Abstract
Telomeres are engaged in a host of cellular functions, and their length is regulated by multiple genes. Telomere shortening, in the course of somatic cell replication, ultimately leads to replicative senescence. In humans, rare mutations in genes that regulate telomere length have been identified in monogenic diseases such as dyskeratosis congenita and idiopathic pulmonary fibrosis, which are associated with shortened leukocyte telomere length (LTL) and increased risk for aplastic anemia. Shortened LTL is observed in a host of aging-related complex genetic diseases and is associated with diminished survival in the elderly. We report results of a genome-wide association study of LTL in a consortium of four observational studies (n = 3,417 participants with LTL and genome-wide genotyping). SNPs in the regions of the oligonucleotide/oligosaccharide-binding folds containing one gene (OBFC1; rs4387287; P = 3.9 × 10−9) and chemokine (C-X-C motif) receptor 4 gene (CXCR4; rs4452212; P = 2.9 × 10−8) were associated with LTL at a genome-wide significance level (P < 5 × 10−8). We attempted replication of the top SNPs at these loci through de novo genotyping of 1,893 additional individuals and in silico lookup in another observational study (n = 2,876), and we confirmed the association findings for OBFC1 but not CXCR4. In addition, we confirmed the telomerase RNA component (TERC) as a gene associated with LTL (P = 1.1 × 10−5). The identification of OBFC1 through genome-wide association as a locus for interindividual variation in LTL in the general population advances the understanding of telomere biology in humans and may provide insights into aging-related disorders linked to altered LTL dynamics.
Keywords: genes, aging, CXCR4, TERC
Leukocyte telomere length (LTL) undergoes progressive attrition with age and is relatively short in aging-related diseases, most notably atherosclerosis (1, 2). LTL displays high interindividual variation at birth (3, 4) and thereafter (5–7), and it is longer in women than in men (8–11) and longer in African Americans than in whites of European descent (10). Although controversy existed regarding the relation of LTL to survival in the elderly, recent studies in same-sex elderly twins have found that the twin with shorter LTL was more likely to die before the twin with longer LTL (12, 13).
LTL is a complex genetic trait; it is heritable (6, 10, 11, 14, 15) and modified by paternal age at conception (5, 16–18). In addition, several environmental factors, including smoking (7, 8), sedentary living (19), obesity (7, 10, 20), low socio-economic status (21), and unhealthy lifestyle (9), are associated with shorter LTL. Shortened LTL has been observed in rare syndromes, including idiopathic aplastic anemia, idiopathic pulmonary fibrosis, and dyskeratosis congenita (autosomal dominant and X-linked forms) (22–25). These rare syndromes arise from mutations in genes that encode the catalytic (protein) subunit of telomerase reverse transcriptase (TERT), the telomerase RNA component (TERC), which is the template for the synthesis of telomere repeats, and dyskerin (DKC1), a telomerase regulatory protein that binds TERC.
Genome-wide screens and genetic mapping have identified more than 150 genes in yeast that are potentially involved in telomere regulation (26, 27). A number of suggestive loci for LTL in humans have been identified through linkage analyses (14, 15, 28) and genome-wide association (GWA) analysis (29). To date, however, these approaches have not identified loci other than TERC (30) that are associated with LTL in the general population at a genome-wide level of significance. To identify genetic variants associated with LTL, we established a consortium of four population-based cohorts in which LTL was measured and genome-wide genotyping was performed. We report results of the meta-analysis of GWA of LTL coupled with de novo genotyping and in silico replication of SNPs at the top loci in our GWA study.
Results
Clinical Characteristics.
The sample size of individuals with genome-wide genotyping and LTL measurement was 3,417. Table 1 provides the clinical characteristics of participants in the four GWA cohorts: the Framingham Heart Study (31), the Family Heart Study (32), the Cardiovascular Health Study (33, 34), and the Bogalusa Heart Study (35).
Table 1.
Clinical characteristics of the genome-wide association study cohorts
| Parameter | Framingham Heart Study | Family Heart Study | Cardiovascular Health Study | Bogalusa Heart Study |
| Number | 1,146 | 877 | 1,061 | 333 |
| Women (%) | 51 | 51 | 62 | 42 |
| Age in years (range) | 59 (33–86) | 62 (30–93) | 75 (67–95) | 35 (20–48)* |
| BMI (kg/m2) | 28.0 ± 5.0 | 29.1 ± 5.6 | 26.6 ± 4.4 | 28.0 ± 6.7* |
BMI, body mass index ± SD.
*These values include two timepoints for each individual.
GWA Results.
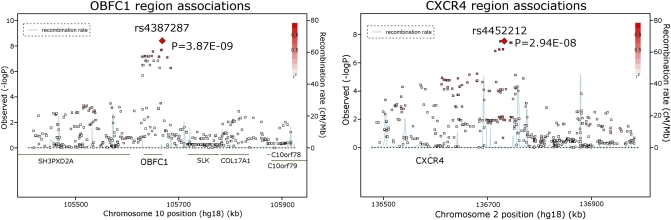
Results of the meta-analysis of the GWA of LTL among the four cohorts are presented in Table 2 for SNPs with P < 5 × 10−7. For the SNPs reported in Table 2, stratified results are provided in Table S1 for the individual cohorts and Table S2 for men and women. In the overall meta-analysis (Table 2), genome-wide significant associations were observed for two regions: the oligonucleotide/oligosaccharide-binding fold containing one gene (OBFC1; lowest P = 3.9 × 10−9 for rs4387287) and the chemokine (C-X-C motif) receptor 4 gene (CXCR4; lowest P = 2.9 × 10−8 for rs4452212). Fig. 1, generated with SNAP (36) (http://www.broadinstitute.org/mpg/snap/), displays associations in the regions of OBFC1 and CXCR4. Mean LTL values by genotype of the top OBFC1 and CXCR4 SNPs are provided in Table S3. OBFC1 was associated with LTL both in men and women (Table S2). In contrast, CXCR4 was more strongly associated with LTL in women than in men (Wald test for sex difference; P < 0.0001). Table 3 displays gene regions associated with LTL in sex-stratified analyses at stratum-specific P < 5 × 10−7 (Wald test for sex differences; P < 0.0001 for all SNPs in Table 3). In men, a genome-wide significant association was observed in the region of DDX18 (P = 8.2 × 10−9 for rs6712766); suggestive association was present for CCBE1, CCDC93, and PODXL. In women, genome-wide significant association was observed in the region of CNTNAP5 (P = 3.8 × 10−8 for rs10189889); suggestive signals were found for TNFSF4, VPS13C, and CXCR4.
Table 2.
Meta-analysis of genome-wide association results for all SNPs with P < 5 × 10−7
| SNP | Chr | Position | Closest gene | Coded allele | Noncoded allele | Coded allele frequency | β | SE | P |
| rs4387287 | 10 | 105667887 | OBFC1 | A | C | 0.08 | 0.12 | 0.02 | 3.87E-09 |
| rs9420907 | 10 | 105666455 | OBFC1 | A | C | 0.91 | −0.11 | 0.02 | 2.01E-08 |
| rs9419958 | 10 | 105665936 | OBFC1 | T | C | 0.08 | 0.11 | 0.02 | 2.08E-08 |
| rs2487999 | 10 | 105649816 | OBFC1 | T | C | 0.05 | 0.13 | 0.02 | 2.69E-08 |
| rs4452212 | 2 | 136732461 | CXCR4 | A | G | 0.65 | −0.08 | 0.01 | 2.94E-08 |
| rs4072435 | 2 | 136730371 | CXCR4 | T | C | 0.65 | −0.08 | 0.01 | 2.96E-08 |
| rs13024450 | 2 | 136731081 | CXCR4 | T | C | 0.65 | −0.08 | 0.01 | 2.96E-08 |
| rs13018756 | 2 | 136724705 | CXCR4 | T | C | 0.58 | −0.08 | 0.01 | 2.99E-08 |
| rs13004902 | 2 | 136744150 | CXCR4 | A | G | 0.36 | 0.08 | 0.01 | 3.71E-08 |
| rs12691881 | 2 | 136746138 | CXCR4 | A | G | 0.35 | 0.08 | 0.01 | 3.76E-08 |
| rs10883943 | 10 | 105641406 | OBFC1 | T | G | 0.58 | −0.08 | 0.01 | 6.09E-08 |
| rs10786775 | 10 | 105647306 | OBFC1 | C | G | 0.95 | −0.12 | 0.02 | 6.13E-08 |
| rs10786774 | 10 | 105634313 | OBFC1 | C | G | 0.99 | −0.12 | 0.02 | 6.29E-08 |
| rs4918068 | 10 | 105634726 | OBFC1 | T | G | 0.05 | 0.12 | 0.02 | 6.33E-08 |
| rs11598840 | 10 | 105635171 | OBFC1 | A | G | 0.05 | 0.12 | 0.02 | 6.43E-08 |
| rs10883942 | 10 | 105641376 | OBFC1 | T | C | 0.42 | 0.08 | 0.01 | 6.49E-08 |
| rs11191849 | 10 | 105640870 | OBFC1 | A | T | 0.59 | −0.08 | 0.01 | 6.50E-08 |
| rs11191848 | 10 | 105640827 | OBFC1 | T | C | 0.42 | 0.08 | 0.01 | 6.52E-08 |
| rs10883941 | 10 | 105640772 | OBFC1 | T | C | 0.58 | −0.08 | 0.01 | 6.54E-08 |
| rs2488000 | 10 | 105661099 | OBFC1 | T | G | 0.05 | 0.12 | 0.02 | 6.81E-08 |
| rs2488001 | 10 | 105661122 | OBFC1 | A | G | 0.05 | 0.12 | 0.02 | 6.96E-08 |
| rs911547 | 10 | 105629411 | OBFC1 | A | G | 0.90 | −0.11 | 0.02 | 6.98E-08 |
| rs1265165 | 10 | 105661517 | OBFC1 | T | C | 0.05 | 0.12 | 0.02 | 7.01E-08 |
| rs2756116 | 10 | 105661794 | OBFC1 | T | C | 0.05 | 0.12 | 0.02 | 7.05E-08 |
| rs11591710 | 10 | 105677622 | OBFC1 | A | C | 0.92 | −0.11 | 0.02 | 7.87E-08 |
| rs2736428 | 6 | 31951903 | SLC44A4 | T | C | 0.29 | 0.08 | 0.01 | 8.59E-08 |
| rs10221893 | 2 | 136730076 | CXCR4 | T | C | 0.63 | −0.08 | 0.02 | 1.12E-07 |
| rs6430612 | 2 | 136722668 | CXCR4 | T | C | 0.65 | −0.08 | 0.02 | 1.15E-07 |
| rs1975174 | 19 | 22307091 | ZNF676 | T | G | 0.47 | 0.07 | 0.01 | 1.20E-07 |
| rs9325507 | 10 | 105635612 | OBFC1 | T | C | 0.58 | −0.07 | 0.01 | 1.39E-07 |
| rs7259376 | 19 | 22299545 | LOC148198 | A | G | 0.53 | −0.07 | 0.01 | 1.44E-07 |
| rs11598018 | 10 | 105651305 | OBFC1 | A | C | 0.58 | −0.07 | 0.01 | 1.44E-07 |
| rs12765878 | 10 | 105659612 | OBFC1 | T | C | 0.43 | 0.07 | 0.01 | 1.45E-07 |
| rs4954585 | 2 | 136714864 | CXCR4 | T | C | 0.75 | −0.08 | 0.02 | 1.45E-07 |
| rs2273698 | 10 | 105639148 | OBFC1 | A | G | 0.58 | −0.07 | 0.01 | 1.54E-07 |
| rs3814220 | 10 | 105637290 | OBFC1 | A | G | 0.43 | 0.07 | 0.01 | 1.55E-07 |
| rs8081000 | 17 | 39070759 | MEOX1 | A | G | 0.49 | 0.08 | 0.01 | 1.59E-07 |
| rs351089 | 4 | 132636028 | PCDH10 | T | C | 0.99 | −0.41 | 0.08 | 2.16E-07 |
| rs11668269 | 19 | 22305800 | LOC148198 | A | G | 0.47 | 0.07 | 0.01 | 2.33E-07 |
| rs808373 | 19 | 22261073 | ZNF676 | C | G | 0.54 | −0.07 | 0.01 | 2.37E-07 |
| rs8111537 | 19 | 22327385 | LOC148198 | C | G | 0.56 | −0.07 | 0.01 | 3.06E-07 |
| rs1980653 | 10 | 105644154 | OBFC1 | A | G | 0.43 | 0.07 | 0.01 | 3.09E-07 |
| rs11191841 | 10 | 105629601 | OBFC1 | T | C | 0.42 | 0.07 | 0.01 | 3.11E-07 |
| rs7100920 | 10 | 105630968 | OBFC1 | T | C | 0.58 | −0.07 | 0.01 | 3.25E-07 |
Fig. 1.
Locus-specific association maps for LTL. Locus-specific (−log10 P values) maps in the meta-analysis of LTL for the OBFC1 (Upper) and CXCR4 (Lower) loci. The OBFC1 locus is represented by sentinel SNP rs4387287, and the CXCR4 locus is represented by rs4452212. For each locus, the sentinel SNP (lowest P value) is depicted by a red diamond, and pairwise linkage disequilibrium in the HapMap CEU with the sentinel SNP is indicated by the color scale. Superimposed on the plot are gene locations (green) and recombination rates (blue).
Table 3.
Meta-analysis of genome-wide association results for men and women for SNPs with P < 5 × 10−7 in either sex
| Meta-GWA study results for women | Meta-GWA study results for men | Wald test* | |||||||||||
| SNP | Chr | Position | Closest gene | Coded allele | Coded allele frequency | β | SE | P | β | SE | P | T value | P |
| rs6712766 | 2 | 118328259 | DDX18 | A | 0.07 | −0.04 | 0.04 | 3.66E-01 | −0.21 | 0.04 | 8.22E-09 | 34.12 | <0.0001 |
| rs1791285 | 18 | 55517163 | CCBE1 | T | 0.80 | 0.03 | 0.03 | 3.70E-01 | −0.16 | 0.03 | 1.25E-07 | 28.78 | <0.0001 |
| rs13432259 | 2 | 118361546 | CCDC93 | A | 0.04 | −0.06 | 0.04 | 1.27E-01 | −0.20 | 0.04 | 2.04E-07 | 29.27 | <0.0001 |
| rs12534413 | 7 | 131104504 | PODXL | T | 0.17 | 0.02 | 0.03 | 5.96E-01 | −0.15 | 0.03 | 2.21E-07 | 27.17 | <0.0001 |
| rs10189889 | 2 | 125161934 | CNTNAP5 | A | 0.03 | 0.34 | 0.06 | 3.77E-08 | −0.06 | 0.06 | 3.47E-01 | 31.18 | <0.0001 |
| rs11583523 | 1 | 171429823 | TNFSF4 | A | 0.99 | −0.98 | 0.19 | 1.61E-07 | −0.18 | 0.16 | 2.57E-01 | 28.73 | <0.0001 |
| rs9920256 | 15 | 59986872 | VPS13C | A | 0.03 | 0.37 | 0.07 | 2.66E-07 | 0.04 | 0.07 | 6.01E-01 | 26.79 | <0.0001 |
| rs4452212 | 2 | 136732461 | CXCR4 | A | 0.65 | −0.10 | 0.02 | 4.27E-07 | −0.05 | 0.02 | 9.84E-03 | 32.15 | <0.0001 |
*Men vs. women.
We examined GWA study results for SNPs in the regions of TERC and TERT, two genes implicated in monogenic diseases associated with shortened LTL. We found evidence of association in the region of TERC (rs3772190; P = 1.13 × 10−5) (Table S4); The TERC locus was recently reported to be associated with LTL in a meta-analysis that included the TwinsU.K. data (30). Meta-analysis of GWA results for the TwinsU.K. top SNP (rs12696304) compared with those from our four cohorts yielded a P value of 5.5 × 10−8. We did not detect a signal in the region of TERT. We additionally examined associations for SNPs in the region of 18q12.2 and other SNPs recently reported to be associated with LTL (29), but we found no association signal (lowest P = 0.02) (Table S5).
Replication Through de Novo Genotyping.
Results of the replication genotyping of SNPs in the OBFC1 and CXCR4 loci are presented in Table 4. For the OBFC1 locus, we genotyped three of the SNPs most strongly associated with LTL (rs4387287, rs9419958, and nonsynonymous rs2487999) in 1,893 additional participants in the Family Heart Study (Table S6). A fourth SNP, rs9420907, was in complete linkage disequilibrium with rs9419958 and therefore, was not genotyped. We found evidence of replication for OBFC1 in whites (P = 0.013 for rs4387287 and P = 0.027 for rs9419958) and the combined sample of whites and African Americans (P = 0.0026 for rs4387287 and P = 0.0032 for rs9419958). For the CXCR4 locus, we genotyped rs4452212 and rs4954585 in additional samples from the Family Heart Study but were unable to replicate the GWA findings (Table 4).
Table 4.
Replication results for OBFC1 and CXCR4
| OBFC1 genotype* | 1 | 2 | 3 | P |
| Whites of European descent (n = 1,319) | ||||
| rs4387287 (C/A) | 6.80 ± 0.024 | 6.88 ± 0.038 | 7.03 ± 0.083 | 0.013 |
| rs9419958 (C/T) | 6.80 ± 0.023 | 6.90 ± 0.040 | 7.09 ± 0.112 | 0.027 |
| rs2487999 (C/T) | 6.81 ± 0.023 | 6.92 ± 0.044 | 7.08 ± 0.20 | 0.21 |
| African Americans (n = 574) | ||||
| rs4387287 (A/C) | 7.08 ± 0.042 | 7.03 ± 0.042 | 7.01 ± 0.071 | 0.34 |
| rs9419958 (T/C) | 7.10 ± 0.055 | 7.06 ± 0.042 | 6.96 ± 0.053 | 0.062 |
| rs2487999 (C/T) | 6.99 ± 0.042 | 7.11 ± 0.041 | 7.07 ± 0.073 | 0.33 |
| Both races (n = 1,893) | ||||
| rs4387287 (C/A) | 6.89 ± 0.028 | 6.96 ± 0.029 | 7.04 ± 0.038 | 0.0026 |
| rs9419958 (C/T) | 6.89 ± 0.025 | 6.99 ± 0.029 | 7.06 ± 0.049 | 0.0032 |
| rs2487999 (C/T) | 6.91 ± 0.023 | 7.03 ± 0.030 | 7.02 ± 0.067 | 0.12 |
| CXCR4 genotype* | ||||
| Whites of European descent (n = 1,593) | ||||
| rs4452212 (A/G) | 6.85 ± 0.031 | 6.83 ± 0.026 | 6.83 ± 0.039 | 0.77 |
| rs4954585 (T/C) | 6.83 ± 0.027 | 6.84 ± 0.025 | 6.82 ± 0.046 | 0.86 |
| African Americans (n = 597) | ||||
| rs4452212 (G/A) | 7.06 ± 0.036 | 7.02 ± 0.049 | 6.84 ± 0.115 | 0.098 |
| rs4954585 (C/T) | 7.04 ± 0.046 | 7.09 ± 0.039 | 6.95 ± 0.065 | 0.26 |
Mean LTL (kb) according to genotype.
*Genotype 1 indicates zero copies, genotype 2 indicates one copy, and genotype 3 indicates two copies of the minor allele. The minor allele is listed second next to the rs number for each SNP. Note that the minor alleles may differ by race. P values reflect an additive genetic model.
Replication Through in Silico Lookup.
Replication of SNPs at our top loci also was undertaken through lookup of results within the LTL GWA of women from the TwinsU.K. Study (30), which had genome-wide genotyping and LTL data for 2,876 women. Six SNPs from the combined sex analysis of Table 2 (rs351089, rs4452212, rs2736428, rs1975174, rs8081000, and rs4387287) were looked up in the TwinsU.K. Study and meta-analyzed in conjunction with the results from the four GWA cohorts; the total sample size was 6,293. The OBFC1 locus replicated (rs4387287, P = 8.9 × 10−4 in TwinsU.K. Study; P = 2.3 × 10−11 in the meta-analysis of our GWA plus TwinsU.K. Study). Table 5 summarizes these results.
Table 5.
Replication through in silico lookup and meta-analysis with TwinsU.K. Study
| TwinsU.K. women only (n = 2,876) | Meta-analysis of all studies (n = 6,293) | ||||||||||
| SNP | Chr | Position | Nearest gene | Coded allele | Noncoded allele | β | SE | P | β | SE | P |
| rs1975174 | 19 | 22307091 | ZNF676 | T | G | 0.02 | 0.02 | 2.40E-01 | 0.05 | 0.01 | 2.05E-06 |
| rs2736428 | 6 | 31951903 | SLC44A4 | T | C | 0.02 | 0.02 | 3.64E-01 | 0.05 | 0.01 | 3.24E-06 |
| rs351089 | 4 | 132636028 | PCDH10 | T | C | −0.06 | 0.08 | 4.25E-01 | −0.23 | 0.06 | 2.86E-05 |
| rs4387287 | 10 | 105667887 | OBFC1 | A | C | 0.08 | 0.03 | 8.92E-04 | 0.10 | 0.02 | 2.33E-11 |
| rs4452212 | 2 | 136732461 | CXCR4 | A | G | −0.02 | 0.02 | 3.31E-01 | −0.05 | 0.01 | 1.68E-06 |
| rs8081000 | 17 | 39070759 | MEOX1 | A | G | 0.00 | 0.02 | 9.26E-01 | 0.04 | 0.01 | 1.43E-04 |
Discussion
Our GWA meta-analysis identified genome-wide significant associations of LTL with regions containing OBFC1 and CXCR4. Our association results for OBFC1 were replicated through de novo genotyping of 1,893 additional individuals and through silico analysis of 2,876 individuals with existing LTL GWA data; however, we were unable to replicate our findings for CXCR4. We also were able to confirm a recently reported association for the TERC locus (30). The potential implications of these findings are discussed based on the postulate that LTL dynamics (birth LTL and its age-dependent shortening afterward) mirror telomere dynamics in hematopoietic stem cells (HSCs) from which leukocytes are derived (37).
Theoretical considerations suggest that genes associated with LTL belong to two general categories (i.e., genes directly engaged in telomere maintenance and genes that impact the turnover rate of HSCs). Based on the biology of the genes implicated in our study, we contend that OBFC1 and TERC fall into the former category and CXCR4 falls into the latter category.
OBFC1 is the human homolog of yeast Stn1 (38, 39), a protein specifically involved in the replication and capping of telomeres (40). Yeast Stn1 regulates synthesis of the telomeric G-rich strand by both protecting terminal telomeric DNA and negatively regulating telomerase action on telomeres (41, 42). In addition, Stn1 coordinates DNA replication of the opposing telomeric C strand. Importantly, it has been recently shown that OBFC1 interacts and colocalizes with telomeric proteins in human cells, suggesting that OBFC1 regulates telomere length and/or function in humans (38, 39). Supporting this is the observation that overexpression of an OBFC1 truncation mutation causes telomere elongation in cancer cells (38).
Interrogation of SNP databases revealed that two of the top OBFC1 SNPs in our LTL GWA results are nonsynonymous and in tight-linkage disequilibrium (rs2487999, P = 2.7 × 10−8; rs10786775, P = 6.1 × 10−8; pairwise r2 = 1.0). They are predicted by PolyPhen (http://genetics.bwh.harvard.edu/pph/) to be benign individually, but the effects of two shared nonsynonymous amino acid substitutions in the coded protein are not known.
Because OBFC1 is a major gene that is directly engaged in telomere biology, we genotyped the top GWA results for OBFC1 in participants from the Family Heart Study and conducted in silico analysis with GWA results from the TwinsU.K. Study in an effort to replicate our findings. We found evidence of replication for OBFC1 in the genotyped individuals from the Family Heart Study (Table 4) and in the in silico analysis of women from the TwinsU.K. Study (Table 5). In our genotyped replication samples, on average, the difference in LTL across OBFC1 genotypes of rs4387287 and rs9419958 amounted to 230 bp and 290 bp in whites, respectively (Table 4). In the meta-analysis, the mean LTL difference across genotypes was 400 bp in men and 180 bp in women for rs437287 (Table S3). These differences in telomere length by genotype are substantial, given that the rate of LTL shortening in the general population is about 20–30 bp/year (43).
Of note, the Framingham Heart Study previously reported an association of the OBFC1 locus (rs3814219; P = 9.5 × 10−7) with brachial-artery basal blood flow, which is a functional index of the vascular endothelium (44). However, the top SNP in the GWA of vascular function was weakly correlated with the top OBFC1 SNPs in this investigation (rs3814219 and rs4387287, r2 = 0.02; other pairwise r2 values did not exceed 0.34).
Although we were not able to replicate our CXCR4 results, several lines of evidence point to a connection between CXCR4 and LTL-related biology. First, CXCR4 is a key regulator of the trafficking of neutrophils between the bone marrow and blood (45, 46). Thus, CXCR4 variants might impact HSC turnover and consequently, LTL. Second, shortened LTL is associated with atherosclerosis (1, 2), an inflammatory disease of the vascular endothelium, marked by a damage-repair feedback loop between the bone marrow (i.e., HSCs) and vascular endothelium. This feedback is mediated by chemotactic signals and endothelial progenitor cells. At its center are chemokines, specifically, CXCR4 and its cognate ligand CXCL12 (47, 48). Moreover, the CXCL12 locus has been found to be associated with myocardial infarction at a genome-wide level of significance (49). Despite these enticing physiological observations, our inability to replicate the association of CXCR4 with LTL deserves further investigation to determine if the LTL association with CXCR4 in our meta-analysis is a false-positive result.
Finally, mutations in TERT, TERC, and DKC1 have been identified in relatively rare monogenic diseases that are associated with shortened telomeres. A lookup of our association results identified an association signal in the TERC locus (rs3772190; P = 1.1 × 10−5); this SNP is in strong linkage disequilibrium with a newly reported GWA finding at this locus (rs12696304, P value in our meta-analysis = 1.6 × 10−4; pairwise r2 = 0.91) (Table S4) (30). Combining the results from the TwinsU.K. Study and our study produced a P value of 5.5 × 10−8, confirming that this locus is associated with LTL.
Given that shortened LTL denotes susceptibility to aging-related diseases, principally atherosclerosis, in the general population, the associations of LTL with OBFC1 and TERC (and potentially, CXCR4 if replicated by other studies) bolster the tenet that telomere biology is an important pathway in human aging. Although many factors are at work in fashioning human LTL, our findings are arguably a major line of evidence that genes engaged in telomere maintenance (and perhaps, HSC turnover) are directly involved in this process and explain, in part, interindividual variation in LTL in the general population.
Materials and Methods
The LTL Consortium includes cohorts from four studies with genome-wide genotyping and LTL measurements: the Framingham Heart Study, the Family Heart Study, the Cardiovascular Health Study, and the Bogalusa Heart Study (SI Text). The consortium established a consensus on covariate selection a priori and an analytical plan for within-study GWA and meta-analysis of results across the studies. Each study received institutional review-board approval of its research procedures, procedures for DNA collection, and use for genetic research. All participants in each cohort study gave written informed consent for participation in the parent study and for the conduct of genetic research using DNA.
LTL Measurement.
LTL was measured by Southern blot analysis of the mean length of terminal restriction fragments as described previously (50, 51). The overlay method was used for the Framingham Heart Study and the Family Heart Study (50), whereas the standard method was used for the Cardiovascular Health Study and the Bogalusa Heart Study (51). These two approaches are highly correlated (50). The laboratory measuring LTL was blind as to the characteristics of DNA donors in all four studies.
Genome-Wide Genotyping.
The platforms used for genome-wide genotyping are provided in Table S7. All physical positions use National Center for Biotechnology Information (NCBI) build 36.3 as the reference.
Framingham Heart Study.
Genotyping was conducted on 9,274 Framingham Heart Study participants using the Affymetrix 500K mapping array and the Affymetrix 50K gene-focused molecular imprinted polymer array (SNP Health Association Resource at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v2.p1). Individuals were excluded from association analyses when the call rate across all genotyped SNPs was less than 97%. There were 503,551 SNPs available for analysis after exclusions. The genotyped sample fulfilling eligibility criteria with LTL measurements and full covariate information was 1,146 individuals. Researchers imputed the genotype data for 2.5 million SNPs using MACH (version 1.0.15) software for autosomal SNPs. The reference panel for the imputation was the publicly available phased haplotypes from HapMap (release 22, build 26, CEU population). The final genotyping data for analysis are composed of 2,543,887 SNPs in HapMap using allele dosage (0–2) values. Because family members were included within the offspring cohort, significance of the association between SNPs and LTL was tested using the linear mixed-effects regression (LME) model, which takes into consideration random individual effects correlated within pedigree according to kinship relationships.
Family Heart Study.
All subjects from the Family Heart Study were genotyped on the Illumina HumMap 550 chip. Of these, 33 (3.3%) were excluded because of technical errors, call rates below 98%, or discrepancies between reported sex and sex-diagnostic markers. There was no significant plate-to-plate variation in allele frequencies. Because the Framingham site of the Family Heart Study was involved in both the Framingham Heart Study and the Family Heart Study, subjects who were in both studies were removed from the Family Heart Study dataset before analysis. For the replication studies, subjects were genotyped after excluding overlap with Framingham subjects and those without LTL measurements or other analysis variables. There were 547,353 SNP markers available for analysis in the Family Heart Study. After exclusions for deviations from Hardy–Weinberg equilibrium (P < 10−6), minor allele frequency < 1%, and markers not in the HapMap, 456,293 markers were used as a framework map for imputation. MACH (version 1.0.15) was used to impute up to 2,543,887 SNPs using the publicly available phased haplotypes from HapMap (release 22, build 26, CEU population) as a reference population. A random sample of 200 (100 males and 100 females) unrelated individuals, excluding those with the highest rates of missing genotypes, was used to estimate parameters that were then applied to the remaining subjects. Genotype dosage was output and used in the analysis. An additive linear-regression model using PLINK (http://pngu.mgh.harvard.edu/∼purcell/plink/) was used for the association analysis of each SNP with LTL. Although some of the 877 subjects were related, the correlation of P values for two chromosomes with and without adjustment for relatedness was over 0.95 with no difference in mean P value. Therefore, no adjustment for relatedness was performed for the analysis of the 877 subjects. For the replication samples, which were family-based, a mixed model was used to account for relatedness separately for whites and African Americans (52).
Cardiovascular Health Study.
Genotyping was performed using the Illumina 370CNV BeadChip system on 3,980 Cardiovascular Health Study (CHS) participants who were free of cardiovascular disease at baseline. Participants were excluded if they had a call rate ≤95%. Because the other cohorts were predominantly white, the African-American participants were excluded from this analysis. To date, genotyping has been successful in 3,291 of the attempted 3,397 white participants. A total of 1,080 samples from individuals of European ancestry were available with both LTL measurements and GWA analysis. The following exclusions were applied to identify a final set of 306,655 autosomal SNPs: call rate < 97%, Hardey-Weinberg Equilibrium P < 10−5, more than two duplicate errors or Mendelian inconsistencies (for reference Centre d’Etude du Polymorphisme Humain trios), heterozygote frequency = 0, or SNP not found in HapMap. Imputation was then performed using BIMBAM v0.99 with reference to HapMap CEU using release 22, build 36 with one round of imputations and the default expectation-maximization warm-ups and runs. During the analysis, SNPs were excluded for variance on the allele dosage ≤0.01.
Bogalusa Heart Study.
Individuals were genotyped on the Illumina Human610 Genotyping Beadchip (53, 54). Genotypes were called using the BeadStudio clustering algorithm. Samples were filtered based on the 10th and 50th percentile GenCall score and overall call rates (<99%). Additionally, some related individuals were present in the sample and were identified using Identity-by-Descent measures in PLINK (55). Individuals were filtered such that there were no pairs with pi-hat > 0.10. This resulted in a total of 333 individuals. SNPs were filtered based on call rates (<90%). Cluster plots for SNPs with call rates between 90% and 95% or with cluster separation scores < 0.30 were manually inspected. By genotyping 29 samples in duplicate (18 known replicates and 11 blind replicates), we observed reproducibility >99.99%. There were 626,145 genotyped SNPs used in the reproducibility analysis. Imputation was performed with MACH (v. 1.0.16) using phased CEU haplotypes from HapMap Phase 2 (release 22) and 550,798 genotyped SNPs. A random sample of 200 individuals was used to estimate recombination and error rates before imputing the entire sample. This resulted in imputing 2,543,887 SNPs. By masking 0.1% of the genotypes before imputation, we observed allelic error rates of 1.6% and genotypic error rates of 3.1%.
Telomere length was measured at two timepoints for each individual in the Bogalusa Heart Study (BHS). We modeled and tested associations between SNPs and the two LTL measures using a linear mixed model as implemented in the nlme R package (56). A covariance structure for the two LTL measurements was chosen by testing all spatial structures with the model, and the structure that resulted in the lowest Akaike Information Criterion score was used (in this study, the best covariance was exponential). If an SNP had been genotyped, the genotype value (0, 1, or 2) was used as a predictor, whereas if the SNP was imputed, the estimated dosage was used.
GWA Analysis.
In each cohort, GWA was conducted for the continuous variable LTL (in kb pairs) using an additive genetic model with adjustment for age, age2, sex, cigarette smoking, and body mass index (BMI). GWA was conducted with 2.5 million HapMap SNPs in conjunction with measured genotypes or allele dose when genotypes were imputed. In the Framingham Heart Study where family members were included within the offspring cohort, significance of the association between SNPs and LTL was tested using the linear mixed-effects regression (LME) model, which takes into consideration random individual effects correlated within pedigree according to kinship relationships. Meta-analysis was performed using inverse variance weighting, and a level of statistical significance was established at 5 × 10−8. A P value between 5 × 10−7 and 5 × 10−8 was considered strongly suggestive of association.
Replication Genotyping.
Genotyping was performed using MGB Taqman probe assays from Applied Biosystems. Primer and probe sequences are available on request. Specifically, the reaction mix in a final volume of 5 μl included 10 ng genomic DNA, 4.5 pmol of each primer, 1.25 pmol of each probe, 1× PCR buffer from Qiagen, 2× Q solution from Qiagen, 500 pmol dNTP, and 0.15 units of Qiagen DNA polymerase. PCR cycling included 55 cycles of a two-step PCR (95 °C for 15 seconds and 60 °C for 1 min) after an initial 2 min at 95 °C. PCR plates were read on an ABI PRISM 7900 HT instrument for genotype assignment. Duplicates of 22 DNA samples and water controls were genotyped for quality control. The laboratory technician was blinded as to which samples were duplicates or water controls. The order of the DNA samples in the 384-well plates was randomized to ensure balance in study conditions across covariates. Genotyping call rates ranged from 95% to 99%, and duplicate concordance rates were higher than 99%. For replication analyses, a P value of 0.05 divided by the number of SNPs tested for replication was considered statistically significant.
In Silico Replication.
The TwinsU.K. Study (29, 30) conducted GWA of LTL measured by the Southern blot method (51). Methods of genome-wide genotyping and GWA have been summarized previously (29). For replication purposes, in silico lookup was carried out in TwinsU.K. women for the top SNPs from the GWA meta-analysis of the four primary cohorts of this investigation. The sample size of TwinsU.K. women for this analysis was 2,876. These results were meta-analyzed with the primary GWA results for these SNPs in our GWA. The total sample size for this combined analysis was 6,293.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911494107/-/DCSupplemental.
References
- 1.Aviv A. Telomeres and human somatic fitness. J Gerontol A Biol Sci Med Sci. 2006;61:871–873. doi: 10.1093/gerona/61.8.871. [DOI] [PubMed] [Google Scholar]
- 2.Samani NJ, van der Harst P. Biological ageing and cardiovascular disease. Heart. 2008;94:537–539. doi: 10.1136/hrt.2007.136010. [DOI] [PubMed] [Google Scholar]
- 3.Okuda K, et al. Telomere length in the newborn. Pediatr Res. 2002;52:377–381. doi: 10.1203/00006450-200209000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Akkad A, et al. Telomere length in small-for-gestational-age babies. BJOG. 2006;113:318–323. doi: 10.1111/j.1471-0528.2005.00839.x. [DOI] [PubMed] [Google Scholar]
- 5.Njajou OT, et al. Telomere length is paternally inherited and is associated with parental lifespan. Proc Natl Acad Sci USA. 2007;104:12135–12139. doi: 10.1073/pnas.0702703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slagboom PE, Droog S, Boomsma DI. Genetic determination of telomere size in humans: A twin study of three age groups. Am J Hum Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- 7.Valdes AM, et al. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366:662–664. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 8.Nawrot TS, Staessen JA, Gardner JP, Aviv A. Telomere length and possible link to X chromosome. Lancet. 2004;363:507–510. doi: 10.1016/S0140-6736(04)15535-9. [DOI] [PubMed] [Google Scholar]
- 9.Bekaert S, et al. Telomere length and cardiovascular risk factors in a middle-aged population free of overt cardiovascular disease. Aging Cell. 2007;6:639–647. doi: 10.1111/j.1474-9726.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 10.Hunt SC, et al. Leukocyte telomeres are longer in African Americans than in whites: The National Heart, Lung, and Blood Institute Family Heart Study and the Bogalusa Heart Study. Aging Cell. 2008;7:451–458. doi: 10.1111/j.1474-9726.2008.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeanclos E, et al. Telomere length inversely correlates with pulse pressure and is highly familial. Hypertension. 2000;36:195–200. doi: 10.1161/01.hyp.36.2.195. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M, et al. Telomore length and mortality: A study of leuckocytes in elderly Danish twins. Am J Epidemiol. 2008;167:799–806. doi: 10.1093/aje/kwm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakaysa SL, et al. Telomere length predicts survival independent of genetic influences. Aging Cell. 2007;6:769–774. doi: 10.1111/j.1474-9726.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- 14.Andrew T, et al. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am J Hum Genet. 2006;78:480–486. doi: 10.1086/500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasa-Nicotera M, et al. Mapping of a major locus that determines telomere length in humans. Am J Hum Genet. 2005;76:147–151. doi: 10.1086/426734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unryn BM, Cook LS, Riabowol KT. Paternal age is positively linked to telomere length of children. Aging Cell. 2005;4:97–101. doi: 10.1111/j.1474-9728.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 17.De Meyer T, et al. Paternal age at birth is an important determinant of offspring telomere length. Hum Mol Genet. 2007;16:3097–3102. doi: 10.1093/hmg/ddm271. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M, et al. Offspring's leukocyte telomere length, paternal age, and telomere elongation in sperm. PLoS Genet. 2008;4:e37. doi: 10.1371/journal.pgen.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherkas LF, et al. The association between physical activity in leisure time and leukocyte telomere length. Arch Intern Med. 2008;168:154–158. doi: 10.1001/archinternmed.2007.39. [DOI] [PubMed] [Google Scholar]
- 20.Gardner JP, et al. Rise in insulin resistance is associated with escalated telomere attrition. Circulation. 2005;111:2171–2177. doi: 10.1161/01.CIR.0000163550.70487.0B. [DOI] [PubMed] [Google Scholar]
- 21.Cherkas LF, et al. The effects of social status on biological aging as measured by white-blood-cell telomere length. Aging Cell. 2006;5:361–365. doi: 10.1111/j.1474-9726.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- 22.Mason PJ, Wilson DB, Bessler M. Dyskeratosis congenita—a disease of dysfunctional telomere maintenance. Curr Mol Med. 2005;5:159–170. doi: 10.2174/1566524053586581. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 24.Tsakiri KD, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 26.Askree SH, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatbonton T, et al. Telomere length as a quantitative trait: Genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangino M, et al. A regulatory SNP of the BICD1 gene contributes to telomere length variation in humans. Hum Mol Genet. 2008;17:2518–2523. doi: 10.1093/hmg/ddn152. [DOI] [PubMed] [Google Scholar]
- 29.Mangino M, et al. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J Med Genet. 2009;46:451–454. doi: 10.1136/jmg.2008.064956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Codd V, et al. Common variants near TERC are associated with mean telomere length. Nat Genet. 2010;42:197–199. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 32.Higgins M, et al. NHLBI Family Heart Study: Objectives and design. Am J Epidemiol. 1996;143:1219–1228. doi: 10.1093/oxfordjournals.aje.a008709. [DOI] [PubMed] [Google Scholar]
- 33.Fried LP, et al. The Cardiovascular Health Study: Design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 34.Tell GS, et al. Recruitment of adults 65 years and older as participants in the Cardiovascular Health Study. Ann Epidemiol. 1993;3:358–366. doi: 10.1016/1047-2797(93)90062-9. [DOI] [PubMed] [Google Scholar]
- 35.Berenson GS, et al. Cardiovascular Risk Factors in Children—The Early Natural History of Atherosclerosis and Essential Hypertension. New York: Oxford University Press; 1980. pp. 47–123. [Google Scholar]
- 36.Johnson AD, et al. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sidorov I, Kimura M, Yashin A, Aviv A. Leukocyte telomere dynamics and human hematopoietic stem cell kinetics during somatic growth. Exp Hematol. 2009;37:514–524. doi: 10.1016/j.exphem.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Wan M, Qin J, Songyang Z, Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyake Y, et al. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 41.Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, et al. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aviv A. The epidemiology of human telomeres: Faults and promises. J Gerontol A Biol Sci Med Sci. 2008;63:979–983. doi: 10.1093/gerona/63.9.979. [DOI] [PubMed] [Google Scholar]
- 44.Vasan RS, et al. Genome-wide association of echocardiographic dimensions, brachial artery endothelial function and treadmill exercise responses in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S2. doi: 10.1186/1471-2350-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eash KJ, Means JM, White DW, Link DC. CXCR4 is a key regulator of neutrophil release from the bone marrow under basal and stress granulopoiesis conditions. Blood. 2009;113:4711–4719. doi: 10.1182/blood-2008-09-177287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furze RC, Rankin SM. Neutrophil mobilization and clearance in the bone marrow. Immunology. 2008;125:281–288. doi: 10.1111/j.1365-2567.2008.02950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zernecke A, Shagdarsuren E, Weber C. Chemokines in atherosclerosis: An update. Arterioscler Thromb Vasc Biol. 2008;28:1897–1908. doi: 10.1161/ATVBAHA.107.161174. [DOI] [PubMed] [Google Scholar]
- 48.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 49.Kathiresan S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasan RS, et al. Association of leukocyte telomere length with circulating biomarkers of the renin-angiotensin-aldosterone system: The Framingham Heart Study. Circulation. 2008;117:1138–1144. doi: 10.1161/CIRCULATIONAHA.107.731794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benetos A, et al. Telomere length as an indicator of biological aging: The gender effect and relation with pulse pressure and pulse wave velocity. Hypertension. 2001;37:381–385. doi: 10.1161/01.hyp.37.2.381. [DOI] [PubMed] [Google Scholar]
- 52.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 53.Eberle MA, et al. Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genet. 2007;3:1827–1837. doi: 10.1371/journal.pgen.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keating BJ, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinheiro J, et al. nlme: Linear and Nonlinear Mixed Effects Models. 2009 R package Version 3. Available at http://cran.r-project.org/web/packages/nlme/citation.html. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.