in multiple measures within the same principal component.
Results
Data Reduction
Many of the measures collected in the open field are significantly correlated (Table 1) so we conducted separate factor analyses in the parental strains and the F2 to examine the underlying structure of the data. In both the parental strains and the F2 population, two factors with eigenvalues greater than one were extracted. These two factors account for 79% and 71% of the variance in the parental strains and the F2 respectively. We designated one factor as “Activity” because it includes the measures of distance traveled, number of ambulatory episodes and percent time resting, and the second factor as “Anxiety” since it includes percent time spent in the center and average velocity (Table 2). Rearing behavior did not load well onto either factor.
Table 1.
Correlations among behaviors measured in the open field with P-values in parentheses. Values in bold are significant based on Bonferroni correction.
| Pct Center | Total Distance | Total Rearing | Ambulatory Episodes | Average Velocity | |
|---|---|---|---|---|---|
| Total Distance | 0.081 (P>0.05) | ||||
| Total Rearing | −0.005 (P>0.05) | 0.279 (P<0.0001) | |||
| Ambulatory Episodes | 0.140 (P<0.01) | 0.963 (P<0.0001) | 0.291 (P<0.0001) | ||
| Average Velocity | 0.482 (P<0.0001) | 0.243 (P<0.0001) | 0.148 (P<0.01) | 0.302 (P<0.0001) | |
| Percent Resting | −0.213 (P<0.0001) | −0.788 (P<0.0001) | −0.131 (P<0.05) | −0.784 (P<0.0001) | −0.051 (P>0.05) |
Table 2.
Results of factor analyses in both parental strains and F2 mice.
| Parental Strains | B6.C58 F2 | |||
|---|---|---|---|---|
| Activity | Anxiety | Activity | Anxiety | |
| Total Distance | 0.971 | 0.067 | 0.966 | 0.081 |
| Ambulatory Episodes | 0.946 | 0.232 | 0.959 | 0.153 |
| Percent Time Resting | −0.943 | −0.030 | −0.875 | −0.046 |
| Average Velocity | 0.124 | 0.882 | 0.147 | 0.852 |
| Percent Time Center | −0.047 | 0.882 | 0.046 | 0.858 |
| Total Rearing | 0.386 | 0.505 | 0.370 | 0.057 |
| Eigenvalue | 3.15 | 1.61 | 2.92 | 1.36 |
| Pct Variance | 52.5 | 26.9 | 48.7 | 22.6 |
Open Field Behavior in Parental Strains and F1s
An ANOVA with both sex and strain as independent variables for the parental strains yielded a main effect of strain only and indicates that B6 mice spend more time resting (F(1,43)=4.8;p=0.03) and more time in the center of the open field (F(1,43)=6.6;p=0.05). A significant main effect of sex (F(1,43)=5.4;p=0.02) was observed for rearing with C58 mice rearing significantly more than B6 mice. No significant main effects were seen for strain or sex for any of the other behaviors measured and no interaction effects were observed for any behavior.
Although sex differences exist only for rearing behaviors in the parental strains, the F2 population showed significant sex differences for all open field behavioral measures. Analyses of variance indicate that measures of total distance traveled, (F(1,360)=13.9;p=0.0002) and percent time spent in the center of the arena (F(1,360)=16.5; p=0.0001) are significantly higher in females. Females exhibit fewer rearings (F(1,361)=5.7; p=0.02) than males (Table 3).
Table 3.
Means and standard deviations (in parentheses) sexes combined and separately for behaviors measured in the open field in both C57BL/6J and C58/J strains.
| Total Distance | Percent Resting | Rearing | Average Velocity | Ambulatory Episodes | Percent Time in Center | |
|---|---|---|---|---|---|---|
| C57BL/6J | 3001.2 (531.9) | 56.8 (6.6) | 80.0 (34.7) | 34.7 (3.8) | 102.5 (18.6) | 17.5 (4.8) |
| M (N=12) | 2863.5 (575.1) | 57.1 (6.4) | 89.2 (37.5) | 35.1 (4.2) | 97.3 (19.1) | 18.6 (5.4) |
| F (N=12) | 3138.9 (468.4) | 56.6 (7.1) | 70.8 (30.4) | 34.4 (3.6) | 107.7 (17.3) | 16.5 (4.0) |
| C58/J | 3716.9 (1787.5) | 51.1 (11.1) | 66.6 (27.1) | 33.6 (3.8) | 109.6 (43.9) | 13.3 (6.7) |
| M (N=11) | 4141.2 (1680.9) | 48.1 (9.9) | 77.0 (25.9) | 35.0 (3.4) | 121.3 (37.3) | 14.3 (6.0) |
| F (N=12) | 3254.1 (1862.7) | 54.4 (11.7) | 55.2 (24.4) | 32.1 (3.7) | 96.8 (48.6) | 12.2 (7.5) |
| B6.C58 F2 | 3491.0 (869.5) | 53.0 (6.9) | 89.7 (36.5) | 36.0 (4.8) | 117.3 (28.4) | 19.9 (8.7) |
| M (N=166) | 3309.4 (793.8) | 53.9 (6.8) | 94.6 (36.5) | 34.9 (4.5) | 112.2 (28.1) | 18.0 (8.6) |
| F (N=196) | 3644.8 (902.5) | 52.2 (6.9) | 85.5 (36.0) | 36.9 (4.9) | 121.7 (28.0) | 21.5 (8.5) |
Relatively few sex differences were identified in B6.C58 F1 mice. A significant main effect of sex was observed only for percent time resting (F(1,25)=4.7;p=0.04) and average velocity (F(1,25)=5.9;p=0.02) indicating that females spend less time resting and move more quickly. Absence of overall sex differences in the F1 generation make it unlikely that QTL for these behaviors reside on the Y chromosome.
Main genome scan
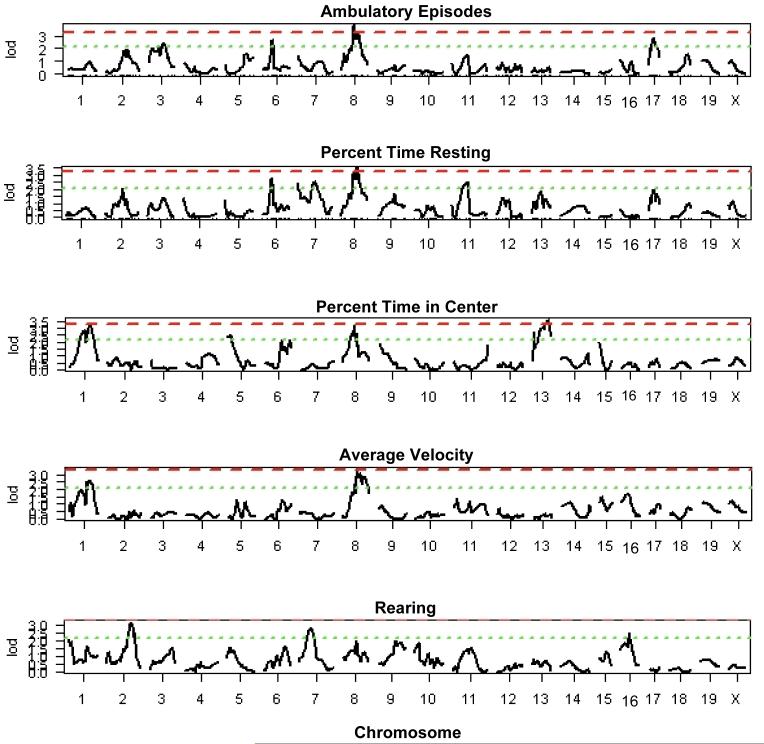
A genome scan for main effect QTL was conducted on all variables with sex as an additive covariate. The results of the genome scan are depicted in Figure 1; suggestive and significant thresholds are indicated by a dotted or dashed line, respectively. A significant QTL on Chr 8 is associated with multiple behaviors including total distance (LOD=4.1, peak 49Mb), ambulatory episodes (LOD=3.8, peak 62Mb), percent time resting (LOD=3.5, peak 89Mb) and average velocity (LOD=3.3, peak 78Mb). A suggestive locus exists in the same region for percent time spent in center of the open field (LOD=3.2, peak 58Mb).
Figure 1.
Genome-wide scans for all behaviors. Dashed line indicates corrected genomewide significance threshold of LOD 3.2 at P=0.05. Dotted line indicates suggestive threshold of LOD 2.1 at P=0.63.
Two additional QTL were identified that associate with multiple phenotypes. A locus on Chr 6 is significant for total distance (LOD=3.3, peak 47Mb), and suggestive for both ambulatory episodes (LOD=2.7, peak 49Mb) and percent time resting (LOD=2.8, peak 49Mb). A QTL on Chr 1 (LOD=3.3, peak 160Mb) is significantly associated with percent time spent in the center of the open field and suggestive for average velocity (LOD=2.6, peak 160Mb). In addition to these loci that were found to associate with multiple measures, a significant QTL was also identified on Chr 13 (LOD=3.6, peak 77Mb) that associates specifically with percent time spent in the center of the open field. A number of suggestive QTL were also identified; their genomic positions and peak LOD scores are listed in Table 4.
Table 4.
Summary of suggestive and significant QTL (bold) identified and allelic affects at peak LOD score position. Genome-wide corrected significant and suggestive LOD scores are 3.2 and 2.1 respectively. Significant QTL are bold.
| Chr | Location (Mb) | LOD | P-valueπ | Increasing Allele | |
|---|---|---|---|---|---|
| Total Distance | 3 | 23.4 | 2.4 | 0.49 | C58 |
| 6 | 47.4 | 3.4 | 0.04 | B6/C58* | |
| 8 | 61.9 | 4.1 | 0.00 | B6 | |
| 17 | 46.0 | 2.8 | 0.25 | C58 | |
| Ambulatory Episodes | 3 | 86.0 | 2.5 | 0.42 | C58 |
| 6 | 49.0 | 2.7 | 0.28 | B6/C58* | |
| 8 | 66.6 | 3.8 | 0.01 | B6 | |
| 17 | 46.0 | 2.8 | 0.24 | C58 | |
| Percent Time Resting | 6 | 49.0 | 2.8 | 0.25 | B6 |
| 7 | 60.9 | 2.6 | 0.34 | B6 | |
| 8 | 89.5 | 3.5 | 0.03 | C58 | |
| 11 | 71.6 | 2.5 | 0.39 | B6 | |
| Percent Time in Center | 1 | 160.1 | 3.3 | 0.05 | B6 |
| 5 | 5.8 | 2.5 | 0.42 | C58 | |
| 6 | 96.3 | 2.1 | 0.62 | B6&C58† | |
| 8 | 57.7 | 3.2 | 0.10 | C58 | |
| 13 | 76.6 | 3.6 | 0.03 | B6 | |
| Average Velocity | 1 | 160.1 | 2.6 | 0.32 | B6 |
| 8 | 77.7 | 3.3 | 0.04 | C58 | |
| Rearing | 2 | 135.2 | 3.2 | 0.10 | C58 |
| 7 | 33.3 | 2.8 | 0.25 | B6 | |
| 16 | 56.1 | 2.5 | 0.41 | B6 |
Heterozygotes display increased total distance and more ambulatory episodes than either homozygote
Both B6 and C58 homozygotes spend more time in center than heterozygotes
P-values are adjusted for genomewide significance
To determine allelic effects, we examined trait means for F2 animals of each genotype at each of the suggestive and significant QTL we identified (data not shown, results summarized in Table 1). For activity-related traits (total distance, ambulatory episodes and percent time resting), homozygosity for the B6 allele at Chr 8 results in increased activity and ambulatory episodes and decreased percent time resting. At the Chr 6 locus, no significant difference is observed between animals that are homozygous for either B6 or C58, however, heterozygotes show increased activity and ambulatory episodes and decreased time resting as compared to either homozygote. For anxiety-related traits (percent time in center, average velocity), mice homozygous for B6 at the Chr 8 QTL spend less time in the center of the open field and show decreased velocity. In contrast, homozygosity for the C58 allele at the Chr 1 locus decreases percent time spent in the center and average velocity as compared to B6 homozygotes (for average velocity) or heterozygotes (for percent time in center). Homozygosity for the C58 allele at the Chr 13 QTL also results in decreased percent time spent in the center. Allelic effects for suggestive loci are summarized in Table 4.
Pairwise Genome Scan
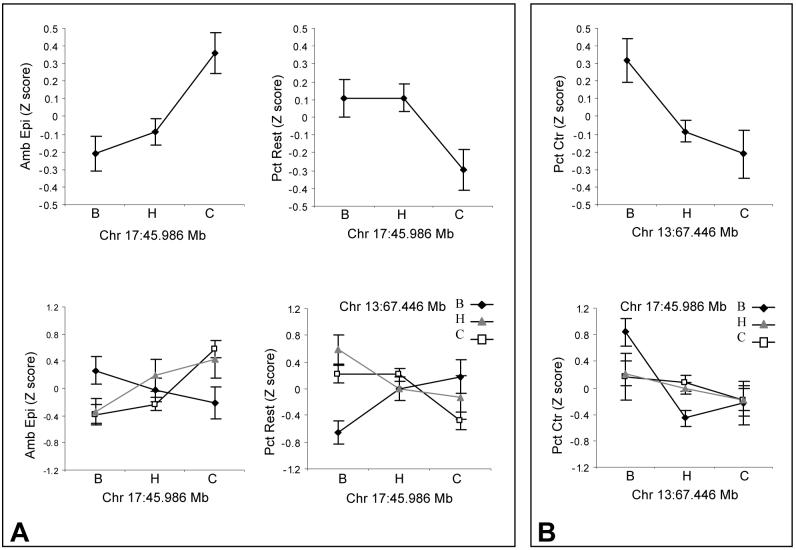
Epistatic interactions were investigated using a genome-wide simultaneous search strategy. Significance thresholds were calculated by permutation analysis using 600 permutations. The main genome scan identified a single locus on Chr 13 that was significant for percent time spent in the center (LOD=3.6) and a single locus on Chr 17 (LOD=2.8) that was suggestive for the activity-related traits but that was not associated with percent time in the center. Interestingly, the pairwise genome scan detected a significant interaction between these two loci that influences almost all behaviors collected in the open field. Percent time resting had a full LOD score of 8.7, exceeding the genomewide-corrected level of significance (LOD=8.4). Ambulatory episodes, percent time in the center and total distance had full LOD scores (8.4, 8.1 and 7.9 respectively) exceeding the suggestive threshold of 6.6. Based on the significant full LOD score of percent time resting and confirmatory evidence from the other three behaviors, interaction tests were run and indicate that total distance, ambulatory episodes and percent time resting exceed the genomewide-corrected suggestive interaction LOD score of 4.3 with interaction LOD scores of 4.6, 4.6 and 5.1 respectively. The direction of the effect for the Chr 17 locus for activity related traits indicates an increase in activity and decrease in percent time in the center for F2 mice carrying two C58 alleles. The interaction between the two loci is such that homozygosity for B6 alleles at Chr 13 eliminates this Chr 17 effect (Figure 2A). Conversely, homozygosity for B6 alleles on Chr 13 confers decreased anxiety (more time spent in the center of the open field) but either one or two C58 alleles at the Chr 17 locus eliminate this effect (Figure 2B).
Figure 2.
Interaction effects for Chr 13 and Chr 17 loci. A) Main effect of Chr 17 QTL at 45.986Mb for activity related traits and interaction with Chr 13 locus and B) main effect of Chr 13 QTL at 67.446Mb for anxiety trait and interaction with Chr 17 locus. Error bars are standard error of the mean.
Multiple Regression Analysis
All significant QTL, suggestive QTL that were identified across multiple behaviors and interactions were entered into a multiple regression model for each behavior with sex as a covariate (Table 5). These analyses indicated that for the anxiety-like behaviors (percent time in center and average velocity) the QTL on Chr 1 near 160Mb accounts for approximately 2.5 percent of the variance. For all of the activity-like behaviors and percent time in the center, the combination of the Chr 8 QTL and the interaction between Chr 13 and Chr 17 controlled most of the variance with individual contributions for each loci and the interaction ranging from 7 to 10 percent depending on the behavior. The variance accounted for by the combined effect of these QTL ranged from 10.8 to 30 percent depending on the behavior (Table 5).
Table 5.
Multiple regression analysis of open field behavioral data in B6 × C58 F2 intercross.
| Behavior | Variable | F value | p value | Variance (%)a |
|---|---|---|---|---|
| Total Distance | Sex | 11.8 | 6.9×10−4 | 2.6 |
| Chr3@23.4Mb | 6.6 | 1.6×10−3 | 2.9 | |
| Chr6@47.4Mb | 6.0 | 2.8×10−3 | 2.7 | |
| Chr8@61.9Mb | 8.5 | 2.4×10−4 | 3.8 | |
| Chr13@76.6Mb | 4.3 | 3.3×10−4 | 5.7b | |
| Chr17@46.0Mb | 6.3 | 2.4×10−6 | 8.5b | |
| Chr13@76.6Mb:Chr17@46.0Mb | 6.2 | 7.9×10−5 | 5.5 | |
| % VARIANCE EXPLAINEDc | 23.2 | |||
| Ambulatory Episodes | Sex | 9.7 | 2.0×10−3 | 2.2 |
| Chr3@91.3Mb | 5.7 | 3.7×10−3 | 2.6 | |
| Chr6@47.4Mb | 4.9 | 7.9×10−3 | 2.2 | |
| Chr8@66.6Mb | 8.0 | 4.1×10−4 | 3.6 | |
| Chr13@72.9Mb | 4.6 | 1.5×10−4 | 6.3b | |
| Chr17@46.0Mb | 6.7 | 9.4×10−7 | 9.1b | |
| Chr13@72.9Mb:Chr17@46.0Mb | 6.1 | 9.9×10−5 | 5.5 | |
| % VARIANCE EXPLAINEDc | 21.6 | |||
| Percent Time Resting | Sex | 6.3 | 1.2×10−2 | 1.5 |
| Chr8@89.5Mb | 9.1 | 1.5×10−4 | 4.3 | |
| Chr13@72.9Mb | 5.5 | 1.8×10−5 | 7.9b | |
| Chr17@46.0Mb | 6.1 | 3.8×10−6 | 8.8b | |
| Chr13@72.9Mb:Chr17@46.0Mb | 6.8 | 2.9×10−5 | 6.5 | |
| % VARIANCE EXPLAINEDc | 16.2 | |||
| Percent Time in Center | Sex | 27.7 | 2.5×10−7 | 6.3 |
| Chr1@160.1Mb | 5.7 | 3.8×10−3 | 2.6 | |
| Chr8@57.7Mb | 5.1 | 6.8×10−3 | 2.3 | |
| Chr13@76.6Mb | 6.3 | 2.5×10−6 | 8.6b | |
| Chr17@27.2Mb | 3.9 | 8.4×10−4 | 5.3b | |
| Chr13@76.6:Chr17@27.2 | 5.6 | 2.4×10−4 | 5.1 | |
| % VARIANCE EXPLAINEDc | 20.8 | |||
| Average Velocity | Sex | 20.0 | 1.1×10−5 | 5.0 |
| Chr1@160.1Mb | 4.9 | 8.3×10−3 | 2.4 | |
| Chr8@77.7Mb | 6.8 | 1.2×10−3 | 3.4 | |
| % VARIANCE EXPLAINEDc | 10.9 | |||
| Rearing | Sex | 6.3 | 1.3×10−2 | 1.6 |
| Chr2@135.2Mb | 6.1 | 2.6×10−3 | 3.0 | |
| Chr7@33.3Mb | 6.5 | 1.7×10−3 | 3.2 | |
| Chr16@56.1Mb | 5.9 | 3.1×10−3 | 2.9 | |
| % VARIANCE EXPLAINEDc | 11.5 |
Variance indicates percentage of total F2 phenotypic variance associated with the behavior being measured
Percentage of variance explained includes interaction effect
Percent variance explained fully adjusted based on Type III sums of squares and not sequential (Type I)
Discussion
Traditional QTL mapping strategies have relied on the use of distantly-related inbred strains with widely divergent phenotypes to maximize the phenotypic and genetic variance for the trait under investigation. While B6 and C58 strains do show some phenotypic differences, they are by no means widely divergent. Existing data on SNP frequencies between strains indicate that the genetic variation between B6 and C58 is about half of that seen in strains that are typically used in comparison to B6 in QTL studies such as DBA/2J, A/J and 129S1/SvImJ (Pletcher et al., 2004). The relative genetic and phenotypic similarity between the C57BL/6J and C58/J strains would seem to make them poor choices for these types of mapping studies. We selected C58 as a mapping partner for an ENU-induced mutant line generated on a B6 background and performed the intercross we described as a control for that mapping cross. This choice was based on the expectation that using a closely-related inbred strain would decrease the potential for epistatic interactions between our mutation and the C58 background that might mask our mutant phenotype. However, it is not necessary for two parental strains to differ in phenotypic values associated with complex traits to be able to identify QTL in their intercross progeny (Gora-Maslak et al., 1991). Similar phenotypes in different strains may result from distinct genetic interactions and novel combinations of increasing and decreasing parental alleles and interactions may lead to extreme phenotypes not observed in the parental strains, a phenomenon known as transgressive segregation {reviewed in (Rieseberg et al., 1999)}. Similarly, alleles with effects that were masked in the parental strains by modifiers may segregate from them in the progeny allowing detection of their phenotypic effects.
To our knowledge, QTL studies using phenotypically similar strains have not previously been reported. As such, we present the first experimental evidence that it is possible to identify QTL using such populations. It may even be advantageous to do so since limiting background effects may make it easier to identify complex trait genes of relatively small effect. As reported here, although these strains are phenotypically and genotypically similar, we did identify significant QTL for open field behavior and interacting loci in their intercross progeny. Our results highlight the genetic interactions that can occur even between comparatively closely related and phenotypically similar strains. These results serve as a warning to those engaged in mapping ENU-induced mutants. Careful evaluation of the chosen mapping partner prior to starting a mapping project is necessary and the use and analysis of concurrent control crosses should be considered. However, these results also uphold the utility of mixed genetic populations such as the collaborative cross (Churchill et al., 2004) to identify genes and genetic interactions resulting from transgressive segregration.
The QTL we identified on chromosome 8 exhibits significant overlap with two previously reported open-field behavioral QTL identified in B6-derived recombinant inbred (RI) lines. Neiderheiser et al. (Neiderhiser et al., 1992) identified a QTL at 33 cM (˜64 Mb) in CXB RI animals that is associated with rearing and behavior in the open field and exploratory behavior on an open runway. A QTL in the same region was also identified in AXB/BXA RI lines and is associated with total locomotor activity in the open field (Gill & Boyle, 2005). These overlapping QTL may reflect association of the same genomic locus with open field behavioral measures in these three B6-derived populations, however, the size of the genomic regions encompassed by the QTL are large and await independent replication.
A rodent's behavior in the open field assay is the result of the stress and anxiety induced by a novel environment as well as overall differences in locomotion. Factor analysis has been used by several groups to study the underlying architecture of anxiety-related behavioral responses in rodents (Fernandes et al., 1999, Liu & Gershenfeld, 2003, Ramos et al., 1998, Rodgers & Johnson, 1995) and the results we obtained are similar to those reported in other studies. Detailed ethological analyses of open field behavior indicate that speed in the open field is indicative of location in the arena such that higher velocity is associated with activity in the center of the arena and lower velocity is observed with movement along the wall (Lipkind et al., 2004). This is reflected in our factor analysis since both percent time in the center and average velocity loaded positively onto the anxiety factor, which indicates that animals that spend more time in the center are also moving more quickly than those who spend more time near the walls and in corners. Others have noted that rearing or vertical movements in the open field, as well as in other assays, form a separate class of movements, which corresponds to our findings (Henderson et al., 2004) but a connection between rearing response to a novel environment and open field locomotion has also been shown (Van Abeelen, 1977). It is possible that strain specificity accounts for these differing results since van Abeelen analyzed a B6 by DBA/2J cross and Henderson et al. utilized a heterogeneous cross between 8 different inbred strains.
Although the results of our factor analysis seem to indicate a separation between anxious behavior and locomotion in the open field, this difference is only partially supported by the QTL results. By examining all traits, a clear pattern emerges in which a core set of QTL - mainly the QTL on Chr 8 near 34-46 cM and the interaction between Chrs 13 and 17 - account for the largest portion of the variance explained by all QTL for multiple behaviors. These core QTL, along with several QTL for specific behaviors, comprise a set of significant and suggestive loci that must be verified in replication experiments and further characterized. A locus on Chr 1 that is significantly associated with percent time in center and suggestive for the average velocity behavior is not present in any of the activity-related behaviors, which indicates that this locus may be related more to anxiety rather than overall activity in the open field. A QTL for locomotor behavior at the distal end of Chr 1 has been identified in multiple previous studies (De Ledesma et al., 2006, Flint et al., 1995, Gershenfeld et al., 1997, Henderson et al., 2004, Kelly et al., 2003, Mott et al., 2000, Singer et al., 2005, Talbot et al., 1999, Turri et al., 2004). Our failure to detect as association with this region and locomotor behavior may reflect an artifact in the data, inadequate sample size or relatedness of the parental strains. Alternatively, the distal chromosome 1 locus we identified may be the same QTL, but its effect in the B6/C58 F2 animals may be altered by the presence of genetic modifiers. It is also possible that this QTL is a novel locus that specifically influences anxiety-related behavior but not locomotor activity in offspring from these two strains. The elimination of rearing behavior from either the anxiety or activity factor is also supported by the QTL results since neither the Chr 8 QTL nor the Chr 13:Chr17 interaction effect was identified for this behavior.
The presence of a main effect QTL on Chr 13 for percent time in the center of the open field is an example of a locus that may be specific to anxiety behavior. However, the interaction between this locus and a locus on Chr 17 that is suggestive for several activity-related traits implies that these two loci both exert an influence on activity and anxiety but only when specific combinations of alleles from both loci are present. It is possible that the Chr 13 QTL modulates only anxiety, and that this effect is mitigated by the Chr 17 QTL that influences locomotor activity. However, the mechanism for this effect is unclear since alleles that increase activity behaviors such as ambulatory episodes, would be expected to increase time in the center based on their positive correlation. It is also possible that the Chr 17 and Chr 13 QTL individually influence both activity and anxiety-related behaviors but that the population size in this study was too small to detect both QTL for all behaviors. Further investigation of the interaction effect between these two loci and the mechanism by which it is acting will require additional studies using a larger population of animals.
Based on the theory of transgressive segregation, interactions like these among two strains that are phenotypically and genotypically similar are exactly what one might expect. Unfortunately, without a clear understanding of the biology of behavior and with no immediate knowledge of the quantitative trait genes underlying QTL, it is difficult to explain the underlying mechanisms that drive these types of complex interactions. However, the array of resources that are quickly becoming available - phenotypic data on a wide array of inbred strains (MPD) and ever-increasing SNP density between inbred strains (Szatkiewicz et al., 2008) - will make possible the use of genetic crosses between inbred strains that were once considered too similar, thereby expanding the information that can be gained from QTL studies and leading to the eventual identification of quantitative trait genes.
Acknowledgements
The authors thank George Vogler for suggestions on the use of factor analysis and Arimantas Lionikas for helpful comments on the manuscript. Research described in this article was supported by a Novartis Grant SFP-1407 and funding from the NIH NIDA Grant DA 022392 to LMT.
References
- Bolivar VJ, Caldarone BJ, Reilly AA, Flaherty L. Habituation of activity in an open field: A survey of inbred strains and F1 hybrids. Behav Genet. 2000;30:285–293. doi: 10.1023/a:1026545316455. [DOI] [PubMed] [Google Scholar]
- Bothe GW, Bolivar VJ, Vedder MJ, Geistfeld JG. Behavioral differences among fourteen inbred mouse strains commonly used as disease models. Comp Med. 2005;55:326–334. [PubMed] [Google Scholar]
- Broadhurst PL. Experiments in Personality. Routledge and Kegan Paul; London: 1960. Application of biometrical genetics to the inheritance of behaviour; pp. 3–102. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O'Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- de Ledesma AM, Desai AN, Bolivar VJ, Symula DJ, Flaherty L. Two new behavioral QTLs, Emo4 and Reb1, map to mouse Chromosome 1: Congenic strains and candidate gene identification studies. Mamm Genome. 2006;17:111–118. doi: 10.1007/s00335-005-0107-y. [DOI] [PubMed] [Google Scholar]
- DeFries JC, Gervais MC, Thomas EA. Response to 30 generations of selection for open-field activity in laboratory mice. Behav Genet. 1978;8:3–13. doi: 10.1007/BF01067700. [DOI] [PubMed] [Google Scholar]
- Fernandes C, Gonzalez MI, Wilson CA, File SE. Factor analysis shows that female rat behaviour is characterized primarily by activity, male rats are driven by sex and anxiety. Pharmacol Biochem Behav. 1999;64:731–738. doi: 10.1016/s0091-3057(99)00139-2. [DOI] [PubMed] [Google Scholar]
- Flint J. The genetic basis of neuroticism. Neurosci Biobehav Rev. 2004;28:307–316. doi: 10.1016/j.neubiorev.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, Collins AC. A simple genetic basis for a complex psychological trait in laboratory mice. Science. 1995;269:1432–1435. doi: 10.1126/science.7660127. [DOI] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Mathis C, Crawley JN, Li X, Paul SM. Mapping quantitative trait loci for open-field behavior in mice. Behav Genet. 1997;27:201–210. doi: 10.1023/a:1025653812535. [DOI] [PubMed] [Google Scholar]
- Gill KJ, Boyle AE. Quantitative trait loci for novelty/stress-induced locomotor activation in recombinant inbred (RI) and recombinant congenic (RC) strains of mice. Behav Brain Res. 2005;161:113–124. doi: 10.1016/j.bbr.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Gora-Maslak G, McClearn GE, Crabbe JC, Phillips TJ, Belknap JK, Plomin R. Use of recombinant inbred strains to identify quantitative trait loci in psychopharmacology. Psychopharmacology (Berl) 1991;104:413–424. doi: 10.1007/BF02245643. [DOI] [PubMed] [Google Scholar]
- Gray J, Mcnaughton N. The neuropsychology of anxiety. Oxford University Press; Oxford: 2000. [Google Scholar]
- Hall C. Emotional behavior in the rat. I Defecation and urination as measures of individual differences in emotionality. Journal of Comparative Psychology. 1934;18:385–403. [Google Scholar]
- Hall C. Emotional behaviour in the rat: III. The relationship between emotionality and ambulatory activity. Journal of Comparative Psychology. 1936;22:345–352. [Google Scholar]
- Henderson ND, Turri MG, DeFries JC, Flint J. QTL analysis of multiple behavioral measures of anxiety in mice. Behav Genet. 2004;34:267–293. doi: 10.1023/B:BEGE.0000017872.25069.44. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001;158:1568–1578. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Prescott CA, Kendler KS. Genetic and environmental sources of covariation between generalized anxiety disorder and neuroticism. Am J Psychiatry. 2004;161:1581–1587. doi: 10.1176/appi.ajp.161.9.1581. [DOI] [PubMed] [Google Scholar]
- Jardine R, Martin NG, Henderson AS. Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol. 1984;1:89–107. doi: 10.1002/gepi.1370010202. [DOI] [PubMed] [Google Scholar]
- Kao CH, Zeng ZB, Teasdale RD. Multiple interval mapping for quantitative trait loci. Genetics. 1999;152:1203–1216. doi: 10.1093/genetics/152.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MA, Low MJ, Phillips TJ, Wakeland EK, Yanagisawa M. The mapping of quantitative trait loci underlying strain differences in locomotor activity between 129S6 and C57BL/6J mice. Mamm Genome. 2003;14:692–702. doi: 10.1007/s00335-003-2273-0. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. Panic disorder in women: a population-based twin study. Psychol Med. 1993;23:397–406. doi: 10.1017/s003329170002849x. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lipkind D, Sakov A, Kafkafi N, Elmer GI, Benjamini Y, Golani I. New replicable anxiety-related measures of wall vs center behavior of mice in the open field. J Appl Physiol. 2004;97:347–359. doi: 10.1152/japplphysiol.00148.2004. [DOI] [PubMed] [Google Scholar]
- Liu X, Gershenfeld HK. An exploratory factor analysis of the Tail Suspension Test in 12 inbred strains of mice and an F2 intercross. Brain Res Bull. 2003;60:223–231. doi: 10.1016/s0361-9230(03)00033-9. [DOI] [PubMed] [Google Scholar]
- McClearn GE. The genetics of mouse behavior in novel situations. J. Comp. Physiol. Psychol. 1959;52:62–67. doi: 10.1037/h0044664. [DOI] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiderhiser JM, Plomin R, McClearn GE. The use of CXB recombinant inbred mice to detect quantitative trait loci in behavior. Physiol Behav. 1992;52:429–439. doi: 10.1016/0031-9384(92)90328-y. [DOI] [PubMed] [Google Scholar]
- Nettleton D, Doerge RW. Accounting for variability in the use of permutation testing to detect quantitative trait loci. Biometrics. 2000;56:52–58. doi: 10.1111/j.0006-341x.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, Mural RJ, Paigen B, Wiltshire T. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prut L, Belzung C. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. Eur J Pharmacol. 2003;463:3–33. doi: 10.1016/s0014-2999(03)01272-x. [DOI] [PubMed] [Google Scholar]
- Ramos A, Mellerin Y, Mormede P, Chaouloff F. A genetic and multifactorial analysis of anxiety-related behaviours in Lewis and SHR intercrosses. Behav Brain Res. 1998;96:195–205. doi: 10.1016/s0166-4328(98)00023-0. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation and speciation. Heredity. 1999;83(Pt 4):363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Rodgers RJ, Johnson NJ. Factor analysis of spatiotemporal and ethological measures in the murine elevated plus-maze test of anxiety. Pharmacol Biochem Behav. 1995;52:297–303. doi: 10.1016/0091-3057(95)00138-m. [DOI] [PubMed] [Google Scholar]
- Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JB, Hill AE, Nadeau JH, Lander ES. Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics. 2005;169:855–862. doi: 10.1534/genetics.104.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg LC, Valdar W, Gauguier D, Nunez G, Taylor A, Burnett S, Arboledas-Hita C, Hernandez-Pliego P, Davidson S, Burns P, Bhattacharya S, Hough T, Higgs D, Klenerman P, Cookson WO, Zhang Y, Deacon RM, Rawlins JN, Mott R, Flint J. A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm Genome. 2006;17:129–146. doi: 10.1007/s00335-005-0112-1. [DOI] [PubMed] [Google Scholar]
- Sugiyama F, Churchill GA, Higgins DC, Johns C, Makaritsis KP, Gavras H, Paigen B. Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics. 2001;71:70–77. doi: 10.1006/geno.2000.6401. [DOI] [PubMed] [Google Scholar]
- Sugiyama F, Churchill GA, Li R, Libby LJ, Carver T, Yagami K, John SW, Paigen B. QTL associated with blood pressure, heart rate, and heart weight in CBA/CaJ and BALB/cJ mice. Physiol Genomics. 2002;10:5–12. doi: 10.1152/physiolgenomics.00002.2002. [DOI] [PubMed] [Google Scholar]
- Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2008 doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot CJ, Nicod A, Cherny SS, Fulker DW, Collins AC, Flint J. High-resolution mapping of quantitative trait loci in outbred mice. Nat Genet. 1999;21:305–308. doi: 10.1038/6825. [DOI] [PubMed] [Google Scholar]
- Thompson WR. The inheritance of behaviour: Behavioural differences in fifteen mouse strains. Canad. J. Psychol. 1953;7:145–155. doi: 10.1037/h0083586. [DOI] [PubMed] [Google Scholar]
- Turri MG, DeFries JC, Henderson ND, Flint J. Multivariate analysis of quantitative trait loci influencing variation in anxiety-related behavior in laboratory mice. Mamm Genome. 2004;15:69–76. doi: 10.1007/s00335-003-3032-y. [DOI] [PubMed] [Google Scholar]
- Turri MG, Henderson ND, DeFries JC, Flint J. Quantitative trait locus mapping in laboratory mice derived from a replicated selection experiment for open-field activity. Genetics. 2001;158:1217–1226. doi: 10.1093/genetics/158.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turri MG, Talbot CJ, Radcliffe RA, Wehner JM, Flint J. High-resolution mapping of quantitative trait loci for emotionality in selected strains of mice. Mamm Genome. 1999;10:1098–1101. doi: 10.1007/s003359901169. [DOI] [PubMed] [Google Scholar]
- van Abeelen JH. Rearing responses and locomotor activity in mice: single-locus control. Behav Biol. 1977;19:401–404. doi: 10.1016/s0091-6773(77)91826-0. [DOI] [PubMed] [Google Scholar]
- Wiltshire T, Pletcher MT, Batalov S, Barnes SW, Tarantino LM, Cooke MP, Wu H, Smylie K, Santrosyan A, Copeland NG, Jenkins NA, Kalush F, Mural RJ, Glynne RJ, Kay SA, Adams MD, Fletcher CF. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc Natl Acad Sci USA. 2003;100:3380–3385. doi: 10.1073/pnas.0130101100. [DOI] [PMC free article] [PubMed] [Google Scholar]