Abstract
Methylation of cytosine residues within the CpG dinucleotide in mammalian cells is an important mediator of gene expression, genome stability, X-chromosome inactivation, genomic imprinting, chromatin structure, and embryonic development. The majority of CpG sites in mammalian cells is methylated in a nonrandom fashion, raising the question of how DNA methylation is distributed along the genome. Here, we focused on the functions of DNA methyltransferase-3b (Dnmt3b), of which deregulated activity is linked to several human pathologies. We generated Dnmt3b hypomorphic mutant mice with reduced catalytic activity, which first revealed a deregulation of Hox genes expression, consistent with the observed homeotic transformations of the posterior axis. In addition, analysis of deregulated expression programs in Dnmt3b mutant embryos, using DNA microarrays, highlighted illegitimate activation of several germ-line genes in somatic tissues that appeared to be linked directly to their hypomethylation in mutant embryos. We provide evidence that these genes are direct targets of Dnmt3b. Moreover, the recruitment of Dnmt3b to their proximal promoter is dependant on the binding of the E2F6 transcriptional repressor, which emerges as a common hallmark in the promoters of genes found to be up-regulated as a consequence of impaired Dnmt3b activity. Therefore, our results unraveled a coordinated regulation of genes involved in meiosis, through E2F6-dependant methylation and transcriptional silencing in somatic tissues.
Keywords: DNA methylation, immunodeficiency, centromeric instability, facial anomalies, E2F family, hypomorphic mutation, hox genes
Methylation of cytosines, predominantly within CpG dinucleotides, is a key epigenetic mark of vertebrate DNA (reviewed in ref. 1). The reaction is catalyzed by a family of DNA methyltransferases (DNMT), among which DNMT1 is specialized in the maintenance of DNA methylation patterns after DNA replication, whereas DNMT3A and DNMT3B are responsible for the de novo establishment of methylation during development and gametogenesis (review in ref. 2). CpGs are not evenly distributed across the genome, with regions of high CpG density, known as CpG islands, mainly localized at promoters and transcriptional start sites. Yet, genome-wide analysis of DNA methylation in human cells revealed that the majority of CpG islands at promoters are unmethylated in normal cells. In contrast, germ-line-specific genes, imprinted or X-linked genes, as well as repetitive DNA, are methylated in somatic cells (3–7).
The recruitment of individual DNMT to different genomic regions in vivo, particularly to gene regulatory regions, and the establishment of intact genomic methylation patterns in development, is known to require the interaction of regulatory factors. DNMT3L, which lacks the conserved catalytic domain characteristic of cytosine methyltransferases, is necessary for maternal methylation imprinting, possibly by interacting with and stimulating the activity of DNMT3A and DNMT3B (8, 9). LSH, a protein related to the SNF2 family of chromatin-remodeling ATPases, is required for efficient DNA methylation in mammals, especially at centromeric repeats (10, 11). Sequence specificity during developmental or lineage choice is thought to result from interactions between DNMT and transcription factors that would target methylation to specific DNA sequences in gene regulatory regions (12–15).
DNA methylation is essential for mammalian development, as revealed by the lethality of DNMT deficiencies in mice (16, 17). DNA methylation is generally associated with a repressed chromatin state and the inhibition of promoter activity (reviewed in refs. 18 and 19). However, less is known about other functions in developmentally regulated gene expression and genome integrity. In turn, alteration of DNA methylation patterns is a hallmark of several human diseases, including cancer, thus revealing the crucial role of DNMT in normal physiological processes (reviewed in refs. 20 and 21). Notably, global hypomethylation causes inappropriate expression of certain sequences, like genes that are normally restricted to germ cells, leading to their classification as cancer-testis genes (22). Hypomethylation also affects repetitive sequences associated with enhanced chromosomal instability (23). Recent data show that deregulated expression of DNMT3B in cancer cells can contribute to tumorigenesis (24–27). Another interesting case is the ICF syndrome (Immunodeficiency, Centromeric instability, Facial anomalies; OMIM #242860), a genetic disease arising from germ-line mutations within the DNMT3B gene (28, 29), also characterized by hypomethylation of satellite DNA and chromosomal instability (30, 31).
Given the close connection between DNMT3B dysfunctions and human diseases, we decided to shed light on the genes that are potentially directly regulated through DNMT3B-mediated DNA methylation, and on the molecular mechanisms that target Dnmt3b to specific genomic sequences. In the mouse, it is known that centromeric minor satellite DNA repeats are specifically methylated by Dnmt3b but not by Dnmt3a (17, 32). However, little is known about the discrete genes, expression of which could be silenced through Dnmt3b-mediated DNA methylation.
Microarray analysis of deregulated expression programs, in the hypomorphic Dnmt3b mutant mice we generated and described here, combined with an analysis of the molecular mechanisms involved in the illegitimate activation of a specific set of genes, revealed the existence of a functional interplay between Dnmt3b and the transcriptional repressor E2F6 in the methylation of several germ-line-specific genes and their normal transcriptional repression in somatic tissues.
Results
Dnmt3b Hypomorphic Mutant Mice Exhibit Homeotic Transformations.
Because Dnmt3b knockout is lethal early in embryonic development, we generated hypomorphic Dnmt3b mutant mice to explore its role in both development and transcriptional silencing. We focused on compound heterozygote mice (mEx3/mEx24) (Fig. S1), with a particular combination of mutations that exists in the human DNMT3B gene, in patients affected by ICF syndrome (28). We provide evidence, detailed in SI Materials and Methods, that the hypomorphic Dnmt3b mutant mice present the expected molecular features that strongly reflect an alteration in Dnmt3b functions (Figs. S1 and S2). In particular, Dnmt3b mutant mice show hypomethylation of centromeric minor satellite repeats (Fig. S3 C and D). This finding is in contrast to major satellite repeats that remain methylated despite the mutation (Fig. S3 E and F), but is consistent with minor satellite DNA being a specific target of Dnmt3b (17).
Heterozygote mice mEx3/WT and mEx24/WT appeared undistinguishable in size from their WT littermates. In contrast, compound heterozygotes mEx3/mEx24 mice were smaller compared with control littermates of the same age (Fig. 1A). To further analyze phenotypic defects in Dnmt3b compound heterozygotes, we stained the skeleton of 5-month-old mice with alizarin red to reveal skull and skeleton anomalies (Fig. 1 B and C). In contrast to WT or mEx3/WT littermates, compound heterozygote mice exhibited a shorter nose, a larger distance between the terminations of coronal suture on the frontal bone, and a larger interparietal bone (Fig. 1B and Fig. S4A). The frontal bone of mEx3/mEx24 mice is enlarged, resulting in a dome-shaped head with abnormal coronal suture morphology (Fig. 1B). More interestingly, morphological defects also affected the axial skeleton, which exhibited posterior homeotic transformations (Fig. 1C). We observed two types of transformations, coexisting in some mice, consisting in a transformation of the thoracic vertebra T13 into a lumbar vertebra L1, characterized by the absence or shorter floating ribs, and a transformation of the lumbar vertebra L6 into the sacral vertebra S1, characterized by its association with the iliac bones (Fig. 1C and Fig. S4B).
Fig. 1.
Dnmt3b mutant mice exhibit developmental defects of the skull and homeotic transformations of the skeleton. (A) Gross morphology of adult mEx3/WT vs. mEx3/mEx24 mice. (B) Schematic representation of a normal mouse skull with the main annotated distances (Left) and dorsal view of skulls stained with alizarin red from 2 different mEx3/mEx24 (m3/m24) mice (#1 and #2) compared with a mEx3/WT (m3/WT) littermate. The histogram represents variations of the measured intervals. d(CS), coronal suture distance; IB, ibnterparietal one; ICD, inner canthal distance; N, nasal bone length; NL, nose length; SH, skull height; SL, skull length; SW, skull width. (C) Ventral view of axial skeleton of adult mEx3/WT compared to two mEx3/mEx24 mice (#1 and #2) stained with alizarin red. Arrows show the positions of the defects identified as homeotic transformations. L, lumbar vertebra; S, sacral vertebra; T, thoracic vertebra. (D) RT-PCR analysis for the detection of the indicated Hox genes expression in mEx3/WT (3/WT) and mEx3/mEx24 (3/24) MEF. GAPDH RT-PCR was used as a normalization control.
Thus, analysis of developmental defects in the hypomorphic Dnmt3b mutant mice revealed not only severe defects of the skull, but also posterior transformations. Such alterations often result from the deregulated expression of Homeotic genes, key players in establishing positional identity along the antero-posterior axis. RT-PCR analysis of Hoxa11 and Hoxa13 mRNA levels confirmed a deregulated expression of these genes in mEx3/mEx24 murine embryonic fibroblasts (MEF) compared with WT MEF, consistent with their role as global regulators of the lumbo-sacral region of the axial skeleton (33) and the aforementioned homeotic transformations resulting from Dnmt3b impaired activity (Fig. 1D).
Taken together, our data provide in vivo evidence for a major role of Dnmt3b in the development of axial skeleton.
Expression Profiling in Dnmt3b Mutant Embryos Reveals Illegitimate Activation of Germ-Line Genes.
Centromeric minor satellite DNA is preferentially methylated by Dnmt3b (17) (Fig. S3 C and D). However, little is known about genes silenced through Dnmt3b targeting to regulatory regions. To identify other potential Dnmt3b target genes, we examined deregulated expression programs in cells deficient for Dnmt3b activity by microarray analysis. We compared the expression profiles of mEx3/mEx24 and WT 18.5 days post coitum (dpc) embryos in the thymus, an organ that shows the highest Dnmt3b protein expression levels at this stage of development (Fig. S3B). We predicted that a subset of genes would be up-regulated in mutant relative to WT embryos as a direct consequence of DNA hypomethylation.
Microarray expression profiling identified 25 genes that were up-regulated in mEx3/mEx24 thymus compared with WT thymus (fold >1.2 and P < 0.001) as a consequence of reduced Dnmt3b activity in compound embryos (Fig. S5). Thirteen of these genes were confirmed to be up-regulated in a DNA microarray analysis of mEx3/mEx24 MEF compared with WT MEF (shaded in Fig. S5). Gene ontology analysis of transcript profiling data revealed an overrepresentation of genes normally expressed in testis (21/25; P = 7.08e-03) (Fig. S5). Out of these 25 genes, 17 contained CpG-rich regions in their promoter (68%) (Fig. S6). In addition, transcription factor binding sites for retinoid-X receptor (RXR), zinc binding protein (ZBP), early-growth-response (EGR), and E2F families of transcription factors were identified in about 85% of these 25 promoters (Fig. S7A), with an occurrence higher (odds ratio > 1) than in all of the promoters of the mouse genome (extracted from Genomatix), suggesting common pathways for the regulation of these genes.
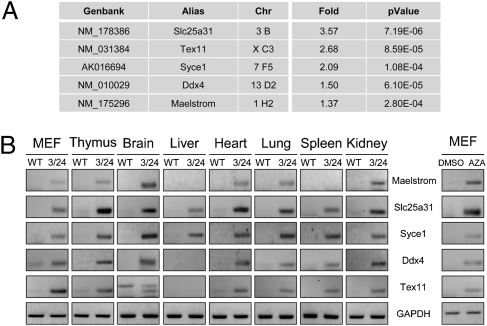
We chose to focus our study on the unexpected set of germ-line-specific genes showing illegitimate expression in both MEF and thymus derived from Dnmt3b mutant embryos (in bold in Fig. 2A and Fig. S5). RT-PCR analysis of Maelstrom, Slc25a31, Syce1, Ddx4, and Tex11 mRNA confirmed that expression of these genes was undetectable in WT somatic tissues, whereas it was activated in MEF and the thymus, but also in the majority of tested tissues derived from mEx3/mEx24 embryos at 18.5 dpc (Fig. 2B, Left). Interestingly, these five genes were not activated in control mEx3/WT littermates, in which one allele of Dnmt3b still encodes a normal protein. The promoter region of these five genes contains a CpG-rich region (Fig. S6), strongly suggesting that illegitimate activation in somatic tissues most likely results from impaired methylation at their regulatory regions. Treatment of WT MEF with the demethylating agent 5-azacytidine (AZA) for 4 d was sufficient to activate the expression of all five germ-cell-specific genes.
Fig. 2.
Germ-line genes are aberrantly expressed in Dnmt3b mutant embryos. (A) Germ-line gene expression changes in thymus derived from 18.5 dpc mEx3/mEx24 embryos. GenBank ID, symbol name (alias), chromosomal locus (Chr), fold-change expression, and corresponding P value (n = 5) are indicated. (B) RT-PCR analysis for detection of expression of the indicated germ-line gene expression (Maelstrom, Slc25a31, Syce1, Ddx4, Tex11) from mEx3/mEx24 (3/24) and WT MEF (12.5 dpc) or thymus, brain, liver, heart, lung, spleen, and kidney (18.5 dpc), and from untreated WT MEF or treated with AZA at 5 μM for 4 d. Mouse GAPDH transcripts were used as a normalization control.
These data demonstrate, as suggested in previous studies (34–36), that DNA methylation has a dominant role in the silencing of germ-line genes in somatic cells.
Germ-Line Genes Are Direct Targets of Dnmt3b in Somatic Tissues.
We examined global methylation of the proximal promoter region of the five selected germ-line genes by methylation-sensitive restriction enzyme-coupled PCR assay (MSRE), using the methylation-sensitive HpaII and HhaI enzymes for which several restriction sites reside in the proximal promoter region of the considered genes (Fig. S8A), and by genomic bisulfite sequencing assay in the thymus from WT and mEx3/mEx24 18.5 dpc embryos (Fig. 3A). In MSRE experiments, amplification of a promoter region using specific primers is permitted if the restriction sites are methylated and not cleavable. The Xlr amplified region, which does not contain HpaII/HhaI restriction sites, was used as an uncleavable control. This assay showed that the promoter regions of Maelstrom, Slc25A31, Ddx4, and to a lower extent, the promoters of Tex11 and Syce1, which contain far fewer restriction sites, clearly contained methylated restriction sites in untreated somatic compared with AZA-treated cells (Fig. S8A). Similarly, these promoter regions were not cleaved in WT MEF, thymus, or carcasses isolated at 12.5 and 18.5 dpc, consistent with their silencing through DNA methylation. In contrast, an alteration in the methylation of CpG sites residing in HpaII/HhaI sites was observed at the promoters of germ-line genes in mEx3/mEx24 tissues, indicative of a loss of methylation in mutant embryos (Fig. S8A). To confirm and quantify the methylation status observed by MSRE, genomic bisulfite sequencing was used, examining the same CpG regions in the proximal promoters of the five selected genes. After genomic bisulfite sequencing analysis, we found almost complete methylation of the proximal promoter regions in WT thymus and marked loss of CpG methylation in thymus isolated from mEx3/mEx24 embryos (from 56% for Maelstrom and Tex11 to over 75% for Slc25A31, Syce1, and Ddx4), for all five genes analyzed (Fig. 3A). ChIP analysis using antibodies against Dnmt3b to precipitate chromatin prepared from WT and mEx3/mEx24 MEF clearly demonstrated the occupancy of their proximal promoter by Dnmt3b in both WT and mutant embryos (Fig. 3B and Fig. S8B). This result also indicates that loss of methylation in the promoter region of the germ-line genes in mutant mice is not the result of a mislocalization of the mutated Dnmt3b protein, but rather, because of an alteration of its activity.
Fig. 3.
Germ-line genes are repressed by Dnmt3b-mediated methylation. (A) Methylation analysis of the proximal promoter region of the indicated germ-line genes. Genomic DNA derived from WT and mEx3/mEx24 thymus was subjected to genomic bisulfite sequencing and methylation status examined at proximal promoters of the indicated genes. Methylated CpG are represented by black circles and unmethylated sites by open circles. The arrows represent the transcriptional start site for each gene and the numbers above the line indicate the extent of the region analyzed relative to the transcriptional start site. (B) ChIP assays performed on chromatin prepared from WT or mEx3/mEx24 MEF, using Dnmt3b or E2F6 specific antibodies followed by real-time PCR using primers amplifying segments in the proximal promoters of indicated genes. ChIP signals were normalized to input signal, and subtracted for background signal in an IgG control. Results are mean and SEM of two to three independent ChIP experiments analyzed in duplicate. Histograms represent the fold-enrichment over the signal generated by amplification of a control tubulin gene (set at 1), which does not contain CpG islands or consensus E2F6 binding sites. (C) Real-time PCR analysis of expression of the indicated genes in MEF from mEx3/mEx24 embryos compared with their WT littermate (Left) or MEF from E2F6−/− embryos compared with their WT littermates (Right). Histograms represent the averaged fold-changes relative to WT control from replicates in two to three independent experiments. GAPDH PCR signal was used as a normalization control. Error bars represent SEM.
Together, these data revealed that silencing of germ-line-specific Maelstrom, Slc25a31, Syce1, Ddx4, and Tex11 genes in somatic tissues is driven by common mechanisms involving the catalytic functions of Dnmt3b.
Dnmt3b-Mediated Silencing of Germ-Line-Specific Genes Requires E2F6 Binding to Their Proximal Promoter.
How de novo DNMT are recruited to specific genomic regions is still unclear, but is assumed to require binding partners. Notably, the chromosomal location of deregulated genes in Dnmt3b embryos is distinct, indicating that their deregulation is not likely to result from a wide effect of the mutation on a particular region of the genome (Fig. 2A and Fig. S5). As mentioned above, in silico analysis of the proximal promoter regions of the 25 up-regulated genes led to the identification of consensus binding sites for several families of transcription factors, at higher occurrence than expected (Fig. S7A). Although the impact of these families of transcription factors on the recruitment of Dnmt3b to promoters of germ-line genes deserves further investigation, we specifically focused on the E2F family of transcriptional regulators because, strikingly, homeotic transformations found in Dnmt3b compound heterozygotes mice (Fig. 1C) are reminiscent of transformations described after invalidation of one of its members, the transcriptional repressor E2F6 (37). More importantly, among the E2F family members, E2F6 was predicted to have specific binding sites in 16 out of the 25 genes, an occurrence 10-times higher than that found in the promoters of all mouse genes (Fig. S7B). In addition, among the genes we have identified as potential Dnmt3b target genes, Slc25a31 and Hoxa11 were already known to be targeted and repressed by E2F6 (38, 39). We therefore focused on this particular factor and investigated the interplay between the transcriptional repressor E2F6 and Dnmt3b in the silencing of germ-line genes in somatic tissues. Importantly, levels of E2F6 mRNA was unaltered in Dnmt3b mutant cells (Fig. S8E), and proximal promoters of the tested genes were occupied by the transcriptional repressor E2F6 in WT cells, except for Ddx4, as revealed by ChIP assays (Fig. 3B and Fig. S8C). We first confirmed the activation of Slc25a31 in E2F6−/− tissues (Fig. 3C and Fig. S8D). We then found that Maelstrom, Syce1, and Tex11, but not Ddx4, were activated in E2F6−/− tissues (Fig. 3C and Fig. S8D), therefore revealing the loss of their silent state in both E2F6 null and Dnmt3b hypomorphic mutant embryos.
We then investigated whether E2F6 could recruit Dnmt3b to repress the expression of germ-line target genes. We first showed that exogenous E2F6 and WT or mutated Dnmt3b proteins could be coimmunoprecipitated from transiently transfected cells (Fig. 4A, Upper). We found that endogenous E2F6 and Dnmt3b belonged to the same insoluble subnuclear fraction of primary MEFs, from which they can be coimmunoprecipitated (Fig. S9A), and that the mutation in the catalytic domain of Dnmt3b (Dnmt3bm24) did not disrupt their interaction (Fig. S9B). ChIP experiments were then performed from WT and E2F6−/− cells using Dnmt3b or E2F6 antibodies. These experiments confirmed that the already known E2F6 target gene, Slc25A31 (38), was efficiently and specifically amplified from the precipitated chromatin of WT cells, and further revealed that Dnmt3b targeting to the germ-line genes was lost in E2F6−/− cells (Fig. 4B). Ddx4, which is not targeted by E2F6 (Fig. 3B) and the expression of which is unaffected in the absence of E2F6 (Fig. 3C), served as a control to show that, in that case, the absence of E2F6 did not perturb the binding of Dnmt3b to its target germ-line genes (Fig. 4B). Thus, these data provide strong evidence that Dnmt3b is recruited through E2F6 interaction to mediate the silencing of Maelstrom, Slc25a31, Syce1, and Tex11 germ-line genes.
Fig. 4.
Targeting of Dnmt3b to deregulated germ-line genes requires E2F6 binding. (A) Co-immunoprecipitation of Dnmt3b with E2F6 from transiently transfected HEK-293 cells with Dnmt3b, E2F6, or both expression vectors, using specific antibodies against Dnmt3b and E2F6. Precipitated proteins were analyzed by Western blotting and revealed using E2F6- and Dnmt3b-specific antibodies. (B) ChIP assays performed on chromatin prepared from WT and E2F6−/− MEF, using Dnmt3b- and E2F6-specific antibodies, followed by RT-PCR analysis using primers amplifying segments in the proximal promoters of the indicated genes. IN, input.
Taken together, these data suggest that E2F6 binding is crucial for the maintenance of Dnmt3B-mediated DNA methylation and silencing of certain germ-line-specific genes.
Discussion
We reported here that mutations known to impair Dnmt3b DNA methyltransferase catalytic activity result in homeotic gene deregulation associated with skeleton posterior transformations, as well as in the illegitimate expression of germ-line-specific genes in somatic cells. We provide evidence that targeting of Dnmt3b to the promoter region of these germ-line genes and maintenance of their silent state in somatic tissues requires the transcriptional repressor E2F6. Together, our data add a unique example of coordinated gene regulation, whereby genes involved in the same pathway, namely meiosis, contain a common transcription factor-binding site in their promoter, which serves as a sequence-specific factor for recruitment of DNA methylation and maintenance of their silencing in somatic cells.
The Dnmt3b hypomorphic mutant mice we generated exhibit growth defects, skull anomalies, high rate of mortality at birth, and hypomethylation of minor satellite sequences, similar to what was observed in a previously described mouse model (40). However, a deeper analysis of the skeleton revealed severe skull defects and posterior homeotic transformations that often reflect Hox genes deregulation (33). We confirmed a significant up-regulation of Hoxa11 and Hoxa13 transcripts in Dnmt3b mutant mice that may account for the observed transformations. Indeed, Hoxa10-13 expression has been linked to the establishment of the thoracic and lumbar vertebrae identity (33). Likewise, a recent study highlighted the implication of LSH protein in the control of DNA methylation and silencing of other Hox genes, Hoxa5-7, during development (41). Therefore, although it may implicate distinct coregulatory factors at different stages of development, DNA methylation is likely to participate in the spatiotemporal regulation of homeotic genes.
Germ-line-specific gene expression is typically restricted to germ cells, but is also illegitimately activated in a wide range of human tumors (22), and as shown in our study, in somatic cells with impaired catalytic activity of the murine Dnmt3b DNA methyltransferase. The proximal promoter of these genes appears to be methylated and occupied by Dnmt3b in normal somatic cells, consistent with their silent state in these cells. In contrast, mutations that impair Dnmt3b catalytic activity do not affect the ability of Dnmt3b to bind to promoters of germ-line genes, but result in their hypomethylation and subsequent aberrant activated transcription. Other examples exist in which DNA methylation was shown to be necessary to prevent inappropriate expression of this class of genes (34–36). We have now demonstrated that silencing of a subset of germ-line genes in somatic tissues requires Dnmt3b catalytic activity.
How the various DNMT are directed to specific genomic sites in vivo is not well understood. Molecular mechanisms may include direct interactions between DNMT and diverse regulatory factors, as well as the involvement of histone-modifying enzymes, and proteins involved in the biogenesis of small RNA (42). In addition, DNMT have been shown to interact with transcription factors, the innate specificity of which for defined DNA sequences could participate in the preferential targeting of DNA methylation to specific gene promoters (15). Our analysis of the proximal promoters of deregulated germ-line genes following impaired Dnmt3b catalytic activity allowed the identification of putative signature motifs for Dnmt3b target genes, with an over representation of binding sites for E2F6 factor. We confirmed that the germ-line genes we identified as Dnmt3b target genes were also E2F6 target genes. Of note, and in agreement with gel-shift data (43), promoters of these genes are occupied by E2F6 in WT cells, indicating that E2F6-mediated DNA methylation and silencing does not prevent its binding. Coherent with this factor's involvement in transcriptional repression, we report that repression of Dnmt3b-target germ-line genes in WT cells requires E2F6 binding to their regulatory sequences. This adds unique target genes to the previously reported germ-line genes that are derepressed in somatic tissues as a consequence of invalidation of E2F6 in mice (38, 44, 45). Importantly, binding of E2F6 is required for Dnmt3b binding to the promoter region of these germ-line genes, as well as essential to maintain their silent state, in four of five Dnmt3b target germ-line genes, providing evidence that these genes are silenced in somatic cells through a Dnmt3b-dependant DNA methylation, through recruitment by the E2F6 transcriptional repressor. The functional in vivo interaction between Dnmt3b and E2F6 provided here confirms the previously reported specific partnership between E2F6 and Dnmt3b, but not Dnmt3a, in cell-free systems (15).
DNA methylation may not be a general or a dominant mechanism involved in E2F6-mediated gene silencing, in agreement with the reported DNA methylation-independent silencing of the E2F6 target STAG3 gene (44). Other previously identified E2F6 target germ-line genes tend to be up-regulated in our microarray analysis, although with a less significant P value, probably indicative of the variability of their deregulated expression among embryos. Alternatively, silencing of a subset of germ-line genes might require a different DNMT, such as Dnmt1, shown to be implicated in silencing of a specific set of germ-line genes (35). However, our data support a role for the E2F6 transcription factor as a platform for the recruitment of Dnmt3b-mediated DNA methylation at germ-line genes. In addition, our data show that both E2F6 and Dnmt3b are strongly associated with chromatin, and specifically recruited to gene promoters that need to be maintained as methylated in nongerm cells. As suggested earlier (46, 47), the de novo DNMT Dnmt3b, in complex with E2F6 in the particular case of germ-line genes could be involved in maintenance of gene silencing at genes that need to be permanently methylated and repressed in somatic cells.
The association between DNA hypomethylation and disease is well recognized in the case of cancer and the genetic ICF syndrome (23, 30). The functional deregulation of DNMT3B seems to play a central role in these pathologies, as suggested by the striking similarities at the molecular level between cancer cells and cells derived from ICF patients. Indeed, elevated expression of truncated catalytically inactive splice variants of Dnmt3b has been described in many cancers and associated with the typical centromeric hypomethylation and chromosomal instability (24–26), similar to what has been described in ICF patients (23). In addition, aberrant activation of germ-line genes has been described in different types of tumors (22) and, as reported here, in a mouse model with hypomorphic Dnmt3b mutations characteristic of mutations described in ICF patients. Similarly, aberrant expression of some germ-line genes was found in lymphocytes from patients affected by ICF syndrome (48, 49). Although a potential predisposition of ICF patients to cancer is still under investigation, the classification of some germ-cell genes as cancer-testis genes raises the question of their role in oncogenesis. Interestingly, Slc25a31, Syce1, Tex11, and Ddx4 genes have been implicated recently, by loss or gain of function experiments, in DNA repair processes (50–53). In addition, aberrant expression of meiosis-specific genes can lead to mitotic catastrophe (22). Along the same lines, we have shown in a previous work that deregulated transcription of centromeric minor satellite repeats, associated with their hypomethylation, could lead to aberrant chromosome segregation and aneuploidy (54). Thus, it appears that the potential contribution of aberrant transcription of centromeric repeats or meiotic genes, as a consequence of DNA hypomethylation to chromosome instability, represents a promising field of investigation.
In essence, our data will lay the ground for further studies on elucidating the function of Dnmt3b in the context of maintenance of germ-line gene silencing in somatic cells, in normal and disease situations.
Materials and Methods
Generation of Dnmt3b Mutant Mice.
Dnmt3b mutant mice were generated at the Mouse Clinical Institute – Institut Clinique de la Souris facility, Illkirch, France (http://www-mci.u-strasbg.fr). Details are provided in SI Materials and Methods.
Cells, Plasmids, and Antibodies.
Primary cells, cell lines, plasmids, and antibodies used in this study are listed in SI Materials and Methods.
Preparation of RNA, cDNA, and Genomic DNA.
Nucleic acids were extracted using standard procedures detailed in SI Materials and Methods.
Microarray.
For transcriptome analysis, microarrays with 24,109 spotted mouse oligonucleotides were used (55) and hybridized with RNA extracted from mutant or WT embryonic thymus at 18.5 dpc or MEF isolated from 12.5-dpc embryos. The detailed procedure and the Web sites of the different software and databases used for the analysis are provided in SI Materials and Methods.
Skeleton Staining.
Emptied carcasses of adult mice were fixed in 70% ethanol (48 h) and acetone (48 h) and the remaining tissues were digested in 1% NaOH and 2 mL of Alizarine red saturated solution during a period of 24 h. Carcasses were washed once in 1% NaOH and successively placed over 4 weeks in 0.1% NaOH solutions with increasing glycerol concentration (from 10 to 50%).
Analysis of DNA Methylation.
Analysis of DNA methylation was performed using classical procedures, by genomic bisulfite sequencing or MSRE, followed by Southern blot analysis in the case of repetitive DNA or PCR in the case of unique gene. Detailed procedures are provided in SI Materials and Methods.
ChIP, Immunoprecipitations, and Western Blot Analysis.
Nuclei were isolated as previously described (56) from MEF cells after cross-linking with 1% formaldehyde for 10 min at room temperature for ChIP analysis. Detailed procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Malek Djabali and Claudine Schiff for constructive discussion about this work, Jorg Storre and Stefan Gaubatz for providing the murine embryonic fibroblasts from E2F6 knockout embryos, Paul Danielian and Jacqueline Lees for the preparation of cDNA from WT and E2F6−/− cortex, Pierre-Antoine Defossez for providing the mouse Dnmt3b cDNA, and Yvan Lallemand for his advices on the skeleton staining technique. We also thank Fabienne Nigon and Damien Ulveling for technical help and Emma Walton for editing the manuscript. This work was supported by Agence Nationale pour la Recherche Grant ANR-06-MRAR-040-01. Work in C.F.’s laboratory is supported by research funding from Institut National de la Santé et de la Recherche Médicale and grants from Association pour la Recherche sur le Cancer network, Ligue Nationale Contre le Cancer, and Association Française contre les Myopathies. F.H. was supported by Association Française contre les Myopathies and Fondation pour la Recherche Médicale fellowships.
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarrray data deposited at Gene Expression Omnibus (GEO) is accessible at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=xfgbfiuqmiascxq&acc=GSE19597.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000473107/-/DCSupplemental.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 4.Rollins RA, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 6.Shen L, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 9.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 10.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YA, et al. DNA methyltransferase-3a interacts with p53 and represses p53-mediated gene expression. Cancer Biol Ther. 2005;4:1138–1143. doi: 10.4161/cbt.4.10.2073. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki M, et al. Site-specific DNA methylation by a complex of PU.1 and Dnmt3a/b. Oncogene. 2006;25:2477–2488. doi: 10.1038/sj.onc.1209272. [DOI] [PubMed] [Google Scholar]
- 15.Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–499. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- 16.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 18.Bird AP, Wolffe AP. Methylation-induced repression—Belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 19.Robertson KD, Jones PA. DNA methylation: Past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 20.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 21.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 22.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 23.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775(1):138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Saito Y, et al. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc Natl Acad Sci USA. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostler KR, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopalakrishnan S, et al. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic DNA methylation patterns and displays altered DNA binding. Mol Cancer Res. 2009;7:1622–1634. doi: 10.1158/1541-7786.MCR-09-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisenberger DJ, et al. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2(1):62–72. [PubMed] [Google Scholar]
- 28.Xu GL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 29.Hansen RS, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrlich M, et al. ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity. 2008;41:253–271. doi: 10.1080/08916930802024202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matarazzo MR, De Bonis ML, Vacca M, Della Ragione F, D'Esposito M. Lessons from two human chromatin diseases, ICF syndrome and Rett syndrome. Int J Biochem Cell Biol. 2009;41(1):117–126. doi: 10.1016/j.biocel.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 32.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 34.De Smet C, Lurquin C, Lethé B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maatouk DM, et al. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- 36.Rodić N, et al. DNA methylation is required for silencing of ant4, an adenine nucleotide translocase selectively expressed in mouse embryonic stem cells and germ cells. Stem Cells. 2005;23:1314–1323. doi: 10.1634/stemcells.2005-0119. [DOI] [PubMed] [Google Scholar]
- 37.Storre J, et al. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 2002;3:695–700. doi: 10.1093/embo-reports/kvf141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kehoe SM, et al. A conserved E2F6-binding element in murine meiosis-specific gene promoters. Biol Reprod. 2008;79:921–930. doi: 10.1095/biolreprod.108.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Courel M, Friesenhahn L, Lees JA. E2f6 and Bmi1 cooperate in axial skeletal development. Dev Dyn. 2008;237:1232–1242. doi: 10.1002/dvdy.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueda Y, et al. Roles for Dnmt3b in mammalian development: A mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- 41.Xi S, et al. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci USA. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci USA. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storre J, et al. Silencing of the meiotic genes SMC1beta and STAG3 in somatic cells by E2F6. J Biol Chem. 2005;280:41380–41386. doi: 10.1074/jbc.M506797200. [DOI] [PubMed] [Google Scholar]
- 45.Pohlers M, et al. A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr Biol. 2005;15:1051–1057. doi: 10.1016/j.cub.2005.04.060. [DOI] [PubMed] [Google Scholar]
- 46.Jeong S, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao Q, et al. Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum Mol Genet. 2002;11:2091–2102. doi: 10.1093/hmg/11.18.2091. [DOI] [PubMed] [Google Scholar]
- 49.Jin B, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 50.Adelman CA, Petrini JH. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 2008;4:e1000042. doi: 10.1371/journal.pgen.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimoto H, et al. Germ cell specific protein VASA is over-expressed in epithelial ovarian cancer and disrupts DNA damage-induced G2 checkpoint. Gynecol Oncol. 2008;111:312–319. doi: 10.1016/j.ygyno.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Brower JV, Lim CH, Jorgensen M, Oh SP, Terada N. Adenine nucleotide translocase 4 deficiency leads to early meiotic arrest of murine male germ cells. Reproduction. 2009;138:463–470. doi: 10.1530/REP-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolcun-Filas E, et al. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5:e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Brigand K, et al. An open-access long oligonucleotide microarray resource for analysis of the human and mouse transcriptomes. Nucleic Acids Res. 2006;34:e87. doi: 10.1093/nar/gkl485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Méndez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.