Abstract
Sirtuin 1 (SIRT1) is a class III histone deacetylase that deacetylates histone and nonhistone proteins to regulate gene transcription and protein function. Because SIRT1 regulates very diverse responses such as apoptosis, insulin sensitivity, autophagy, differentiation, and stem cell pluripotency, it has been a challenge to reconcile how it orchestrates such pleiotropic effects. Here we show that SIRT1 serves as an important regulator of Wnt signaling. We demonstrate that SIRT1 loss of function leads to a significant decrease in the levels of all three Dishevelled (Dvl) proteins. Furthermore, we demonstrate that SIRT1 and Dvl proteins complex in vivo and that inhibition of SIRT1 leads to changes in gene expression of Wnt target genes. Finally, we demonstrate that Wnt-stimulated cell migration is inhibited by a SIRT1 inhibitor. Because the three mammalian Dvl proteins serve as key messengers for as many as 19 Wnt ligands, SIRT1-mediated regulation of Dvl proteins may explain the diverse physiological responses observed in different cellular contexts. Previously, SIRT1 had only been shown to mediate the epigenetic silencing of Wnt antagonists. In contrast, here we report that SIRT1 regulates Dvl protein levels and Wnt signaling in several cellular contexts. These findings demonstrate that SIRT1 is a regulator of transient and constitutive Wnt signaling.
Keywords: breast cancer, colon cancer, cambinol, β-catenin, sirtuin 2
Sirtuin 1 (SIRT1) is an NAD+-dependent histone deacetylase (HDAC) that regulates a very broad and complex array of physiological processes. As such, it has been the source of some controversy, as it has been difficult to reconcile the role it plays in the coordination of cellular responses and gene expression in both normal and pathophysiological settings. For example, SIRT1 has been shown to inhibit the maturation of preadipocytes (1, 2), antagonize p53-dependent apoptosis in response to stress (3, 4), and promote chemoresistance to conventional chemotherapeutic agents (5, 6), and is associated with microsatellite instability and CpG island methylator phenotype in human colorectal cancer (7). Furthermore, reports demonstrate that SIRT1 coordinates diverse metabolic responses to changes in nutrient availability (8), regulates autophagy (9), and controls key stages of spermatogenesis and germ stem cell proliferation and function (10, 11). Given the complex influence of SIRT1 on cell-fate decisions in multiple physiological settings, it is reasonable to anticipate that SIRT1 regulates one or more signaling networks recognized for their influence on these diverse cellular and organismal responses.
It is well-established that Wnt signaling, likewise, regulates diverse processes such as adipogenesis (12), tumorigenesis (13), and stem cell pluripotency (14, 15). When Wnt ligands are present, they transmit signals through specific Frizzled (Fzd) or Fzd/LRP5/6 coreceptor complexes (16). This signal is then propagated via Dishevelled (Dvl) proteins that will direct canonical (β-catenin-dependent) or noncanonical (β-catenin-independent) signaling (17). Most of the mechanistic insights into Wnt signaling have come from extensive studies detailing how β-catenin is regulated, yet precisely how Dvl is regulated is poorly understood despite its crucial function in bridging cytoplasmic Wnt signals with nuclear responses. Early observations demonstrated that Dsh-1 is negatively regulated by NEDL1 (18) and the Naked cuticle-PR72 complex (19), and Dsh-2 and Dsh-3 were nicely shown to be negatively regulated by the KLHL12-Cullin-3 complex (20). Recently, reports have revealed an intriguing requirement of nuclear entry of Dvl for canonical signaling (21, 22). However, why the nuclear entry of Dvl is required for canonical signaling is not known. Here we show that SIRT1 positively regulates the protein levels of all three Dvl family members. We demonstrate that SIRT1 and Dvl proteins complex in vivo and that inhibition of SIRT1 leads to changes in gene expression of classic Wnt/Dvl target genes. Moreover, we show that SIRT1 regulates Wnt3a-induced cell migration. Finally, we propose that SIRT1-mediated regulation of Dvl proteins may help explain the diverse SIRT1-dependent physiological responses observed in different cellular contexts.
Results
The first clue that SIRT1 regulates Wnt signaling came from the observation that SIRT1 localizes to the promoter of the Wnt antagonist secreted frizzled related protein 1 (SFRP1) and directly contributes to its aberrant epigenetic silencing (23). Inhibition of SIRT1 was shown to cause a reexpression of SFRP1. SIRT1 was originally shown to mediate epigenetic silencing of SFRP1 and SFRP2 (23), and later was extended to DKK1 (24), another Wnt antagonist. However, whether this connection of SIRT1 with Wnt signaling extended beyond the repression of Wnt antagonists was unknown. To address this, we systematically analyzed the impact of loss of SIRT1 function on several key mediators of Wnt signaling. First, we determined the expression pattern of Wnt proteins in colorectal cancer, breast cancer, and HEK293 cell lines to assess the extent of autocrine Wnt signaling. We performed reverse transcription (RT) PCR analysis and observed a variable pattern of expression in Wnt ligands (Fig. S2A) indicating potentially different states of constitutive autocrine signaling. We found that the four colon cancer (HT-29, HCT116, RKO, and DLD-1), two breast cancer (T-47D and MDA-MB-231), and HEK293 cell lines each expressed multiple but different Wnt ligands.
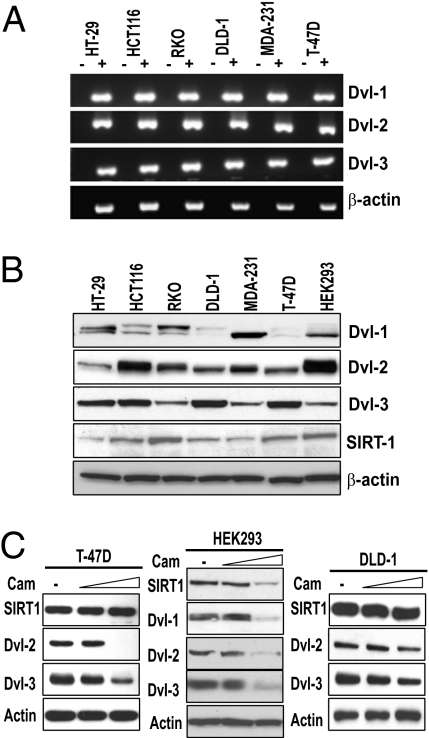
Next, we determined the pattern of expression of all three Dvl proteins. We found that, in contrast to the pattern of mRNA expression of Wnts, Dvl mRNA levels did not vary considerably among the six lines (Fig. 1A). Interestingly, however, we observed a more varied pattern of Dvl protein expression (Fig. 1B). Whereas Dvl-1 protein varied more among the cell lines, Dvl-2 and Dvl-3 expression was consistently higher. Because of the pattern of Wnt ligand expression, and because Dvl proteins translocate to the nucleus and serve as key effectors of extracellular signals (21, 22, 25), we focused on their regulation with respect to SIRT1 loss of function. Of the Dvl family members, we were particularly interested in Dvl-3 because it has been reported to be overexpressed in 36% (33/91) of colon tumors and displayed a marked increase in its nuclear accumulation (25). Additionally, Dvl-3 was shown to be overexpressed in 30% of invasive ductal breast carcinomas (26) and Dvl-2 was shown to contribute to pancreatic cancer malignancy (27). We continued analysis of SIRT1 regulation in cell lines that display constitutive Wnt signaling yet show different patterns of Wnt ligand autocrine stimulation.
Fig. 1.
Dishevelled expression is regulated posttranslationally. (A) Total RNA was isolated from the indicated cell lines and RT-PCR analysis of the three Dishevelled (Dvl) genes was performed using intron-spanning primers. For each amplification, either the reverse transcriptase was excluded (−) or included (+) in the reverse-transcription reaction and β-actin was included as a control. (B) The protein expression patterns of all three Dvl family members, SIRT1, and β-actin were analyzed by Western blotting. (C) T-47D, HEK293, and DLD-1 cells were treated with vehicle control (dimethyl sulfoxide; DMSO) or 50 μM or 100 μM cambinol (SIRT1 inhibitor) for 18 h (T-47D) or 24 h (HEK293 and DLD-1) and the indicated proteins were analyzed by Western blot.
As the first loss-of-function approach, we treated multiple cell lines with a range of doses of cambinol, an inhibitor of SIRT1 and SIRT2 (28), and continued analysis with two doses of cambinol. Strikingly, we observed a robust dose-dependent reduction in the levels of Dvl-2 and Dvl-3 in T-47D cells upon treatment with cambinol for 18 h (Fig. 1C). The levels of Dvl-1 were too low to assess changes in T-47D cells (Fig. 1B). We tested the role of SIRT1 in the regulation of Dvl proteins in HEK293 cells and found that inhibition by cambinol caused a reduction in all three Dvl proteins (Fig. 1C). Furthermore, we observed that cambinol-mediated SIRT1 inhibition in DLD-1 colon cancer cells treated with the same doses caused a reduction in Dvl proteins (Fig. 1C). Additionally, we observed that pharmacologic inhibition caused decreases in all three Dvl proteins in RKO cells (Fig. S1A), a reduction in Dvl-3 in MCF-7 cells (Fig. S1B), and a reduction in Dvl-1 and Dvl-3 in HCT116 cells (Fig. S1C). Only Dvl-3 appeared to be expressed at detectable levels in MCF-7 cells. Together, these data demonstrate that SIRT1 inhibition leads to robust decreases in Dvl proteins in several cellular contexts.
Because cambinol inhibits SIRT1 and SIRT2 (28), we specifically inhibited SIRT1 via shRNA to further verify the impact of SIRT1 inhibition on Dvl regulation. Consistent with the pharmacological inhibition, we found that the shRNA that caused a knockdown of SIRT1 in T-47D cells led to a reduction in Dvl-2 and Dvl-3 protein (Fig. 2A). This trend was also observed in HEK293 cells, where the shRNA constructs that caused a reduction in SIRT1 also caused a reduction in Dvl-3 proteins (Fig. 2B). Finally, this trend was also observed in DLD-1 cells, as inhibition of SIRT1 by siRNA led to a significant reduction in Dvl-3 and a small reduction in Dvl-2 (Fig. 2C). Together, these data suggest that SIRT1 is a positive regulator of Dvl proteins. Because cambinol inhibits both SIRT1 and SIRT2, we also performed siRNA-mediated knockdown of SIRT1 and SIRT2 individually and together. We found that the individual knockdown of SIRT1 and SIRT2 caused a decreased in Dvl-2 and Dvl-3 and the combined knockdown caused a reduction in Dvl-2 and Dvl-3 (Fig. 2 D–F). A recent study demonstrated that the three Dvl proteins heterotrimerize in HEK293 cells (29), and thus the stability of one Dvl family member may influence the stability of another.
Fig. 2.
SIRT1 inhibition leads to a significant reduction in Dvl protein expression. (A) T-47D cells were transfected for 48 h with either control vector (pRetroSuper) or with plasmids encoding two shRNAs specific for SIRT1 (589 or 1091). Expression of Dvl-2 and Dvl-3 was assessed by Western blot. (B) Control or SIRT1 shRNAs were transfected into HEK293 cells and Dvl-3 was analyzed by Western blot. The 1091 shRNA is effective at reducing SIRT1 protein levels and causes a consistent reduction in Dvl-3 protein levels. (C) Control or SIRT1 siRNAs were transfected into DLD-1 cells for 24 h and Dvl-2 and Dvl-3 were analyzed by Western blot. (D–F) T47-D cells were transfected with (D) control siRNA and SIRT1 siRNA, (E) SIRT2 siRNA, or (F) both SIRT1 and SIRT2 siRNA at 100 nM for 72 h to measure Dvl-2, Dvl-3, SIRT1, and/or SIRT2 expression via Western blot. (G) T-47D cells were pretreated with 5 μM lactacystin (Lacta) (left) or 0.5 μM MG132 (right) for 15 min followed by a 22-h treatment with or without 100 μM cambinol. DVL2, DVL3, and actin levels were measured. Control blots for a polyubiquitinated protein (p27) and a loading control (β-actin) were performed on the same membrane.
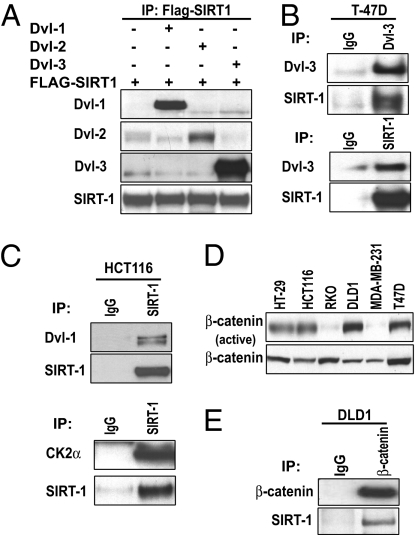
Nuclear localization of Dvl is important for Wnt signaling (21), and because SIRT1 loss of function leads to decreased Dvl protein levels, we next asked whether SIRT1 inhibition leads to changes in Dvl mRNA levels. We did not observe any changes in Dvl mRNA upon SIRT1 inhibition in either T-47D, DLD-1, or HCT116 cells (Fig. S1E) at the same doses that caused a reduction in protein. To further address the mechanism, we pretreated T-47D cells with proteasome inhibitors MG132 or lactacystin followed by treatment with or without cambinol for 22 h (Fig. 2G). The level of both Dvl-2 and 3 decreased with cambinol alone yet MG132 or lactacystin inhibition did not restore Dvl protein levels. This suggests that SIRT1 might be imposing a translational control on Dvl and hence maintains pro-tein levels high in cancer cells. Consistent with this finding, a report by Shimazu et al. demonstrated that SIRT1 and SIRT2 deacetylate 3′-end mRNA-processing machinery and that the acetylation state regulates the nuclear localization of the poly(A) polymerase (30). Next, we asked whether SIRT1 complexes with Dvl proteins. HEK293 cells were transiently cotransfected with expression plasmids encoding SIRT1-Flag and Dvl-1, Dvl-2, or Dvl-3. Interestingly, we found that SIRT1 bound to all three Dvl family members (Fig. 3A). The greatest association appeared to occur between SIRT1 and Dvl-3. We also extended this analysis to determine whether endogenous SIRT1 and Dvl associate. We observed that endogenous SIRT1 and Dvl-3 associate in T-47D cells (Fig. 3B). We attempted coimmunoprecipitation with SIRT1 and Dvl-2, but were limited in reagent availability. We next wanted to determine whether endogenous SIRT1 and Dvl-1 formed a complex. We found that endogenous SIRT1 and Dvl-1 associate in HCT116 cells (Fig. 3C Upper) and in RKO cells (Fig. S2C). Taken together, these data demonstrate that SIRT1 forms a complex with Dvl proteins.
Fig. 3.
SIRT1 and Dvl form a complex in vivo. (A) HEK293 cells were transiently transfected with SIRT1-Flag and either an empty vector, Dvl-1, Dvl-2, or Dvl-3. SIRT1-Flag was immunoprecipitated and Western blots for each Dvl family member or SIRT1 was performed. (B) (Upper) Interaction between endogenous SIRT1 and Dvl-3 in T-47D cells was performed by immunoprecipitation of Dvl-3 followed by blots for Dvl-3 and SIRT1. (Lower) SIRT1 was immunoprecipitated followed by blots for Dvl-3 and SIRT1. (C) (Upper) HCT116 cells were harvested, endogenous SIRT1 was immunoprecipitated, and Dvl-1 and SIRT1 were analyzed by Western blot. (Lower) SIRT1 was immunoprecipitated followed by blots for CK2α and SIRT1. (D) The protein expression patterns of active and total β-catenin were analyzed by Western blotting in the indicated lines. (E) Either control IgG or β-catenin was immunoprecipitated followed by blots for β-catenin and SIRT1.
Casein kinase I and II family members regulate cell-fate decisions (31, 32) and control recruitment of Wnt regulators to target genes (33). Recent reports demonstrate that SIRT1 is phosphorylated by casein kinase II (CK2) (34, 35), and this regulates substrate binding and deacetylase activity. Consistent with these reports, we observed that endogenous SIRT1 complexes with CK2α (Fig. 3C Lower). Because CK family members phosphorylate Dvl, β-catenin, and SIRT1, we next began to explore the impact of SIRT1 inhibition on the canonical branch of Wnt signaling. First, we determined the relative steady-state levels of unphosphorylated (active) β-catenin in our panel of cell lines. We found that whereas total β-catenin levels were appreciable in every line, unphosphorylated (active) β-catenin was higher in DLD1, T-47D, and HT-29 cells (Fig. 3D). HCT116 cells express mutant β-catenin, so we chose to focus on the lines with wild-type β-catenin to determine whether endogenous SIRT1 associates with β-catenin. We performed coimmunoprecipitations in DLD-1 cells and found association between endogenous SIRT1 and β-catenin (Fig. 3E). Together, these data demonstrate that SIRT1 physically associates with multiple regulators of Wnt signaling which have previously been shown to complex.
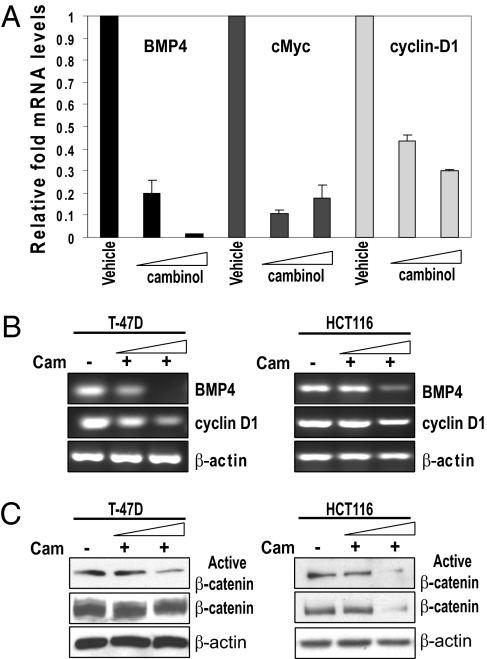
Because Dvl proteins had been shown to heterotrimerize (29) and we observed that SIRT1 binds to each one, we reasoned that SIRT1 would also regulate the expression of Wnt/Dvl target genes. To address this, we treated T-47D and HCT116 cells with the same doses of cambinol described earlier that caused a decrease in Dvl and performed semiquantitative RT-PCR analysis (Fig. 4B and Fig. S3B). We also performed quantitative real-time RT-PCR analysis in HCT116 (Fig. 4A) and MDA-MB-231 cells (Fig. S3A). We found that inhibition of SIRT1 caused a significant reduction in the mRNA levels of three Wnt/Dvl target genes, BMP4 (36), cMyc (25), and cyclin D1 (37, 38), in T-47D, HCT116, and MDA-MB-231 cells (Fig. 4 A and B and Fig. S3). Recent reports demonstrated that BMP4 is one of four previously unreported genes that predict colorectal cancer risk (39). Additionally, BMP4 overexpression has been observed in colonic adenocarcinomas, and it has been shown to promote migration and invasion of HCT116 cells (40). Likewise, alterations in cMyc (41) and cyclin D1 (42) expression have been linked with cancer. Together, these data suggest that SIRT1 not only regulates Dvl protein levels but also results in changes in expression of Wnt/Dvl target genes.
Fig. 4.
SIRT1 regulates Wnt target gene expression. (A) HCT116 cells were treated with either vehicle control (DMSO) or 50 μM or 100 μM cambinol (SIRT1 inhibitor) for 24 h, and total RNA was isolated and quantitative RT-PCR analysis of Wnt target genes (BMP4, cMyc, and cyclin D1) was performed using intron-spanning primers. (B) T-47D or HCT116 cells were treated with either vehicle control (DMSO) or 50 μM or 100 μM cambinol (SIRT1 inhibitor) for 18 h, and total RNA was isolated and semiquantitative RT-PCR analysis of Wnt target genes (BMP4 and cyclin D1) and β-actin was performed using intron-spanning primers. (C) T-47D and HCT116 cells were treated with either vehicle control (DMSO) or 50 μM or 100 μM cambinol and Western blots for unphosphorylated (active) β-catenin, total β-catenin, and β-actin were performed.
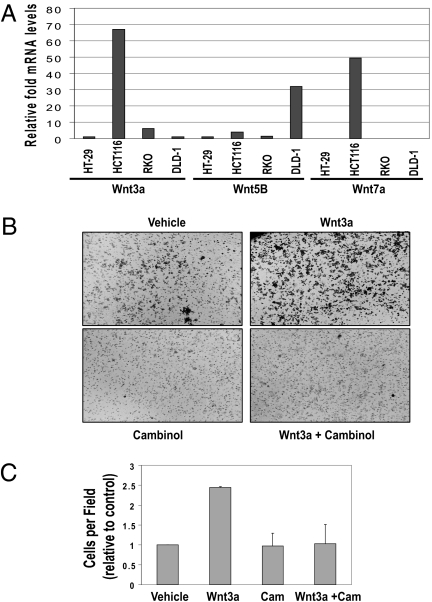
Dvl proteins are critical for both β-catenin-dependent and -independent signaling. Next, we wanted to determine whether SIRT1 inhibition that caused Dvl loss of function also decreased unphosphorylated active β-catenin. We observed that SIRT1 inhibition caused a reduction in the unphosphorylated form of β-catenin and a decrease in total β-catenin levels in both HCT116 and T-47D cells. This trend was also seen in T-47D cells, but there was a smaller reduction in total β-catenin levels (Fig. 4C). To further assess the influence that SIRT1/2 exerts on Wnt signaling, we first determined the relative levels of Wnt ligands (e.g., Wnt3a, Wnt5b, and Wnt7a) known to stimulate different branches of signaling in our candidate cell lines. Wnt3a has been shown to stimulate cell migration and stimulate both canonical and noncanonical branches of Wnt signaling (43). We found that Wnt3a and Wnt7a showed the greatest level of expression in HCT116 cells relative to the other lines, with Wnt3a being expressed the most (Fig. 5A). Because Wnt-mediated signaling is important for cell migration and SIRT1 has been shown to regulate cell migration (44), we examined the influence of SIRT1 or SIRT2 loss of function in regulating Wnt3a-stimulated cell migration. Using transwell assays, HCT116 cells were allowed to migrate for 8 h in response to either a vehicle control (Fig. 5B Upper Left) or purified Wnt3a ligand (Fig. 5B Upper Right). We observed that 8-h Wnt3a stimulation increased migration of HCT116 cells by about 2.5-fold, and cambinol inhibited Wnt3a-induced migration of HCT116 (Fig. 5 B and C). Previous reports demonstrated that whereas cambinol sensitizes cells to chemotherapeutic agents, treatment of HCT116 cells alone (50 μM cambinol) had no effect on cell viability (28) in the absence of a chemotherapeutic agent. We similarly observed no change in HCT116 cell viability under the conditions of migration that could account for cambinol inhibition of Wnt3a-induced mi-gration (Fig. 5C). Additionally, we observed that Wnt3a stimulation over a period of 24 h increased migration of HT-29 cells by about 4-fold. We observed that cambinol inhibited Wnt3a-induced migration of T-47D (Fig. S4 A and B) and HT-29 cells (Fig. S5 A and B). Together, these data suggest that sirtuin proteins are important regulators of Wnt3a-stimulated cell migration.
Fig. 5.
SIRT1 regulates transient Wnt signaling. (A) Total RNA was isolated from four colon cancer lines (HT-29, HCT116, RKO, and DLD-1) and the relative levels of three candidate Wnt genes (Wnt3a, Wnt5b, and Wnt7a) were analyzed by quantitative RT-PCR analysis. (B) HCT116 cells were analyzed in a transwell migration assay in the absence or presence of purified Wnt3a ligand (260 ng/mL) ± cambinol (130 μM). (C) Changes in migration were measured after 8 h and cells were manually counted in a (680 × 512) still image field with an Olympus DP 70 microscope. Three assays were counted with duplicates per condition.
Discussion
In this study, we describe an important link between the SIRT1 and Dvl proteins. We have demonstrated that SIRT1 not only regulates the levels of Dvl proteins but forms a complex with them in vivo. We have shown that SIRT1 loss of function leads to a precipitous drop in Dvl proteins in five different cell lines representing different tissues. Coincident with this reduction in Dvl protein levels, we have demonstrated a change in the activation of Wnt/Dvl target genes. Finally, we have demonstrated that SIRT1 promotes constitutive Wnt signaling and Wnt-induced cell migration. These findings provide insight into previous reports and potentially explain some of the paradoxes of the diverse physiological processes orchestrated by SIRT1. Because of the increased appreciation of the complexity of Wnt signaling, such as the active nuclear export of factors such as TCF-1 (45) and nuclear entry of proteins such as Dvl (21, 22), some of the historical views of “cytoplasmic” versus “nuclear” Wnt signaling are being modified. Moreover, SIRT1 shuttles between the cytoplasm and nucleus (46, 47), but the physiological relevance of this trafficking is unclear.
SIRT1 inhibits the maturation of preadipocytes by inhibition of PPAR-γ (2). Whereas Wnt/β-catenin signaling inhibits adipocyte maturation and fat storage, PPAR-γ has the opposite effect (48). Here we demonstrate that SIRT1 promotes Wnt/Dvl signaling, which may provide a mechanistic framework linking SIRT1, Dvl, and PPAR-γ in specific cellular contexts. Also, controversy has surrounded the role of SIRT1 during tumorigenesis (4, 28, 49, 50). Whereas the majority of reports have demonstrated that Wnt signaling contributes to tumorigenesis, especially in breast and colorectal cancer, a recent report has shown the opposite. This report describes a case where activated Wnt/β-catenin signaling in melanoma is associated with decreased proliferation in patient tumors (51), in contrast to another study demonstrating that Wnt antibody-based therapies induce apoptosis and inhibit tumor growth (52). Moreover, Firestein et al. (50) report that SIRT1 suppresses the ability of β-catenin to drive transcription and proliferation. Although our study focused primarily on Dvl regulation, because SIRT1 inhibition reduces all three Dvl proteins in a cell-type-dependent manner, both β-catenin-dependent and -independent Wnt signaling will be affected. One difference in our studies that assessed mechanism involves the use of different experimental approaches and a different interpretation of the role that β-catenin-independent Wnt signaling may play in tumorigenesis. In our studies, loss of SIRT1 function was achieved by pharmacologic, shRNA, and siRNA inhibition of SIRT1. Firestein et al. overexpressed wild-type and a catalytically dead mutant of SIRT1 (H363Y) and observed that WT SIRT1 caused a reduction in proliferation relative to empty vector in HCT116 and DLD-1 but not RKO cells. First, SIRT1 overexpression in HCT116 cells has been reported to activate autophagy under starved conditions (9), and the extent to which autophagy was activated with SIRT1 overexpression in the Firestein et al. study may de-termine the extent of proliferation. Second, SIRT1 has been shown to deacetylate cortactin and promote cell migration, and SIRT1-null mouse embryonic fibroblasts (MEFs) were shown to be less motile that SIRT1 WT MEFs (44). Zhang et al. (44) also demonstrated that high SIRT1 levels in human breast tumor samples correlated with deacetylated cortactin. One interesting possibility is that the level of SIRT1 expression and the presence of specific Wnt ligands may act as a determinant of whether Dvl will “choose” to activate canonical versus noncanonical signaling. Furthermore, with an overexpression of SIRT1, a decrease in proliferation may be expected if cells are to switch to a program of gene expression that promotes autophagy or migration. Canonical β-catenin-dependent signaling is classically linked with proliferation, whereas noncanonical signaling is linked with cell migration; however, Dvl regulation is important for both branches. Third, because RKO cells have significantly decreased active β-catenin levels relative to HCT116 and DLD-1 cells, SIRT1 overexpression may have less of an impact on switching Wnt signaling from canonical to noncanonical and explain why SIRT1 overexpression did not alter proliferation in RKO cells.
Part of the controversy surrounding SIRT1 in tumorigenesis comes from studies involving different mouse models on different backgrounds and the possibility that cytosolic versus nuclear SIRT1 may play different roles. A recent study reported that cytoplasm-localized SIRT1 was associated with apoptosis and led to increased sensitivity to apoptosis (47) and, using a tissue microarray, Firestein et al. (50) reported that 58% (47/81) of human colon cancers analyzed were positive for SIRT1 in the nucleus. The implication of this is unclear, so many questions remain to be addressed. A recent report analyzing patterns of SIRT1 expression in human colorectal cancer helped to clarify matters (7). Ogino and colleagues analyzed a large number of colorectal tumors (n = 485) from two independent prospective cohort studies and detected SIRT1 overexpression in 37% of the tumors by immunohistochemistry. The large number of cases provided sufficient power for multivariate analysis and it was shown that SIRT1 overexpression was significantly associated with tumors displaying CpG island methylator phenotype and microsatellite instability. SIRT1 positivity was also significantly associated with tumors with a high grade and a mucinous component. Because deregulated Wnt signaling is known to profoundly contribute to colorectal tumorigenesis, it is important to understand how SIRT1 impacts the flow of information from extracellular factors such as Wnt antagonists to intracellular factors such as Dvl proteins that then enter the nucleus. Dvl proteins are key modulators of the type and magnitude of the Wnt signal that is relayed. In addition to colorectal cancer, Dvl proteins have been linked with other cancers. Dvl-3 was shown to be overexpressed in 30% of invasive ductal breast carcinomas (26), in 75% of fresh microdissected non-small-cell lung carcinoma (53), and in primary mesotheliomas (54). Moreover, increased Dvl-1 was observed in 73% of primary cervical squamous cell cancers in comparison with noncancerous uterine squamous cell tissues, and Dvl-1 was shown to be amplified in 54% and up-regulated in 46% of primary breast cancers examined relative to noncancerous breast tissue.
Wnt signals are relayed via the three Dvl proteins, and their pattern of expression influences the propagation of this signal and regulates cell-fate decisions. Although we observed that SIRT1 inhibition led to a significant decrease in all three Dvl proteins, the magnitude of the decrease in specific family members appeared to be cell-type-dependent. Finally, SIRT1 controls key stages of spermatogenesis and germ stem cell expansion and function (10, 11), and Dvl proteins have been shown to regulate spermatogenesis, especially the late stages, which are responsible for spermatid morphological changes ensuring the capability of motility (14). Both mouse models reveal overlapping functions of SIRT1 and Dvl and demonstrate a potential partnership in the regulation of germ stem cell fate. We propose that SIRT1-mediated regulation of Dvl proteins may help explain the diverse SIRT1-dependent physiological responses observed in different physiological and pathological contexts.
Materials and Methods
Cell Culture, Transfection, and Retroviral Infection.
Cell lines were obtained from the American Type Culture Collection. DLD-1 and T-47D cells were cultured in RPMI1640. HEK293 cells were cultured in minimum essential medium, and HCT116 cells were cultured in McCoy's 5A medium. Media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). Transfections were performed with either Lipofectamine (shRNA) or Oligofectamine (siRNA) according to the manufacturer's suggestions. For more details regarding siRNA and shRNA methods, see SI Materials and Methods.
Western Blots and Immunoprecipitations.
Cells were harvested in 50 mM Tris-HCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 50 mM sodium fluoride, 1× Complete protease mixture (Roche). Protein concentrations were measured by BCA protein assay (Pierce). Protein extracts were subjected to polyacrylamide gel electrophoresis using the 4–12% NuPAGE gel system (Invitrogen), transferred to PVDF (Millipore) membranes, and immunoblotted. For more details, see SI Materials and Methods.
Endpoint and Real-Time Quantitative PCR.
Intron-spanning primers specific for each of the targets were designed, and changes in gene expression were measured by endpoint using JumpStart RedTaq (Sigma) and quantitative RT-PCR using Fermentas Maxima SYBR Green, and fluorescence was detected on an ABI PRISM 7900 sequence detector (Applied Biosystems). Total RNA was isolated using TRIzol (Invitrogen), and 2 μg RNA was reverse-transcribed with M-MLV Reverse Transcriptase (Promega). For quantitative PCR, an ABI PRISM 7900 sequence detector was used. For more details on primers and PCR, see SI Materials and Methods.
Migration and Cell-Viability Assays.
Transwell assays were performed with 6.5-mm-diameter inserts, 8.0-μm pore size (Costar), and 24-well cell-culture plates for HCT116, T-47D, and HT-29 cells. For more details on migration and viability assays, see SI Materials and Methods.
Supplementary Material
Acknowledgments
Our research was supported by an intramural grant to K.P. from the Feist-Weiller Cancer Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911325107/-/DCSupplemental.
References
- 1.Qiang L, Wang H, Farmer SR. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-L α. Mol Cell Biol. 2007;27:4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo J, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 4.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Kojima K, et al. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem Biophys Res Commun. 2008;373:423–428. doi: 10.1016/j.bbrc.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 6.Liang XJ, et al. SIRT1 contributes in part to cisplatin resistance in cancer cells by altering mitochondrial metabolism. Mol Cancer Res. 2008;6:1499–1506. doi: 10.1158/1541-7786.MCR-07-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nosho K, et al. SIRT1 histone deacetylase expression is associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2009;22:922–932. doi: 10.1038/modpathol.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 α and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One. 2008;3:e1571. doi: 10.1371/journal.pone.0001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBurney MW, et al. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prestwich TC, Macdougald OA. Wnt/β-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19:612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 14.Ma P, et al. Stage-dependent Dishevelled-1 expression during mouse spermato-genesis suggests a role in regulating spermatid morphological changes. Mol Reprod Dev. 2006;73:774–783. doi: 10.1002/mrd.20468. [DOI] [PubMed] [Google Scholar]
- 15.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 16.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 17.Wharton KA., Jr Runnin’ with the Dvl: Proteins that associate with Dsh/Dvl and their significance to Wnt signal transduction. Dev Biol. 2003;253:1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki K, et al. NEDL1, a novel ubiquitin-protein isopeptide ligase for Dishevelled-1, targets mutant superoxide dismutase-1. J Biol Chem. 2004;279:11327–11335. doi: 10.1074/jbc.M312389200. [DOI] [PubMed] [Google Scholar]
- 19.Creyghton MP, et al. PR72, a novel regulator of Wnt signaling required for Naked cuticle function. Genes Dev. 2005;19:376–386. doi: 10.1101/gad.328905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angers S, et al. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-β-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 21.Itoh K, Brott BK, Bae GU, Ratcliffe MJ, Sokol SY. Nuclear localization is required for Dishevelled function in Wnt/β-catenin signaling. J Biol. 2005;4:3. doi: 10.1186/jbiol20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willert K, Jones KA. Wnt signaling: Is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 23.Pruitt K, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain M, et al. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gan XQ, et al. Nuclear Dvl, c-Jun, β-catenin, and TCF form a complex leading to stabilization of β-catenin-TCF interaction. J Cell Biol. 2008;180:1087–1100. doi: 10.1083/jcb.200710050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad CP, Gupta SD, Rath G, Ralhan R. Wnt signaling pathway in invasive ductal carcinoma of the breast: Relationship between β-catenin, dishevelled and cyclin D1 expression. Oncology. 2007;73:112–117. doi: 10.1159/000120999. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and β-catenin stabilization. Cancer Cell. 2009;15:207–219. doi: 10.1016/j.ccr.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heltweg B, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 29.Lee YN, Gao Y, Wang HY. Differential mediation of the Wnt canonical pathway by mammalian Dishevelleds-1, -2, and -3. Cell Signal. 2008;20:443–452. doi: 10.1016/j.cellsig.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimazu T, Horinouchi S, Yoshida M. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J Biol Chem. 2007;282:4470–4478. doi: 10.1074/jbc.M609745200. [DOI] [PubMed] [Google Scholar]
- 31.Cong F, Schweizer L, Varmus H. Casein kinase Iε modulates the signaling specificities of Dishevelled. Mol Cell Biol. 2004;24:2000–2011. doi: 10.1128/MCB.24.5.2000-2011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tapia JC, Torres VA, Rodriguez DA, Leyton L, Quest AF. Casein kinase 2 (CK2) increases survivin expression via enhanced β-catenin-T cell factor/lymphoid enhancer binding factor-dependent transcription. Proc Natl Acad Sci USA. 2006;103:15079–15084. doi: 10.1073/pnas.0606845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Jones KA. CK2 controls the recruitment of Wnt regulators to target genes in vivo. Curr Biol. 2006;16:2239–2244. doi: 10.1016/j.cub.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 34.Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem Biophys Res Commun. 2009;381:372–377. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- 35.Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS ONE. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JS, et al. Oncogenic β-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 2002;62:2744–2748. [PubMed] [Google Scholar]
- 37.Tetsu O, McCormick F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 38.Shtutman M, et al. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houlston RS, et al. Colorectal Cancer Association Study Consortium; CoRGI Consortium; International Colorectal Cancer Genetic Association Consortium. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng H, et al. Bone morphogenetic protein-4 is overexpressed in colonic adenocarcinomas and promotes migration and invasion of HCT116 cells. Exp Cell Res. 2007;313:1033–1044. doi: 10.1016/j.yexcr.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 42.Esufali S, Charames GS, Pethe VV, Buongiorno P, Bapat B. Activation of tumor-specific splice variant Rac1b by Dishevelled promotes canonical Wnt signaling and decreased adhesion of colorectal cancer cells. Cancer Res. 2007;67:2469–2479. doi: 10.1158/0008-5472.CAN-06-2843. [DOI] [PubMed] [Google Scholar]
- 43.Samarzija I, Sini P, Schlange T, Macdonald G, Hynes NE. Wnt3a regulates proliferation and migration of HUVEC via canonical and non-canonical Wnt signaling pathways. Biochem Biophys Res Commun. 2009;386:449–454. doi: 10.1016/j.bbrc.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28:445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 45.Najdi R, et al. A Wnt kinase network alters nuclear localization of TCF-1 in colon cancer. Oncogene. 2009;28:4133–4146. doi: 10.1038/onc.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 47.Jin Q, et al. Cytoplasm-localized SIRT1 enhances apoptosis. J Cell Physiol. 2007;213:88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- 48.Ross SE, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 49.Huffman DM, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 50.Firestein R, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chien AJ, et al. Activated Wnt/β-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci USA. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You L, et al. An anti-Wnt-2 monoclonal antibody induces apoptosis in malignant melanoma cells and inhibits tumor growth. Cancer Res. 2004;64:5385–5389. doi: 10.1158/0008-5472.CAN-04-1227. [DOI] [PubMed] [Google Scholar]
- 53.Uematsu K, et al. Activation of the Wnt pathway in non small cell lung cancer: Evidence of Dishevelled overexpression. Oncogene. 2003;22:7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 54.Uematsu K, et al. Wnt pathway activation in mesothelioma: Evidence of Dishevelled overexpression and transcriptional activity of β-catenin. Cancer Res. 2003;63:4547–4551. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.