Abstract
Yeast fatty acid synthase (FAS) is a 2.6-MDa barrel-shaped multienzyme complex, which carries out cyclic synthesis of fatty acids. By electron cryomicroscopy of single particles we obtained a three-dimensional map of yeast FAS at 5.9-Å resolution. Compared to the crystal structures of fungal FAS, the EM map reveals major differences and new features that indicate a considerably different arrangement of the complex in solution compared to the crystal structures, as well as a high degree of variance inside the barrel. Distinct density regions in the reaction chambers next to each of the catalytic domains fitted the substrate-binding acyl carrier protein (ACP) domain. In each case, this resulted in the expected distance of ∼18 Å from the ACP substrate-binding site to the active site of the catalytic domains. The multiple, partially occupied positions of the ACP within the reaction chamber provide direct structural insight into the substrate-shuttling mechanism of fatty acid synthesis in this large cellular machine.
Keywords: cryoelectron microscopy, fatty acid synthesis, acyl carrier protein, molecular architecture
Fatty acid synthase (FAS) is the key enzyme for the biosynthesis of fatty acids in living organisms. There are two types of FAS, namely, the type II FAS system in prokaryotes, consisting of a set of individual enzymes (1), and type I FAS found in eukaryotes as a multienzyme complex (2, 3). Type I FAS in mammals is an α2 homodimer (4–7), while type I fungal FAS is an α6β6 hetero-dodecameric complex (8–10). Although fungal and mammalian FAS have very different structures, they conserve all the necessary enzymes found in the FAS type II systems required for fatty acid synthesis (Fig. S1). Mammalian FAS has been studied by cryoelectron microscopy (cryo-EM) (4, 5) and x-ray crystallography (6) and was found to be a highly flexible complex (7). The mobile acyl carrier protein (ACP) domain has not yet been directly observed in the mammalian FAS system.
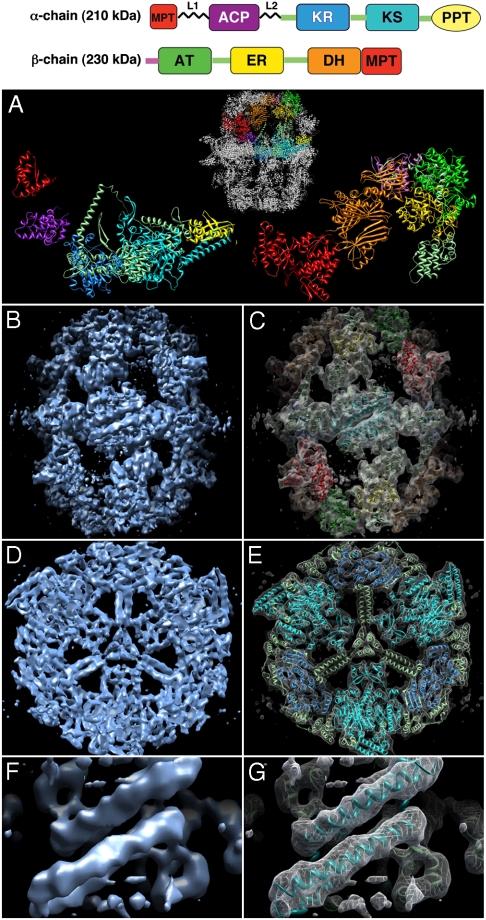
Yeast FAS, which belongs to the fungal type I family, has a total molecular mass of 2.6 MDa. As is known from earlier electron microscopy (EM) studies (8–10), it is a 260-Å × 230-Å barrel of D3 symmetry. Recent x-ray structures of Thermomyces lanuginosus (11) and Saccharomyces cerevisiae FAS at 3.1-Å or 4.0-Å resolution (12–14) are essentially identical. The six α-subunits form an equatorial wheel, which divides the barrel into two separate domes, each consisting of three β-subunits. The α- and β-subunits define three reaction chambers per dome and contain eight catalytic centers. Of these, the α-subunit contributes the phosphopantetheinyl transferase (PPT), ACP, ketoacyl synthase (KS), ketoacyl reductase (KR), and part of the malonyl-palmitoyl transferase (MPT) domain. The β-subunit contributes the acetyl-transferase (AT), enoyl reductase (ER), dehydratase (DH), and the major part of the MPT domain (Fig. 1A). In the fungal FAS, the ACP is tethered by two flexible linkers, which connect it to the MPT domain and the central hub of the equatorial wheel. The linker domains define the radius of action of the ACP, which agrees broadly with the dimensions of the reaction chamber (12). With the exception of the PPT domain, the active sites of all catalytic domains face the reaction chambers in the interior of the FAS barrel (11–14). In addition to the catalytic domains, almost half of the yeast FAS molecular mass is contributed by six structural domains, two in the α-subunit (SD1-2α) and four in the β-subunit (SD1-4β). Previous studies (15) have shown that the reaction chambers work independently of one another.
Fig. 1.
The structure of yeast FAS (A). Domain organization of the FAS α (Left) and β (Right) polypeptides with the domains colored according to the scheme shown above. (B–G) 3D map of yeast FAS at 5.9 Å resolution, without and with the domains of the x-ray structure (PDB ID code 2VKZ) (14) fitted as rigid bodies. The domains are colored according to the scheme shown in A. (B, C) Side view of the α6β6-assembly. (D, E) Central map section showing the α6-wheel. (F, G) The helix pair near the KS dimer on the outside of the α6-wheel showing the helix pitch.
Yeast, which mostly depends on endogenous fatty acid synthesis, has developed a rigid cage-like FAS machinery with six reaction chambers for the increased efficiency of fatty acid synthesis. Here, ACP is proposed to be the only mobile domain performing the substrate shuttling, unlike the open mammalian FAS structure with two reaction chambers where the efficiency of the fatty acid synthesis is determined by the high conformational flexibility of the complex.
The ACP belongs to a class of universal and highly conserved carrier proteins that bind acyl intermediates via the ∼18-Å phosphopantetheine arm and are active in various metabolic pathways, including the biosynthesis of polyketides or fatty acids (16, 17). Both type I and type II ACP have a completely α-helical structure, with a conserved four-helix bundle that harbors the binding site for acyl chain intermediates. However, the ACP domain in fungal FAS has an additional four-helix bundle as compared to that seen in other organisms. The ACP-dependent enzymatic pathways are essential for the cell, making the fungal FAS an important potential drug target (17).
It has been postulated that the ACP domain carries the growing fatty acid chain substrate iteratively to the various catalytic sites in the reaction chamber (12–14). Of these, the AT domain participates in the initiation of fatty acid chain synthesis. The MPT, KS, KR, DH, and ER domains each carry out different steps in the chain elongation cycle. The growing fatty acid chain is attached to the carrier domain, which shuttles between the successive catalytic sites, until it is released into the cytosol by attachment to CoA after having reached a C-16 or C-18 chain length. In yeast, fatty acid synthesis is carried out in the confined environment of a catalytic cage. This makes the process more efficient compared to prokaryotic type II FAS, which relies on diffusion of ACP to the individual enzymes. But so far there has been no direct structural evidence for such a substrate-shuttling mechanism. In the three x-ray structures of yeast FAS (12–14) the ACP is fixed in a single position at the equator near the KS domain. The relative affinities of the catalytic domains for the ACP are unknown.
We present here a 5.9-Å map of yeast FAS obtained by cryo-EM of single particles. The map shows considerable differences in domain organization compared to the four available, largely identical x-ray structures, revealing significant conformational flexibility in the barrel wall. The ACP is found in several alternative locations next to the catalytic domains within the reaction chamber, providing direct evidence for a substrate-shuttling mechanism.
Results
Three-Dimensional Map.
We generated a 3D map of yeast FAS (Fig. 1) from ∼19,000 high-resolution images (Fig. S2). Phase reversals caused by the contrast transfer function (CTF) were corrected by phase flipping and applying a Wiener filter (18), while amplitudes were corrected using low angle x-ray solution scattering data (Fig. S3). The map is similar but not identical to the published x-ray structures of FAS (Fig. S4). The resolution of the final map was measured by the Fourier shell correlation (FSC) method at the FSC = 0.5 criterion as 7.2 Å (19) and 5.9 Å using the FSC = 0.143 criterion (20) (Fig. S2F). Careful inspection suggests that the map resolution is indeed close to 6 Å, as it is possible to distinguish β-sheets in the protein domains (Fig. 1 and Fig. S5), and the helix pitch in some α-helices (Fig. 1F and G).
The cryo-EM map (Fig. 1) resolves clearly all structural features that were seen in the four x-ray structures (11–14). There are, however, significant differences, plus a number of interesting previously undescribed features. To aid map interpretation, the atomic model of yeast FAS (14) was superimposed on the EM map as a rigid body. A good automatic fit as judged by the EM-density correlation factor (21) was achieved after recalibrating the pixel size of the map to 1.14 Å.
Structure of the α6-Wheel.
In the equatorial wheel of the FAS complex, consisting of the six α-chains, all secondary structure elements from the x-ray models match the corresponding densities closely (Fig. 1E). In the three x-ray structures of S. cerevisiae FAS (12–14), the α6-wheel is incomplete, as a part of the α-chain near the KR dimer contact is missing, and the PPT domain is not resolved in two of the three structures. The T. lanuginosus structure (11) shows a four-helix bundle on the outside of the KR dimer, with the helices tilted by ∼45° relative to the long axis of the complex, which is not seen in the yeast x-ray structures. The EM-map density instead indicates a different structure of this part of the α-chain (Fig. S5A).
In one of the yeast FAS structures (13), the PPT domain was found on the outside of the barrel near the KS dimer. A four-helix bundle on the outside of the equatorial α6-wheel, which is part of the KS domain, separates two PPT domains, which are related by 2-fold symmetry. This four-helix bundle is resolved in all x-ray structures, and its density in the EM map is clear (Fig. S5B), although it has moved outward by ∼2 Å (Fig. S4B). The density of the PPT domain in our 6-Å map is comparatively weak, but the recently solved x-ray structure of the individually expressed yeast PPT domain (2WAS) (22) superimposes well (Fig. S5B). With the PPT domain traced and the missing part of the KS domain visualized, the EM map of the yeast FAS shows the complete structure of the α6-wheel.
Different Organization of the Reaction Chamber Wall.
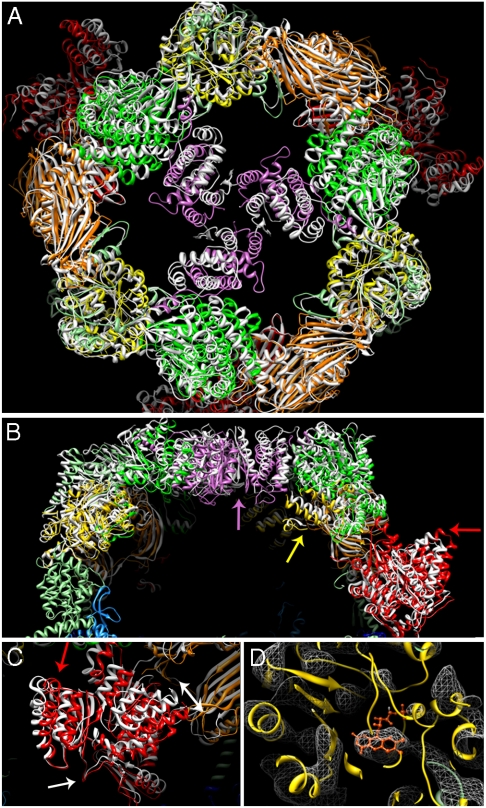
In contrast to the α6-wheel, the global fit of the β3-domes reveals major differences in the position and orientation of the protein domains forming the barrel wall. We remodeled the x-ray structure of yeast FAS by rigid body fits of individual domains into the EM-map density (Fig. 2). A significant radial displacement of the MPT domain is evident (Fig. 2A–C). This domain includes a distinct α-helical hairpin protruding into the solvent, which is shifted by more than one helix diameter toward the center of the barrel in the x-ray structures. While the contact to the KR domain in the equatorial wheel is preserved, the MPT domain rotates ∼20°, resulting in a 9 Å shift. A smaller displacement of ∼5 Å in the same direction is observed for the adjacent DH domain (Fig. 2A and C).
Fig. 2.
Differences between the yeast FAS structure fitted to the EM map and the crystal structure. Fitted domains are colored as in Fig. 1, while the corresponding x-ray structure (2VKZ) is white. (A) Top view of the barrel with the trimerization domain (pink), showing the movement of this domain in the crystal structure with respect to the EM structure. (B) Side view of one dome; arrows indicate the overall movement of domains in the crystal structure relative to the EM map due to compression by crystal contacts at the MPT domain (red arrow). (C) Close-up of the MPT domain seen from the front of the barrel, ∼90° rotated relative to B. The α-hairpin that causes the movement of the MPT domain by ∼10 Å (double-headed arrow) due to crystal contacts is indicated by a red arrow as in B. A white arrow indicates the pivot point at the base of the MPT domain. (D) Clear density is visible for the FMN cofactor (orange) in the ER domain (yellow). The EM map is shown as a white mesh.
The ER domain has a triosephosphate isomerase barrel fold and contains flavin mononucleotide (FMN) as a cofactor. Density for the isoalloxazine ring of FMN is clearly visible in the EM map (Fig. 2D). While the global fit of the x-ray model for this domain was close, it appears to be shifted inward in the EM map by ∼3 Å (Fig. 2A and B), unlike the MPT or DH domains. The AT domain near the top of the barrel was likewise refitted by a ∼9° rotation in a plane parallel to the equator (Fig. 2A and B).
Some of the most striking differences between the EM map and the x-ray models are seen in the structural domain at the apex of the dome around the 3-fold axis (Fig. S4A and E). While the contact to the contiguous AT domain is unaffected, the structural domain is shifted by ∼10 Å downward, and by a similar distance laterally, relative to the 3-fold axis, so that the helix closest to this axis moves by ∼15 Å (Fig. 2A). As a result, the molecular contacts between the three β-subunits are entirely different in the two models: They involve the first strand of the N-terminal β-hairpin (residues 5–15) in the EM model and the α-helix following the hairpin (residues 24–40) in the x-ray model (Fig. 2A).
The refitted EM model indicates a lever-like movement of the β-chain that forms the barrel wall around a pivot point at the MPT domain near its connection to the equatorial wheel (Fig. 2C). This results in an outward displacement of the domains near the α6-wheel, and an inward and downward movement of the upper domains. Overall, the FAS barrel is shorter by ∼20 Å and the dome is wider by up to 18 Å in the EM structure (Fig. 2B and Movie S1).
Multiple Positions of the Acyl Carrier Protein.
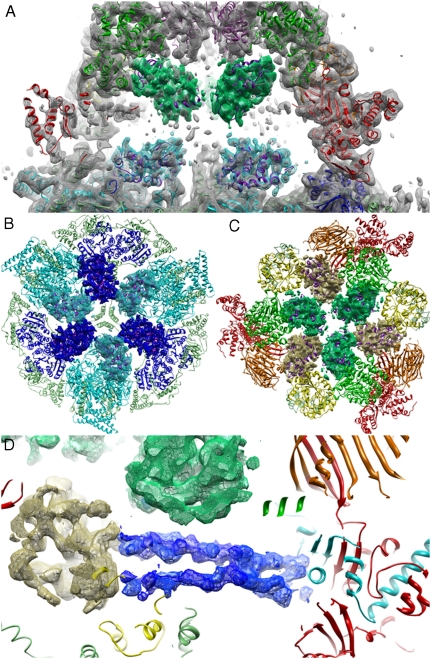
We identified six densities of similar shape and size in the reaction chamber, each next to a catalytic domain. Three of these are comparatively strong (Fig. 3A), one of which is located near the equator adjacent to the KS domain (Fig. 3A and B). The other two densities are located near the AT and ER domains at the top of the barrel (Fig. 3A and C). A fourth, similar density shows up next to the KR domain at the equator (Fig. 3B). Weaker densities are discernible next to the MPT and DH domains. The ACP is not resolved in the T. lanuginosus structure (11). In the three x-ray structures of yeast FAS, the ACP domain is located next to the KS domain, where we find one of the partially occupied map regions. This density was fully accounted for by the ACP model. The atomic model of the ACP domain (12–14) fitted equally well to the three other stronger map regions next to the KR, ER, and AT domains (Fig. 3B and C and Fig. S6). We can therefore assign these map regions to alternative positions of the ACP domain in the reaction chamber (Fig. 4 and Movie S2). The resulting distances between the phosphopantetheine attachment site at Ser180 and the catalytic sites of the KS, KR, ER, and AT domains were measured as approximately 19 Å, 20 Å, 21 Å, and 18 Å, respectively, in excellent agreement with the length of the PPT arm of ∼18 Å (12).
Fig. 3.
Density inside the FAS barrel. Multiple locations of the ACP domain in the reaction chamber are shown in colors matching the nearest catalytic domain (AT: green, ER: yellow, KS: cyan, and KR: blue). The ACP x-ray structure (14) fitted to the EM density is shown as a purple ribbon. (A) Section through two reaction chambers with ACP density next to the AT and the KS domain. (B) View of the central wheel with ACP next to the KR and KS domains. (C) View of the top of the chamber with ACP next to the AT and ER domains. (D) Two ∼64 Å-long densities in the reaction chamber seen directly between the MPT and the ACP density near the ER domain. The densities are not resolved in the x-ray structures and might correspond to the peripheral linker peptide, which connects the ACP (yellow mesh) to the α-chain segment (cyan) of the MPT domain (red).
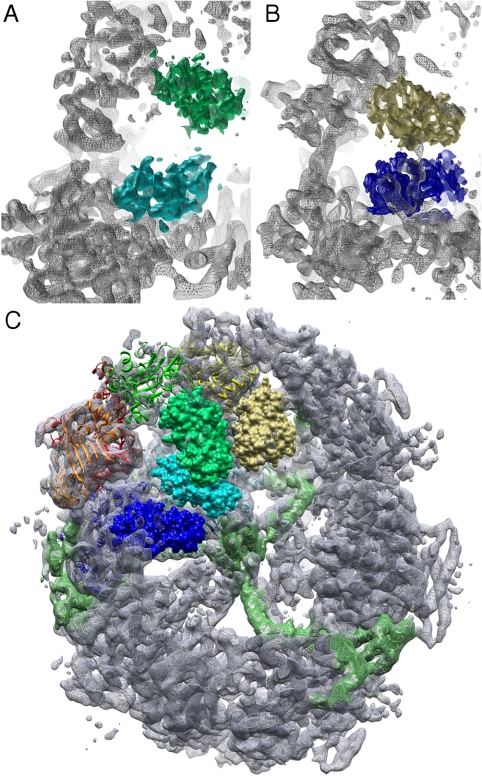
Fig. 4.
Movement of ACP in the reaction chamber. (A) The ACP density near KS (cyan) and AT (green); (B) ACP near KR (blue) and ER (yellow). (C) ACP fitted to the four positions that can be seen in the EM map. Only weak density is seen near the MPT (red) and DH (orange). The three chambers in one dome are delineated by structural domains (light green).
Additionally, we assessed the variability of the 3D volume by a 3D variance map, using a bootstrap algorithm (23, 24). The 3D variance map indicated a high degree of variability throughout the interior of the barrel (Fig. S7). This indicates that the ACP domain is able to move freely inside the reaction chambers (12), though the majority of FAS particles have ACP docked to a catalytic site as indicated by the 3D map.
Relative Occupancies and ACP Docking.
The relative occupancies of ACP sites adjacent to the KS, ER, and AT domains were calculated with respect to one another and estimated from the map densities as ∼30%, while the density next to the KR domain suggests a ∼10% occupancy. The estimated occupancy of the ACP positions next to the MPT and DH domains was below 10%. Considering that the FAS used for our studies is synchronized by adding NADPH and malonyl-CoA and then inhibited with cerulenin (14), and thus not active in fatty acid chain synthesis, the occupancies reflect the affinities of the catalytic domains for ACP, which remains free to move in cerulenin-inhibited FAS. Apparently three domains, KS, AT, and ER, each have roughly equal affinities for the ACP domain, which is about 3-fold higher than that of the KR, while the affinity of the MPT and DH domains for ACP is lower than that of the KR. As no intermediate or overlapping ACP positions were observed in the EM map, this suggests that these occupancies near the three catalytic domains well represent the occupancy of the bound domains (as opposed to simply a generalized overlapping spatial occupancy).
We calculated the electrostatic surface potential of the complete yeast FAS structure and analyzed the surface potentials for the ACP and catalytic domains. The ACP faces the four different catalytic domains with the same surface, which is negatively charged, whereas the interacting surfaces of the KS, KR, ER, and AT domains have a positive surface potential, as have the funnels leading to their catalytic centers. This indicates surface charge complementarity in the region where ACP docks to these domains.
Discussion
Reaction Chamber Conformation and Biosynthetic Activity.
The overall architecture of yeast FAS as seen in the EM map is similar to the x-ray structures (12–14), but significant conformational differences are observed in the positions of the domains in the barrel wall, where the four x-ray structures superimpose exactly onto one another. As the complex in our EM study had been prepared in the same way as in our previous study to solve one of the x-ray structures (14), it is likely that these differences are caused by crystal contacts, which might easily distort the large, hollow FAS barrel. Indeed, a prominent crystal contact involves the loop extending from the MPT domain, which in our map extends ∼10 Å farther into the solvent (Fig. 2B and C). This contact is present in all crystals of yeast or Thermomyces FAS used for structure determination (Fig. S8A). It seems to initiate an inward movement of the MPT and the adjacent DH domain, a concomitant outward movement of the ER domain, plus a displacement of the AT and the structural domain at the N terminus of the β-chain near the 3-fold axis.
As a result of these large domain movements, the reaction chamber has a different shape in the EM solution structure (Fig. 2B). This may be the reason why, so far, all yeast FAS x-ray structures (12–14) show the ACP in a single location at the equatorial KS domain. This is the only one of the observed positions where it is not in contact with the barrel wall and, therefore, would be least affected by changes in shape of the reaction chamber.
The multiple positions occupied by the ACP suggest strongly that our EM map shows a conformation of the complex that is capable of carrying out fatty acid synthesis, which requires the ACP to visit different sites in the chain elongation cycle. This does not appear to be the case in the crystal structures of yeast FAS where the ACP domain is seen stalled in a single location. The EM map shows that it can indeed move and attach to different catalytic centers in the unconstrained, solution conformation.
Substrate-Shuttling Mechanism.
The six reaction chambers in fungal FAS are thought to operate independently (10). Our observation of up to six alternative ACP sites combined with uniform variance in the reaction chambers suggests that the docking events are stochastic in the inhibited enzyme. This, along with the assumed independence of the reaction chambers, implies that several thousand different states of the FAS exist simultaneously in solution, not counting possible intermediates. If only a few different states coexist, as in the translating ribosome (24) or in folding intermediates in the GroEL/ES system (25), these states can be separated into individual class averages by single-particle image processing, but this is clearly not feasible in the case of fungal FAS. Instead, we visualized the mobility of the ACP domain by a variance map, which shows the highest degree of structural variability in the map regions corresponding to the reaction chamber. This indicates that the ACP domain is indeed mobile within the FAS barrel, enabling it to visit successive catalytic sites (Fig. 4).
The relative occupancies of the ACP sites we observe show that no single catalytic domain has a dominant affinity for the ACP domain. For substrate delivery and chain elongation, the ACP would have to spend different amounts of time attached to each catalytic site, depending on the rate of the individual reaction. This may be reflected in the different occupancies of ACP sites in the EM map, which in turn is likely to be governed by the extent of charge and surface complementarity between the ACP and catalytic domains (13).
Linker Peptide.
Two elongated ∼64-Å regions of density stretch across the upper half of the reaction chamber directly between the MPT and the ACP density seen near the ER domain in the reaction chambers (Fig. 3D), roughly parallel to one another and to the equator. Although their density is higher than that for any of the ACP domains, these features are not traced in any of the four x-ray structures of fungal FAS. Their similar appearance and parallel arrangement, together with their apparent partial occupancy, suggest that they might represent two alternative positions of the same feature.
Examination of the yeast FAS sequence indicates only one stretch of unresolved polypeptide in this region that is long enough to account for a feature of this size. This is the 44-amino acid stretch thought to form the unstructured linker peptide L1 (Fig. 1) that tethers the ACP domain to the α-chain segment of the MPT domain (12). Secondary structure prediction (26) shows that most of the sequence is likely to take on a random coil conformation, while a stretch of 12 amino acids at the C terminus and a stretch of 5 amino acids at the N terminus has some propensity for helix formation (Fig. S8B). Although the linker peptide may thus not tend to form an ordered structure when free in solution, it may be able to do so, if only partially, in the confined space of the reaction chamber. A more remote alternative explanation of these striking densities would be other, so far uncharacterized, molecules, possibly related to fatty acid chain binding or transport.
Conclusion
Our 6-Å EM map of yeast FAS presents a virtually complete structure of this 2.6-MDa cellular machine. The map shows all domains in the α-polypeptide chain, including the PPT and KS domains that were previously not or incompletely esolved. The arrangement of catalytic and structural domains of the β-chain forming the barrel wall is significantly different in the unconstrained solution structure observed in our EM map, in which the reaction chamber is both wider and lower than in the crystal structures (12–14). The more constrained conformation of the β-dome in the x-ray structures appears to be due to a crystal contact, which pushes the MPT domain inward, resulting in a change in the position and orientation of all other domains in the β-chain, which move relative to one another as rigid bodies. The shape of the reaction chamber appears to affect the ability of the ACP domain to dock to the catalytic sites, as it is found in only one position in the yeast FAS x-ray structures, whereas in the EM map it is seen in up to six different positions with varying occupancies.
The alternative positions of the ACP domain next to the various catalytic sites in the reaction chamber provide immediate insight into the mechanism of substrate delivery and fatty acid chain elongation. Even though the enzyme complex investigated by us is inhibited and thus unable to synthesize fatty acids, the different occupancies of the ACP domain next to the catalytic centers indicate different binding affinities that are likely related to reaction rates. Moreover, we see a pair of intriguing elongated densities in the reaction chamber that might correspond to a structured linker peptide.
Materials and Methods
Electron Cryomicroscopy.
Yeast (S. cerevisiae) FAS synchronized by addition of NADPH, malonyl-CoA and inhibited by cerulenin was isolated and prepared as described (14). Samples were applied to glow-discharged Quantifoil grids (Quantifoil MicroTools) and vitrified by injection into liquid ethane using a Vitrobot plunge-freezing device (FEI). A first set of ∼15,000 particles was collected on a Tecnai G2 Polara electron microscope operating at 300 kV at a defocus range of 5–7 μm and an electron dose of 10–12 e-/Å2. Images were recorded at a magnification of 39,000× on Kodak SO-163 film and scanned on a Zeiss Photoscan scanner (Intergraph) at a step size of 7 μm on an 8-bit scale. Adjacent pixels were averaged to yield a step size of 3.59 Å at the specimen. For an initial map, roughly 9,000 particles of this first dataset were classified and averaged and an ab initio model generated by the angular reconstitution method in IMAGIC (27). A second set of ∼25,000 particles was collected with the same instrument at 200 kV at a defocus range of 1–3 μm and an electron dose of 20–30 e-/Å2. Images were recorded at a nominal magnification of 59,000× on Kodak SO-163 film and scanned as above on a 12-bit scale with a step size of 1.19 Å at the specimen.
X-Ray Solution Scattering.
X-ray solution scattering experiments were conducted at the Swiss Light Source (SLS) beamline X12SA. FAS samples at concentrations of 10 mg/ml and 2 mg/ml were measured at two detector lengths. The data were background-subtracted using buffer blanks and corrected using established procedures. Data from both length scales were combined into a single scattering profile covering the range from 300 Å to 4 Å resolution.
Data Processing.
Particles were selected using the semiautomatic mode of the BOXER module from EMAN (18). An initial 3D reconstruction from the first dataset using ∼9,000 particles was performed by angular reconstitution, initially with C1 symmetry, using IMAGIC V (27). The model displayed clear D3 symmetry, which was applied for all further steps. The resolution of the initial model was determined by Fourier shell correlation to be 20 Å by the FSC = 0.5 criterion (19).
The phase reversals in the images caused by the CTF were corrected by phase flipping and Wiener filtering (18). The amplitudes were corrected using the low-angle x-ray solution scattering data. A refined 3D reconstruction was obtained with EMAN from ∼19,000 particles (18). This subset of particles, obtained from the original dataset of ∼25,000, was selected using a variance-based similarity measure applied by the dfilt option in EMAN. D3 symmetry was applied throughout. The resolution of the final map was determined by Fourier shell correlation as 7.2 Å by the FSC = 0.5 criterion (19) and as 5.9 Å by the FSC = 0.143 criterion (20).
Estimate of 3D Variance.
A 3D variance map was generated in EMAN (18) from 100 volumes generated from the high-resolution dataset using a random set of ∼18,000 particles. The resultant 3D variance map was divided by the average structure to remove any variance arising from structural alignment errors.
Map Visualization and Analysis.
Molecular graphics images were produced with UCSF Chimera (21).
Fitting of x-ray structures to the EM map was done in Chimera. The yeast FAS structure (2VKZ) (14) and the PPT domain (2WAS) (22) were fitted as rigid bodies, resulting in an excellent fit for the equatorial wheel, but clear displacements of the dome domains. For fitting of the dome, the structure was divided in individually folding domains [the enzymatic domains MPT (β-chain 1,662–2,051 + α chain 1-94), DH (β-chain 1,255–1,421 + 1,524–1,661), ER (β 583–855, AT: β 155–547), and all structural domains of the β-chain linking these enzymatic domains]. Rigid body fits to the EM map were performed for each of these domains individually. This resulted in a clear improvement over the initial overall fit. The atomic model of ACP was fitted into the corresponding EM densities near the KS, KR, ER, and AT domains by rigid body fits; all were highly similar in shape and gross features, though densities near the KR, ER, and AT domains exhibited higher variability. The fit of the ACP near the KS domain was performed automatically. Automated fitting into ACP densities near the other three catalytic domains was possible only for gross features, and finer adjustments were applied manually. To compare the occupancies of different catalytic sites of the ACP domain as seen in the EM map, a sum of strong ACP densities observed near the KS, KR, AT, and ER domains was calculated. A relative occupancy for each site was then calculated by dividing each catalytic site’s total ACP density by the sum of all ACP densities.
For the electrostatic calculations pdb2pqr (28) was used to generate pqr-format files, which were then used in APBS calculations performed using the VMD software package (29). Secondary structure predictions were performed with the PHYRE server (http://www.sbg.bio.ic.ac.uk/phyre) (26).
Supplementary Material
Acknowledgments.
We thank Dieter Oesterhelt who initiated the project. We thank Ronnald Vollrath for purifying the FAS sample, Andreas Menzel at the SLS for help with SAXS data collection, Reinhard Maass and the GWDG for providing computer support, and Patrik Johansson, Rasmus Schroeder, Steven Ludtke, and James Evans and other members of the Stahlberg lab for scientific discussions. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH Grant P41 RR-01081).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.J. is a guest editor invited by the Editorial Board.
Data deposition: The EM map has been deposited in the EM data bank (www.emdatabank.org) under the entry code EMD-1623.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913547107/DCSupplemental.
References
- 1.White SW, Zheng J, Zhang Y-M, Rock CO. The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 2.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Ann Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 3.Smith S, Witkowski A, Joshi AK. Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res. 2003;42(4):298–317. doi: 10.1016/s0163-7827(02)00067-x. [DOI] [PubMed] [Google Scholar]
- 4.Brink J, et al. Quaternary structure of human fatty acid synthase by electron cryomicroscopy. Proc Natl Acad Sci USA. 2002;99(1):138–143. doi: 10.1073/pnas.012589499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asturias FJ, et al. Structure and molecular organization of mammalian fatty acid synthase. Nat Struct Mol Biol. 2005;12(3):225–232. doi: 10.1038/nsmb899. [DOI] [PubMed] [Google Scholar]
- 6.Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 7.Brignole EJ, Smith S, Asturias FJ. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat Struct Mol Biol. 2009;16(2):190–197. doi: 10.1038/nsmb.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoppe W, Schramm HJ, Sturm M, Hunsmann N, Gaßmann J. Three-dimensional electron microscopy of individual biological objects Part III. Experimental results on yeast fatty acid synthetase. Z Naturforsch. 1976;31a:1380–1390. [Google Scholar]
- 9.Stoops JK, Kolodziej SJ, Schroeter JP, Bretaudierre J-P, Wakil SJ. Structure-function relationships of the yeast fatty acid synthase: Negative-stain, cryo-electron microscopy, and image analysis studies of the end views of the structure. Proc Natl Acad Sci USA. 1992;89:6585–6589. doi: 10.1073/pnas.89.14.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolodziej SJ, Penczek PA, Schroeter JP, Stoops JK. Structure-function relationships of the Saccharomyces cerevisiae fatty acid synthase. J Biol Chem. 1996;271(45):28422–28429. doi: 10.1074/jbc.271.45.28422. [DOI] [PubMed] [Google Scholar]
- 11.Jenni S, et al. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science. 2007;316:254–261. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 12.Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 13.Lomakin IB, Xiong Y, Steitz TA. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell. 2007;129:319–332. doi: 10.1016/j.cell.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Johansson P, et al. Inhibition of the fungal fatty acid synthase type I multienzyme complex. Proc Natl Acad Sci USA. 2008;105(35):12803–12808. doi: 10.1073/pnas.0805827105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh N, Wakil SJ, Stoops JK. Yeast fatty acid synthase: Structure to function relationship. Biochemistry. 1985;24:6598–6602. doi: 10.1021/bi00344a044. [DOI] [PubMed] [Google Scholar]
- 16.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 17.Byers DM, Gong H. Acyl carrier protein: Structure-function relationships in a conserved multifunctional protein family. Biochem Cell Biol. 2007;85:649–662. doi: 10.1139/o07-109. [DOI] [PubMed] [Google Scholar]
- 18.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 19.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 20.Rosenthal P, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Pettersen EF, et al. UCSF Chimera: A visualisation system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Johansson P, et al. Multimeric options for the auto-activation of the fungal FAS type I megasynthase. Structure. 2009;17(8):1063–1074. doi: 10.1016/j.str.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Penczek PA, Yang C, Frank J, Spahn CMT. Estimation of variance in single-particle reconstruction using the bootstrap technique. J Struct Biol. 2006;154(1):168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Penczek PA, Frank J, Spahn CMT. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J Struct Biol. 2006;154(1):184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Kanno R, Koike-Takeshita A, Yokoyama K, Taguchi H, Mitsuoka K. Cryo-EM structure of the native GroEL-GroES complex from Thermus thermophilus encapsulating substrate inside the cavity. Structure. 2009;17:287–293. doi: 10.1016/j.str.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 27.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 28.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.