Abstract
A filamentous cytoskeleton largely governs the physical shape and mechanical properties of eukaryotic cells. In bacteria, proteins homologous to all three classes of eukaryotic cytoskeletal filaments have recently been discovered. These proteins are essential for the maintenance of bacterial cell shape and have been shown to guide the localization of key cell-wall-modifying enzymes. However, whether the bacterial cytoskeleton is stiff enough to affect the overall mechanical rigidity of a cell has not been probed. Here, we used an optical trap to measure the bending rigidity of live Escherichia coli cells. We find that the actin-homolog MreB contributes nearly as much to the stiffness of a cell as the peptidoglycan cell wall. By quantitatively modeling these measurements, our data indicate that the MreB is rigidly linked to the cell wall, increasing the mechanical stiffness of the overall system. These data are the first evidence that the bacterial cytoskeleton contributes to the mechanical integrity of a cell in much the same way as it does in eukaryotes.
Keywords: optical trapping, Escherichia coli, MreB
Like all biological organisms, bacteria must constantly interact with their physical environment and endure many different kinds of mechanical stresses. A single cell may be called upon to withstand large forces caused by fluid flow or elastic compression, deal with harsh changes in the external concentration of osmolytes, and contort its body while swimming or gliding, all the while protecting its internal contents and maintaining cellular integrity. In the face of these challenges, it is thought that almost all prokaryotes have adopted a “rigid shell” solution in which the cytoplasm is swelled by turgor pressure and surrounded by a stiff exoskeleton cell wall. However, the recent discovery that bacteria possess homologs of the eukaryotic cytoskeletal proteins actin and tubulin have led some to wonder if these proteins might play a mechanical role in bacteria as they do in eukaryotes (1).
Research over the last several decades has revealed that filamentous, cytoskeletal proteins such as actin bear much of the external load on a eukaryotic cell and largely define its shape and mechanical properties (2). For prokaryotes, however, our knowledge of cellular mechanics is less mature. While experiments have probed the global elasticity of whole bacterial cells and chemically purified sacculi (3), little is known about how the different elastic elements inside bacterial cells bear load.
The actin homolog MreB is necessary for maintaining cell shape in most nonspherical bacteria (4, 5). Depletion of MreB in Escherichia coli leads to the growth of large, malformed cells that have lost their rod shape (6). In vitro, MreB polymerizes into largely straight filament bundles in the presence of ATP or GTP, whereas in vivo, MreB filaments form a helical structure under the inner membrane (7–11). Recent studies have shown that MreB colocalizes with the cell-wall synthesizing enzyme Pbp2 in E. coli and that in Bacillus subtilis, the MreB homolog Mbl determines the spatial patterning of new cell-wall material during elongating growth (12, 13). Based on these observations, it has been proposed that MreB regulates cell shape indirectly by controlling the spatial localization of key cell-wall modulating enzymes (4). However, the possibility that MreB, like actin, might exert mechanical forces and play a structural role in bacterial cells has not been explored experimentally.
Despite this lack of evidence, theoretical modeling of cell growth often requires MreB to be a stiff, force-generating polymer. Lan et al. hypothesized that MreB applies a prestretching force to newly synthesized peptidoglycan strands before they are incorporated into the cell wall during elongation (14). Separate work on Mbl filaments in spiral mutants of B. subtilis used an inextensible Mbl polymer tightly linked to the cell wall to explain a spiral cell shape (15). Regardless of mechanism, for MreB to exert meaningful forces on the cell wall, its elasticity be comparable to that of the wall peptidoglycan. To detect a possible mechanical role for MreB in cells, we probed the contribution of MreB to the bending rigidity of E. coli cells.
Results and Discussion
Depolymerization of MreB Lowers Cell Rigidity.
We bound live cells to a polyethylenimine (PEI)-coated coverslip to measure the bending stiffness. Over time, the Luria Bertani (LB) medium in which we let the bound cells grow blocked the naked PEI-coated surface such that the growing ends of a cell were often unattached. By inducing filamentous growth with the drug cephalexin, a beta-lactam antibiotic that inhibits FtsI and prevents constriction of the Z-ring (16), we were able to identify cells in which one end of the cell was stuck to the surface while the other end remained unattached and susceptible to bending forces. A bending force was then applied with an optical trap by binding a polylysine-coated bead to the tip of a growing cell (Materials and Methods and Fig. 1).
Fig. 1.
Experimental setup. (A) Schematic representation of the experimental setup. The free part and stuck part of the cell are shown in yellow and blue respectively. The optically trapped bead is shown in red. (B) Typical DIC image of an E. coli filament with bound bead.
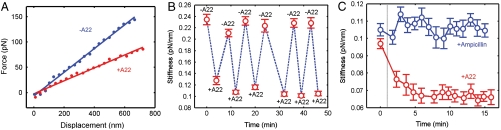
A22, an antibiotic that has been shown to cause the disassembly of MreB polymers by preventing binding of ATP to MreB monomers (9, 17–19), decreased the bending stiffness of cells. For small lateral displacements of the cell tip, the applied force varies linearly with the tip displacement; the slope of this relationship is the bending stiffness (Fig. 2A). When 10 μg/mL of A22 was added to the sample chamber, the bending stiffness of a cell was immediately reduced by approximately 50% (Fig. 2A). This A22-induced stiffness decrease is reversible. Repeated addition and removal of A22 from the media leads to an oscillation in the measured stiffness of a single cell (Fig. 2B).
Fig. 2.
The force-displacement curves and bending stiffness of single E. coli cells. (A) The force-displacement curves of the tip of a cell before and after A22 treatment. (B) The bending stiffness of a cell upon repeated addition and removal of A22. (C) The stiffness of single cells over time after the addition of A22 (Red) or ampicillin (Blue). Strains: BW25113 motA < > AmpR/ pWR20 for A and B; BW25113 motA < > KanR (ampicillin sensitive) for C. Error bars indicate 95% confidence intervals.
In order to compare the bending results from multiple individual cells that have different lengths, we calculated the flexural rigidity, a stiffness parameter that quantifies the resistance to bending and is independent of cell length. The bending stiffness of an elastic rod depends strongly on its length, and each measured cell has a different free-end length. Thus, the distribution of measured bending stiffnesses has a large variance. The bending stiffness, k, of an elastic rod under small-angle deflection is given by k = 3EI/L3, where E is the Young’s modulus, I is the second moment of inertia, and L is the length of the rod. The product EI is the flexural rigidity (20). We developed an image analysis algorithm that uses differential-interference-contrast images taken during a bending experiment to determine the free-end length and shape of a cell under load (Materials and Methods and Fig. S1). As a validation of this approach, we measured the stiffness of a cell over time as it grew (Fig. S2). The measured bending stiffness varied as L-3, suggesting that the flexural rigidity of the cell does not depend on free-end length.
We applied this analysis to the bending of 10 E. coli WA220 cells and calculated their flexural rigidity before and after A22 treatment (Fig. 3). For the population, the flexural rigidity dropped from (2.8 ± 0.5) × 10-20 Nm2 to (2.0 ± 0.4) × 10-20 Nm2 upon the addition of A22. For comparison, the rigidity of E. coli cells is similar to that of a soft rubber rod of the same diameter. On average, the cells exhibited a decrease in flexural rigidity of 30% (Fig. 3D). Before treatment, each cell had a different flexural rigidity. However, the absolute change in flexural rigidity upon A22 addition was proportional to the original rigidity (Fig. 3C), indicating that the drug induces a proportional change in cell bending stiffness.
Fig. 3.
Statistics on the A22-induced decrease in flexural rigidity. (A) The mean and standard error of the flexural rigidity before and after A22 treatment for 10 wild-type WA220 cells. (B) The distribution of the flexural rigidity of the 10 cells in A. (C) The flexural rigidity drop after A22 treatment versus the original flexural rigidity for the 10 wild-type cells and 10 A22 resistant cells. (D) The mean and standard error of the relative flexural rigidity after A22 treatment. (The values are normalized to the flexural rigidity prior to A22 treatment.) Wild type: WA220 (W3110 zhc-12::Tn10 mreB+). A22 resistant strain: WA221 (W3110 zhc-12::Tn10 mreB221). Control: WA220 treated with methanol. **: p < 0.01.
Changes in Cell Shape and Cell Growth Do Not Cause the Decrease in Rigidity.
The observed change of stiffness is not caused by a change in the cell width upon A22 treatment. Using a constitutively expressed cytoplasmic GFP, we used fluorescent imaging to quantify changes in the cell diameter during our experiments. After the addition of A22, cells exhibited a 1.5 ± 0.4% increase in length, due to growth during the experiment, and a negligible 0.7 ± 0.7% decrease in diameter (Fig. S3). For a thin, hollow cylinder of radius R and wall thickness b, the second moment of inertia can be expressed as  . The observed change in width during our experiments corresponds to at most a 2% decrease in the flexural rigidity and cannot explain the reduction in bending stiffness in the presence of A22.
. The observed change in width during our experiments corresponds to at most a 2% decrease in the flexural rigidity and cannot explain the reduction in bending stiffness in the presence of A22.
Several control experiments indicate that the observed A22-induced stiffness decrease is due to MreB disassembly directly rather than through murein remodeling. Repeating the same experimental protocol on an A22-resistant strain of E. coli that contains a single point mutation in the mreB gene (WA221) (21), we observe significantly less rigidity decrease than with the wild-type strain (Fig. 3 C and D). The residual observed decrease in the A22-resistant strain is likely due to an incomplete inhibition of the interaction between A22 and MreB, slightly altered MreB function, or a modified cell-wall architecture. Indeed, WA221 cells have a modestly tapered morphology compared to the purely rod-shaped WA220 strain and have a higher flexural rigidity (Fig. S4). We further verified that the stiffness decrease was not caused (i) by cephalexin treatment using FtsZ-depleted cells (Fig. S5) or (ii) by the addition of the methanol solvent used to dissolve A22 (Fig. 3D). Like the WA221 strain, FtsZ-depleted cells also have a higher flexural rigidity than the cephalexin treated wild type (Fig. S4). Additionally, we verified that the localization of the MreC and MreD proteins, which are believed to lie downstream of MreB, is not affected 5 min after the addition of A22, a long enough time period for us to observe drastic changes in cell stiffness. (Fig. S6).
To test the effect of growth and cell-wall remodeling on our experimental results, we compared the change in stiffness of cells over time after treatment with either A22 or ampicillin, a beta-lactam antibiotic that inhibits transpeptidase activity. The decrease in stiffness upon the addition of A22 happens on a very fast time scale, within the 2 min time window we require to exchange solutions and readjust the optical trap (Fig. 2C, Red). This is consistent with the time needed for depolymerization of MreB by A22 (9), but it is inconsistent with the timescale of cell-wall remodeling, which is likely similar to the time required for the cells to develop a bloated morphology in the presence of A22 or the doubling time (2 h for the 22 °C temperature at which our experiments are performed). After the initial drop in stiffness, cells maintain a roughly constant bending stiffness over tens of minutes (Fig. 2C, Red, and Fig. S7), further suggesting that cell-wall remodeling does not play a role in the observed reduction in cell stiffness.
The addition of 100 μg/mL ampicillin had little effect on cell bending stiffness, even after 15 min (Fig. 2C, Blue, and Fig. S7). This further implies that cell-wall remodeling happens on a much longer time scale than the A22-induced stiffness decrease we observe. We repeated our measurements on starved cells that were no longer growing. As in our previous experiments, these cells exhibited a quick drop in stiffness upon the addition of 10 μg/mL of A22 and recovery of stiffness when the A22 was removed, implying that MreB is able to polymerize in starved cells (Fig. S8). This result is consistent with previous studies that show that starved E. coli cells maintain a significant pool of ATP, about 20%–60% of the total adenylate pool in adhered cells (22). Because murein remodeling is thought to be inhibited in nongrowing cells, these experiments indicate that cell-wall remodeling is not required for the decrease in stiffness upon the addition of A22.
An Elastic Attachment of MreB to the Cell Wall Is Consistent with the Changes in Stiffness.
We built an analytic model of the elasticity of E. coli to better evaluate the different ways in which MreB might affect overall cell stiffness. The model is composed of a stiff helix and an elastic cylinder (a complete description is supplied in SI Text and Fig. S9). A helix lying inside a cylindrical cell will be bent if the cylinder bends. If the helix itself is of a similar stiffness to the cylinder, it will contribute directly to the overall bending stiffness. To date, there has been no measurement of the bending stiffness of MreB filament bundles in vivo or in vitro so that it is hard to calculate this contribution a priori. Alternatively, if MreB is tightly linked to the peptidoglycan, it could increase the stiffness of the cell wall by effectively increasing the crosslink density and stiffness of the cell wall.
We analyzed these two scenarios by examining the bending stiffness for a helix either free within a cylinder or tightly linked to it. Based on our measurements, we can calculate the elastic energy stored in the cylinder and in the helix during bending. Under the no-linkage assumption, the elastic energy in the helix comes from the bending of the helix and we calculate that the addition of A22 would decrease the bending stiffness by a very small amount, 0.00002% to 0.06% (SI Text). When the helix is tightly linked to the cylinder, however, the elastic energy in the helix comes from both the bending and the stretching of the MreB. Under this assumption, the bending stiffness is predicted to change drastically, by 96.2 to 99.7%, upon the addition of A22. These two sets of values lie well above and below the measured effect of A22 on bending stiffness, suggesting that there is a linkage with finite stiffness between the MreB helix and the cell wall. This result is consistent with previous studies showing that MreB interacts with inner-membrane protein MreC, which in turn is thought to bind to the murein synthase Pbp2 (23).
Our results show that MreB, a bacterial actin homologue, contributes nearly as much to the stiffness of a cell as the peptidoglycan cell wall. This is direct evidence that a filamentous cytoskeleton contributes to the mechanical integrity of a bacterial cell in much the same way as it does in eukaryotes, opening up several new lines of inquiry into bacterial cell mechanics. First, bacteria may rely on both the cell wall and the cytoskeleton to actively resist environmental mechanical perturbations, e.g., osmotic pressure changes, by increasing the cell-wall density, thickening the MreB helix, or changing the cross-linking between the cell wall and cytoskeleton. Second, because the bacterial cytoskeleton is stiff relative to the rest of the cell, it can potentially be used to apply significant forces inside bacteria during cell growth and the maintenance of cell shape. Lastly, from an evolutionary point of view, the use of a filamentous cytoskeleton to achieve mechanical goals may have developed in prokaryotes.
Future experiments will be needed to evaluate whether E. coli uses the mechanical rigidity of MreB to apply forces within a cell. Using the experimental setup described here, one can test how E. coli responds to prolonged bending or stretching forces and whether forces alone can complement cell shape changes caused by the disassembly of MreB or other cytoskeletal proteins. In addition, theoretical modeling may also suggest what kind of forces a helical MreB structure apply in vivo and what effect these forces have on the generation and maintenance of a cellular rod shape.
Materials and Methods
Sample Preparation.
E. coli cells were grown in Luria–Bertani broth (LB) medium to exponential phase (OD = 0.2–0.4). The culture was further grown in LB with 50 μg/mL of cephalexin for 15 min and then concentrated 5 times by centrifugation. PEI-coated coverslips were made by flowing 1% polyethylenimine diluted in water into a flow chamber and washed with water after a 5 min incubation. We then flowed the cell culture into the chamber and washed with a mixture of LB and cephalexin (50 μg/mL) after 3 min to remove unattached cells. The chamber was incubated at 37 °C for 30 min to 1 h to let the attached cells grow before placement on the optical trapping instrument. Polylysine-coated beads were made by incubating 0.5-μm-diameter polystyrene beads (Bangs Labs) in 0.1% polylysine diluted in water for 30 min. Beads were then washed 3 times and resuspended in water. The bead solution was diluted twice into LB with cephalexin (50 μg/mL) before being added into the flow chamber. After binding a bead to a cell with the trap, the chamber was washed with cephalexin in LB (50 μg/mL) to remove unattached beads. For A22 treatment, we dilute a 10 mg/mL A22 stock solution (in methanol) 1,000 times into LB with cephalexin (50 μg/mL) to obtain a 10 μg/mL working solution.
For cell starvation, E. coli cells were filamented and set up in the flow chamber as above. Then the chamber was washed with 50 μL of PBS buffer. The cells starved for 10 min before stiffness measurement. A22 was diluted in PBS to obtain a 10 μg/mL working solution. The same cells were imaged 20 min later to verify the lack of cell growth.
For FtsZ depletion, WX7/λGL100 cells were grown at 37 °C in LB containing 0.2% glucose for 2 h. The stiffness measurement was also taken in LB with 0.2% glucose.
Descriptions of the bacterial strains are available in SI Text.
Microscopy and Optical Trapping.
The optical trap is built on a modified Nikon TE2000 microscope and includes both epifluorescence and DIC imaging. A Nd: YVO4 laser (1064 nm, Spectra Physics) is used to generate the trapping potential, while the scattering of an 855-nm diode laser (Bluesky Research) is detected by a position sensitive detector (Newfocus) for position detection. The trap and sample are steered using a closed-loop piezo-driven tip-tilt mirror and stage respectively (Mad City Labs). During a bending experiment, the force and tip displacement were recorded simultaneously with DIC images of the cell shape and saved to disk for offline analysis. Because the cell adjacent to the trapped bead might affect the scattered detection laser light pattern and the resultant position detection, we calibrated the position sensor for each cell with an attached bead.
At the beginning of an experiment, a polylysine-coated bead was held near the tip of a cell until a firm attachment was generated. To bend a cell, the microscope stage was moved in a direction orthogonal to the cell long axis while the optical trap was held stationary. The applied force is proportional to the displacement of the bead from the center of the trap, while the displacement of the cell tip is equal to the magnitude of the stage movement minus the displacement of the bead from the center of the trap. Using this method, we have measured in total 46 cells in various conditions.
The applied force was calculated as a projection of the total force along the direction of bending, i.e., orthogonal to the cell axis. Cells were bent to very small angles, less than 3°, to minimize any stretching forces and ensure that the bending force was greater than 95% of the total force. In order to reduce the potential adverse effects of using very long filamentous cells in our bending experiments, we only included cells with a cell length of less than 17 μm.
Image Analysis.
Our image analysis algorithm is summarized in Fig. S1. First we used the MATLAB Canny edge detector to define the edges of the objects in a grayscale DIC image. We then averaged the two side edges parallel to the cell’s major axis to compute the center line of the cell. The displacement of the center line during bending fit well with the theoretical shape of a bent elastic rod with a stuck end:
 |
where y is the lateral displacement, z is the arc length along the rod starting from the free tip, F is the bending force, A is the flexural regidity, and L is the length of the free cell end.
To quantify the length and width of GFP-expressing cells, we recorded a stack of green fluorescent images with 100-nm axial spacing. An area of interest containing the cell under analysis was manually chosen. We then applied the MATLAB Canny edge detector to define the side edges and the center line of the cell as above. The length of the center line was taken to be the cell length. The distance between the two side edges, calculated along a line orthogonal to the center line, is the cell width.
All errors are reported as standard error of the mean unless otherwise specified.
Supplementary Material
Acknowledgments.
We acknowledge helpful advice from Mingzhai Sun on binding cells to surfaces. We thank Ned Wingreen and Zemer Gitai for valuable discussions and Lawrence Rothfield, Masaaki Wachi, Kenn Gerdes, William Ryu, and Zemer Gitai for supplying strains and plasmids. This research was supported by National Science Foundation CAREER award PHY-0844466, National Institutes of Health Grant P50GM071508, and awards from the Alfred P. Sloan Foundation and the Pew Charitable Trusts to J.W.S.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911517107/-/DCSupplemental.
References
- 1.de Boer PA. Bend into shape. EMBO J. 2009;28(9):1193–1194. doi: 10.1038/emboj.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janmey PA, McCulloch CA. Cell mechanics: Integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 3.Morris DM, Jensen GJ. Toward a biomechanical understanding of whole bacterial cells. Annu Rev Biochem. 2008;77:583–613. doi: 10.1146/annurev.biochem.77.061206.173846. [DOI] [PubMed] [Google Scholar]
- 4.Gitai Z. The new bacterial cell biology: Moving parts and subcellular architecture. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 5.van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 6.Bendezu FO, de Boer PA. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol. 2008;190(5):1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esue O, Wirtz D, Tseng Y. GTPase activity, structure, and mechanical properties of filaments assembled from bacterial cytoskeleton protein MreB. J Bacteriol. 2006;188(3):968–976. doi: 10.1128/JB.188.3.968-976.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vats P, Rothfield L. Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. Proc Natl Acad Sci USA. 2007;104(45):17795–17800. doi: 10.1073/pnas.0708739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Jones LJF, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 11.Bean GJ, Amann KJ. Polymerization properties of the Thermotoga maritimaactin MreB: Roles of temperature, nucleotides, and ions. Biochemistry. 2008;47(2):826–835. doi: 10.1021/bi701538e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vats P, Shih YL, Rothfield L. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol Microbiol. 2009;72(1):170–182. doi: 10.1111/j.1365-2958.2009.06632.x. [DOI] [PubMed] [Google Scholar]
- 13.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 14.Lan G, Wolgemuth CW, Sun SX. Z-ring force and cell shape during division in rod-like bacteria. Proc Natl Acad Sci USA. 2007;104(41):16110–16115. doi: 10.1073/pnas.0702925104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolgemuth CW, et al. How to make a spiral bacterium. Phys Biol. 2005;2:189–199. doi: 10.1088/1478-3975/2/3/006. [DOI] [PubMed] [Google Scholar]
- 16.Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94(2):559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwai N, Nagai K, Wachi M. Novel S-Benzylisothiourea compound that induces spherical cells in Esherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem. 2002;66(12):2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- 18.Karczmarek A, et al. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol Microbiol. 2007;65(1):51–63. doi: 10.1111/j.1365-2958.2007.05777.x. [DOI] [PubMed] [Google Scholar]
- 19.Bean GJ, et al. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48(22):4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landau LD, Lifshitz EM. Theory of Elasticity. 3rd Ed. Oxford; New York: Pergamon Press; 1986. [Google Scholar]
- 21.Kruse T, et al. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Gene Dev. 2006;20:113–124. doi: 10.1101/gad.366606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong Y, Brown DG. Variation in bacterial ATP level and proton motive force due to adhesion to a solid surface. Appl Environ Microbiol. 2009;75(8):2346–2353. doi: 10.1128/AEM.02671-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55(1):78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.