Cytosine methylation is a common DNA modification found in most eukaryotic organisms including plants, animals, and fungi (1, 2). The addition of a methyl group to cytosine nucleotides in DNA does not change the primary DNA sequence, but the covalent modification of DNA by methylation can impact gene expression and activity in a heritable fashion. This type of epigenetic regulation through DNA methylation appears to be a critical process, in an evolutionary sense, with highly conserved enzymes mediating the process (3). In humans, aberrant DNA methylation has been associated with diseases, including cancer (4). To study cytosine methylation patterns across the genome, researchers have used microarray hybridization or direct sequencing of bisulfite-treated DNA (5). However, mapping methylation of individual cytosines in a given genome has been a challenging task and, accordingly, comparative analysis of genome methylation patterns across species has not been performed. Several new studies have addressed this gap in our understanding of the evolution of cytosine methylation (6, 7). Feng et al. in PNAS (7) used next-generation sequencing to investigate the DNA methylation patterns in eight divergent species, including green algae, flowering plants, insects, and vertebrates. Their data allowed a comprehensive comparison of whole-genome methylation profiles across the plant and animal kingdoms, revealing both conserved and divergent features of DNA methylation in eukaryotes.
Although DNA methylation appears to be a widespread epigenetic regulatory mechanism, genomes are methylated in different ways in diverse organisms. In animals, DNA methylation occurs mostly symmetrically (both strands) at the cytosines of a CG dinucleotide. DNA methylation in plant genomes can occur symmetrically at cytosines in both CG and CHG (H = A, T, or C) contexts, and also asymmetrically in a CHH context, with the latter directed and maintained by small RNAs (1). In the model plant Arabidopsis thaliana, levels of cytosine methylation at CG, CHG, and CHH nucleotides are about 24%, 6.7%, and 1.7%, respectively (8, 9). Despite the different methylation sequence contexts, cytosine methylation is established and maintained by a family of conserved DNA methyltransferases (2, 3, 10). Not surprisingly, the absence of DNA methylation in some eukaryotes such as yeast, roundworm, and fruit fly is associated with the evolutionary loss of DNA methyltransferase homologs (3).
In recent years, approaches have been developed to analyze cytosine methylation at a whole-genome level, and this has provided great insights into the biology of DNA methylation. Early approaches used restriction enzymes sensitive to methylated CG sites (11, 12); the level of DNA methylation is determined by enzymatic digestion of methylated DNA, followed by hybridization to high-density oligonucleotide arrays. Another approach captures methylated genomic DNA using immunoprecipitation via an antibody that recognizes 5-methylcytosine, followed by array hybridization or sequencing (13, 14). These approaches were able to determine chromosomal methylation levels and patterns, but had major limitations in their level of resolution, restriction enzyme bias, difficulty in characterizing genome regions rich in repeats, and, most importantly, an inability to detect DNA methylation at a single-nucleotide resolution (5).
Sodium bisulfite treatment converts unmethylated cytosine into uracil, which is replaced by thymine after PCR amplification, while 5-methylcytosine remains unchanged. Therefore, unmethylated or methylated cytosine residues in DNA can be differentiated by bisulfite treatment, DNA sequencing, and comparisons to a reference sequence (15). When combining bisulfite treatment and high-throughput sequencing (Illumina or SBS, 454, SOLiD, etc.), a methylation map can be generated to a single-base-pair resolution across the entire genome. As recently reported from Arabidopsis genome-wide bisulfite sequencing (BS-seq) data (8, 9, 14), cytosine methylation occurs at not only >90% of repetitive sequences and transposons but also within the body of ∼20% of expressed, protein-coding genes. Although CG, CHG, and CHH methylation are all found in repeat-rich pericentromeric heterochromatin, gene-body methylation contains almost exclusively CG methylation. These studies also revealed an interesting, parabolic relationship between gene-body methylation and transcription levels. Whereas modestly expressed genes are more likely to be methylated, genes expressed at the two extremes (lowest and highest levels) are usually less methylated.
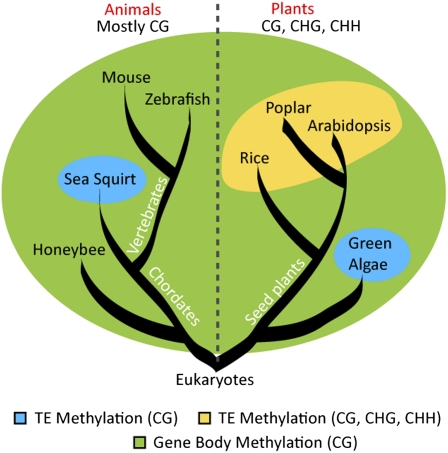
Feng et al. applied BS-seq more broadly than most, profiling DNA methylation patterns in eight diverse eukaryotes (Fig. 1). Besides Arabidopsis, the authors analyzed rice, green algae, and mouse, in which DNA methylation profiles have been previously investigated to varying extents. They added profiles of poplar (a tree), honeybee, sea squirt, and zebrafish, representing a collection of species that span the tree of life from unicellular eukaryotes to multicellular vertebrates. In general, the methylation profiles of flowering plants (Arabidopsis, rice, and poplar) showed similar patterns, with all three contexts (CG, CHG, and CHH) highly enriched in repetitive DNA, transposons, and pericentromeric regions. Methylation occurred almost exclusively in a CG context across the vertebrate genome, except for the unmethylated “CpG islands” near the transcriptional start sites of active genes. Most interestingly, CG methylation within protein-coding genes was preferentially concentrated in the exons, which appears to be a conserved feature in all eukaryotes examined. Gene-body methylation remains apparent in the honeybee genome even though the overall level of genome methylation is very low (∼1% of CG methylation). Taken together, although the function of genic methylation is not fully understood, this conserved methylation pattern is likely an ancient feature, preserved through evolution.
Fig. 1.
Conserved DNA methylation patterns in eukaryotes. Although different methylation contexts are found in animals (CG) and plants (CG, CHG, CHH), gene-body methylation is conserved among eukaryotes. Transposable elements (TEs) are methylated in flowering plants in CG, CHG, and CHH contexts, as well as in a CG context in the green algae and sea squirt genomes. In green algae, non-CG methylation is more enriched in exons of genes compared with TEs and repeats (7). Fungi, not shown in the figure, have genomes generally unmethylated in active genes but heavily methylated at TEs and repeats (6).
Coinciding with the study from Feng et al., Daniel Zilberman's laboratory also reported their data from the comparative DNA methylation profiling by BS-seq of 17 diverse eukaryotes, including plants, animals, and fungi (6). They also conducted transcriptional profiling by RNA sequencing to investigate the functional relationship between DNA methylation and gene expression. Last, they analyzed the presence of a histone variant (H2A.Z) whose distribution pattern was shown to be precisely opposite to that of DNA methylation. Their data agreed with previous observations that gene-body methylation and the depletion of H2A.Z from methylated DNA are evolutionarily conserved, ancient features of the eukaryotic kingdom, predating the divergence of plants and animals (6). However, methylation at transposons and repetitive sequences is less consistently found across species: High levels of transposon methylation are found in land plants and vertebrates, but transposon methylation is not apparent in invertebrates such as silk moth and anemone. These results suggested that the use of DNA methylation to repress deleterious transposons in genomes may have evolved independently in plants and vertebrates, while this function was lost in the invertebrate lineage (6).
High levels of transposon methylation are found in land plants and vertebrates.
Although these latest studies have greatly increased our understanding about evolutionary adaptations and conservation of DNA methylation, inevitably questions remain unanswered. For instance, the function of conserved, genic methylation is still not clear, although it has been proposed to suppress aberrant transcription from cryptic promoters inside the genes (14). Furthermore, we still do not understand the mechanism by which DNA methylation impacts gene transcription levels, especially with regard to the phenomenon of heavier methylation of modestly transcribed genes than those expressed at the extremes. It has been suggested that extreme transcription rates may affect the balance between chromatin disruption and polymerase association, both of which could prevent the generation of aberrant transcripts that might drive methylation via a small RNA-dependent pathway (14). Zilberman et al. also suggest that epigenetic modifications such as DNA methylation and histone modifications could impact nucleosome associations, subsequently interfering with polymerase binding and transcriptional initiation. Interestingly, this assumption is in accordance with periodicity of methylated cytosines in DNA described in Cokus et al. (8), in which a 10- or 167-nucleotide pattern was observed; this correlated with the length of one helical rotation of DNA and the average length of DNA wrapped around a plant nucleosome, respectively. More comprehensive studies in a broad range of genomes will likely provide further insights into the function and evolution of DNA methylation.
Acknowledgments
Work on plant small RNAs and epigenetics in the Meyers laboratory is supported by the National Science Foundation Plant Genome Research Program.
Footnotes
The authors declare no conflict of interest.
See companion article on page 8689 in issue 19 of volume 107.
References
- 1.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lister R, Ecker JR. Finding the fifth base: Genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. April 15, 2010 doi: 10.1126/science.1186366. 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 7.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. April 15, 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: A structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 12.Martienssen RA, Doerge RW, Colot V. Epigenomic mapping in Arabidopsis using tiling microarrays. Chromosome Res. 2005;13:299–308. doi: 10.1007/s10577-005-1507-2. [DOI] [PubMed] [Google Scholar]
- 13.Down TA, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 15.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]