Abstract
The direction of rotation of the Escherichia coli flagellum is controlled by an assembly called the switch complex formed from multiple subunits of the proteins FliG, FliM, and FliN. Structurally, the switch complex corresponds to a drum-shaped feature at the bottom of the basal body, termed the C-ring. Stimulus-regulated reversals in flagellar motor rotation are the basis for directed movement such as chemotaxis. In E. coli, the motors turn counterclockwise (CCW) in their default state, allowing the several filaments on a cell to join together in a bundle and propel the cell smoothly forward. In response to the chemotaxis signaling molecule phospho-CheY (CheYP), the motors can switch to clockwise (CW) rotation, causing dissociation of the filament bundle and reorientation of the cell. CheYP has previously been shown to bind to a conserved segment near the N terminus of FliM. Here, we show that this interaction serves to capture CheYP and that the switch to CW rotation involves the subsequent interaction of CheYP with FliN. FliN is located at the bottom of the C-ring, in close association with the C-terminal domain of FliM (FliMC), and the switch to CW rotation has been shown to involve relative movement of FliN and FliMC. Using a recently developed structural model for the FliN/FliMC array, and the CheYP-binding site here identified on FliN, we propose a mechanism by which CheYP binding could induce the conformational switch to CW rotation.
Keywords: switching, cell motility, signal transduction, molecular motors
Many motile bacteria control the direction of their swimming by regulating the sense of flagellar rotation in response to sensory cues. In the well-studied enteric species Escherichia coli and Salmonella (Salmonella enterica serovar typhimurium), counterclockwise (CCW) rotation allows the several flagellar filaments on a cell to join in a bundle to propel the cell smoothly, whereas clockwise (CW) rotation of one or more flagella disrupts the bundle and causes the cell to tumble. In the absence of external stimuli, a cell executes smooth runs of about a second punctuated by brief tumbles that send it in a new, essentially random, direction; by delaying the switch to CW rotation in response to attractant stimuli, cells prolong runs that happen to be in a favorable direction and so bias their movement toward nutrients, temperatures, or other factors conducive to survival (1, 2).
The direction of flagellar rotation is regulated by a complex at the bottom of the basal body called the switch complex, constructed from the proteins FliG, FliM, and FliN (3). This assembly contains many copies of each protein—about 25 FliG, 35 FliM, and 140 FliN—and is therefore fairly large, having a total mass of about 4 MDa (4–8). In addition to controlling the sense of motor rotation, the switch complex is also essential for flagellar assembly and the generation of torque (3, 9).
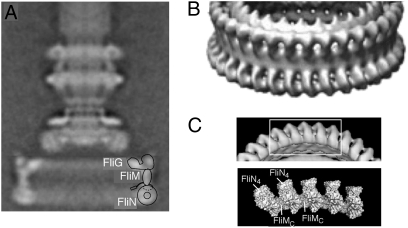
Electron microscopic reconstructions in Salmonella have produced images of the flagellar basal body at resolution sufficient to reveal features the size of protein domains (Fig. 1) (10, 11). The switch complex corresponds to a fairly large (≈50 nm in diameter) drum-shaped feature at the bottom of the basal body, termed the C-ring for its location in the cytosol. In our current structural model of the C-ring (12–17), FliG is located at the top where it accounts for two distinct lobes of electron density seen in the reconstructions (Fig. 1A). FliG is the rotor protein most closely involved in generation of torque, and its C-terminal domain, which we assign to the outer lobe, has been shown to interact with the stator protein MotA (18). The main body of the FliM protein is just below FliG where it forms the relatively thin side-wall of the ring (14), and FliN is at the bottom, organized as an array of ring-shaped tetramers (Fig. 1 B and C) (17, 19). FliM contains a discrete C-terminal domain (FliMC) that is inserted between the FliN tetramers to form the lower part of the C-ring. Targeted cross-linking experiments showed that the switch from CCW to CW rotation is accompanied by a relative movement of FliN and FliMC (15).
Fig. 1.
Electron microscopic images of the flagellar basal body, from studies in Salmonella (10). (A) Single-particle reconstruction of the full basal body, including the LP-ring and a short segment of the rod. The structure is viewed from the side and has been axially averaged. The MS-ring is at the level of the cytoplasmic membrane; the C-ring is in the cytosol. The diameter of the C-ring is about 50 nm. A current working hypothesis for the locations of FliG, FliM, and FliN (12–17) is shown at the right of the C-ring. The two lobes of density at the top of the C-ring are both assigned to FliG, with the outer one corresponding to the C-terminal domain. [In a more fully detailed model, some of the FliM subunits are hypothesized to tilt inward to interact with the inner domain of FliG (13), but this feature is not important in the present context.] The inward-pointing extension on FliM represents the N-terminal segment that is known to interact with CheYP. (B) Detail from a higher-resolution reconstruction (11), showing rings of density at the bottom of the C-ring. [Reproduced with permission from Thomas D, DeRosier DJ (2001) (Copyright 2010, American Society for Microbiology).] (C) The appearance of the bottom of the C-ring as determined in the high-resolution reconstruction and the organization of FliN tetramers and FliMC domains at the bottom of the C-ring as deduced from cross-linking and mutational studies (15).
In the cell, flagellar reversals are controlled by the signaling molecule phospho-CheY (CheYP), which promotes CW rotation (20). CheYP binds to a fairly well-conserved segment near the N terminus of FliM (sequence LSQAEIDALL, in the E. coli protein) (21, 22). A crystal structure showed this FliM segment folded into helical conformation and bound to CheY in a shallow cleft on a face nearly opposite the site of phosphorylation (23). Phosphorylation of CheY is believed to promote binding to the FliM N-terminal segment indirectly via a propagated conformational change (24–26). A recent NMR study gave evidence that CheYP, when bound to the N-terminal segment of FliM, can also interact with the FliM middle domain (FliMM) (27). Last, on the basis of mutational results, we proposed that CheYP might also interact with FliN in the vicinity of a strongly conserved hydrophobic patch (28). Mutations on this surface of FliN caused strongly CCW-biased motor rotation that could be offset, in most cases, by overexpression of CheY. The hypothesized binding of CheYP to FliN has not been confirmed directly.
Whatever the binding target(s) for CheYP, switching must presumably involve movement of the C-terminal domain of FliG, which forms the interface with the stator, where the forces for motor rotation are produced or applied (18, 29). Thus, conformational changes occurring in the lower part of the C-ring, whether they originate primarily in FliN or FliM, must be transmitted “up” through FliMM to induce movement of FliGC. The C-terminal domain of FliG is linked to the rest of the protein by a segment containing a well-conserved Gly–Gly residue pair that could confer flexibility to allow switch-driven movement of FliGC (16). A mutational analysis showed that certain replacements in this segment confer a “rusty hinge” phenotype in which CCW-CW switching is slowed (30). Viable models for switching must also account for a very high degree of cooperativity; the transition to CW rotation depends very sharply on CheYP concentration, exhibiting a Hill coefficient greater than 10 (31). This cooperativity should enhance the responsiveness of the motor to small changes in cellular CheYP concentration and thereby increase the sensitivity of the chemotactic response.
The present study addresses the question of whether CheYP interacts with the rotor protein FliN and whether such an interaction has a role in flagellar direction switching. Using pull-down assays, we show that CheYP can bind to FliN, but this binding occurs only when CheYP is occupied by the N-terminal segment of FliM. Thus, in the normal setting, CheYP is expected to interact with FliN after first being captured by FliM. The binding determinants on both sides of the CheY–FliN interaction were mapped using mutations. FliN mutations that weakened the binding of CheYP were the same as those found previously to cause CCW motor bias (28), indicating that the CheYP–FliN interaction is important for motor direction reversal. The relative movement of FliN and FliMC that accompanies switching (15) is predicted to make the CheY-binding site on FliN more accessible in the CW than in the CCW state, providing a simple structural basis for coupling the binding of CheYP to a conformational change in the switch complex.
Results
CheY Interaction with FliN.
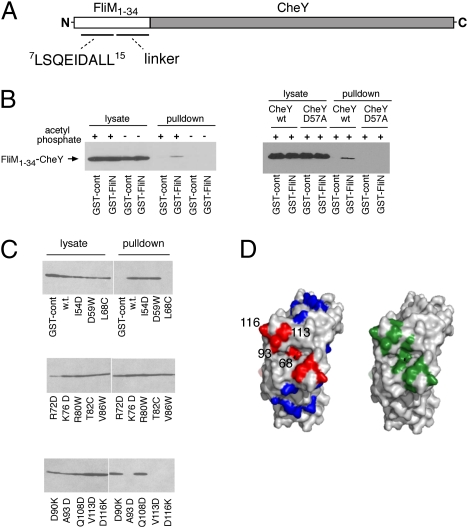
The switch from CCW to CW rotation is triggered by binding of the signaling protein CheYP to the motor. Although the FliM protein of the rotor has usually been considered the main player in switching, a mutational analysis gave evidence that FliN might also play a role, and identified a conserved hydrophobic patch as a probable target of action of CheYP (28). We looked for a CheY–FliN interaction using pull-down assays with a GST–CheY fusion construct but saw no evidence of binding either in the absence or presence of acetyl phosphate, a CheY-phosphorylating agent. In our current working model of the switch complex, the N-terminal segment of FliM that is known to bind CheYP lies near FliN. Accordingly, we hypothesized that CheYP might bind first to this FliM segment and thereby gain binding determinants needed for interaction with FliN. This proposal was tested using a hybrid construct containing 34 residues from the N terminus of FliM fused to the N terminus of CheY (termed M34-Y). This construct contains the conserved CheY-binding segment of FliM as well as a less-conserved segment of about 15 residues that is predicted to have nonregular secondary structure and to function as a linker (Fig. 2A). Upon phosphorylation, the CheY part of this construct should be able to capture the FliM segment and so acquire binding determinants resembling those of FliM-bound CheYP. The binding of this fusion construct to FliN was tested using pull-down assays. Cells expressing the M34-Y fusion protein were mixed with cells expressing GST–FliN, lysed, and mixed with glutathione-Sepharose beads. Beads were washed and treated with glutathione to release GST–FliN and associated pro-teins, and samples were analyzed on immunoblots using a polyclonal anti-FliM antiserum that showed high sensitivity toward the FliM sequences present in the fusion protein. A binding interaction between the M34-Y construct and FliN was readily observed in the pull-down experiment, in the presence but not in absence of acetyl phosphate (Fig. 2B Left). Acetyl phosphate is believed to transfer its phosphoryl group to the physiologically relevant residue Asp-57 of CheY in a reaction facilitated by a nearby protein- bound Mg2+ ion, and is in this sense specific (32). Conceivably, however, acetyl phosphate might react with other positions in the M34-Y construct to induce FliN binding by a mechanism that does not involve Asp-57. To substantiate the role of Asp-57, we carried out the same experiment but using a mutant protein with Asp-57 replaced by alanine. The D57A mutant protein did not bind to FliN, either in the presence or absence of acetyl phosphate (Fig. 2B Right).
Fig. 2.
(A) Schematic of the FliM1–34–CheY fusion construct. The part of the FliM sequence shown explicitly is well conserved across species and is known to bind to CheYP in helical conformation (23). (B Left) Pull-down assay with GST–FliN and the FliM1–34–CheY construct, in presence or absence of the phosphorylating agent acetyl phosphate. (B Right) Effect of the CheY mutation D57A. (C) Binding of the FliM1–34–CheY construct to FliN proteins with mutations in various surface positions. Acetyl phosphate was present in all samples. (D) Comparison of the CheYP-binding region on FliN with positions of previously characterized CCW-biased mutations. (Left) Results of the binding experiment (C) mapped onto the FliN structure (PDB ID code 1yab). Red, positions where mutations eliminated the binding; blue, positions where mutations did not affect binding. (Right) Positions of mutations in FliN that gave CCW motor bias, colored green (data from ref. 27; image made in PyMol).
Mapping Regions Important for the Interaction.
To map the CheY-binding determinants on FliN, the pull-down experiment was repeated using GST–FliN proteins with nonconservative mutations at various surface positions. Of 14 FliN mutations studied, 10 showed no measurable reduction in binding. Binding to the M34-Y protein was eliminated by mutations in FliN residues Leu-68, Ala-93, Val-113, and Asp-116 (Fig. 2C). Mutations that weakened the binding lie in the vicinity of a conserved surface hydrophobic patch, and are the same as those shown previously to cause CCW motor bias (28) (Fig. 2D).
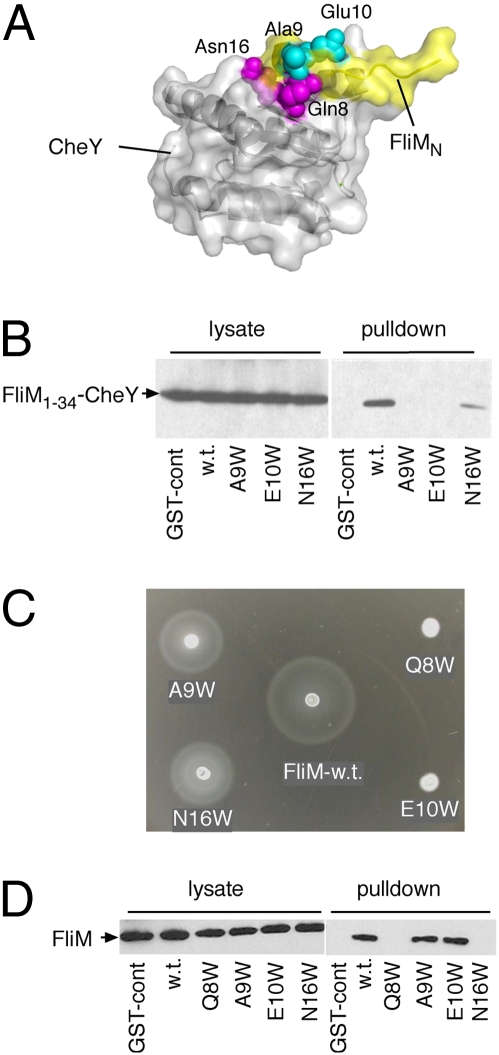
Because the FliM1–34 segment was found necessary for the CheY–FliN interaction, we next tested whether this segment participates directly in the binding. In the crystal structure of a CheY–FliM segment complex (23), residues Ala-9 and Glu-10 of FliM are exposed on the surface where neither side-chain appears to contribute to the interaction with CheY (Fig. 3A). To determine whether these FliM residues are important for the interaction with FliN, they were individually replaced with the larger residue Trp, and binding was measured using the pull-down assay. Binding of the fusion construct to FliN was prevented by both mutations (Fig. 3B). To examine the effects of the A9W and E10W replacements in a more native setting, the same mutations were made in the full-length FliM protein, and function was measured using soft-agar plate assays. Chemotactic migration in soft agar was eliminated by the E10W replacement and reduced to about half of the wild-type rate by the A9W replacement (Fig. 3C). Cells of the E10W mutant swam smoothly in liquid media, and the motors rotated exclusively CCW in a tethering experiment (in which filament stubs are attached to a coverslip to allow monitoring of motor rotation). To rule out an effect of the mutations on the interaction between the FliM segment and CheY, the FliM–CheY interaction was examined using a pull-down assay with GST–CheY. Binding of CheY to FliM in this assay depended on the presence of acetyl phosphate, as reported previously (33). In the pull-down experiment, the binding to CheY was not affected by the A9W or E10W mutations in FliM, consistent with the solvent-exposed positions of these residues in the crystal structure (23).
Fig. 3.
Effect of mutations in the segment of FliM that interacts with CheY. (A) Structure of the complex formed between CheY (light gray) and the FliM segment (yellow) (PDB ID code 1f4v) (23). The solvent-exposed residues Ala-9 and Glu-10 are colored cyan, and residues Gln-8 and Asn-16, which contribute to the FliM–CheY interface, are magenta (image made in PyMol) (B) Interaction of FliN with FliM1–34–CheY constructs with mutations in the FliM segment. The pull-down assay used GST–FliN and the FliM1–34–CheY fusion construct containing the mutations. (C) Effects of the FliM mutations on cell migration in soft agar. The Trp replacements were transferred into the full-length FliM protein for this experiment. (D) Effects of the FliM mutations on the interaction between the FliM segment and CheY, measured using a GST–CheY pull-down assay.
Two other Trp replacements (Q8W and N16W) were made in the FliM segment at more-buried positions that contact CheY and might stabilize the FliM-CheYP binding. Both of these mutations weakened the FliM–CheYP interaction significantly (Fig. 3D). In the soft-agar assay, chemotactic migration was eliminated by the Q8W mutation and decreased to about half of normal by the N16W mutation (Fig. 3C). Cells of the Q8W mutant swam smoothly in liquid media and exhibited only CCW motor rotation in a tethering experiment. The substantial function of the N16W mutant implies that this protein retains some ability to bind CheYP that could not be observed in the pull-down assay. This is not surprising, as the pull-down assay is fairly stringent in the sense that off-rates must be slow for binding to be observed. Consistent with the partial function of the N16W mutant, the M34-Y fusion construct containing this mutation also retained some ability to bind FliN in the pull-down experiment (where the FliM–CheY interaction can occur in cis within the M34-Y fusion; Fig. 3B).
Regulation of CheY Access by the N-Terminal Segment of FliN.
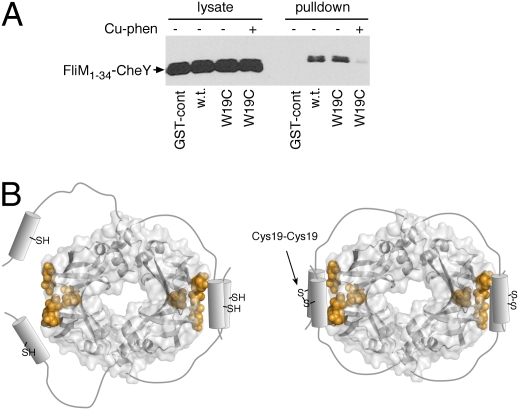
The function of the N-terminal part of FliN is uncertain, as this segment is not required for flagellar assembly and is to a large extent dispensable for motility and chemotaxis (34). A cross-linking study showed that in the FliN tetramer, the N-terminal segments of a FliN dimer interact with each other and with the hydrophobic patch of the other FliN dimer (19). Because CheYP also binds in the region of the hydrophobic patch, the N-terminal segments of FliN might be expected to prevent binding of CheYP. We hypothesized that these segments of FliN might regulate access of CheYP, possibly functioning to prevent CheY binding until the segments are displaced from the patch (upon installation of the FliN tetramer into the flagellum, for example). To test this, we introduced a Cys replacement at FliN residue 19, a position shown previously to allow efficient FliN–FliN cross-linking, and used Cu-phenanthroline to induce disulfide bond formation. Binding of the M34-Y protein to the cross-linked FliN was tested using the pull-down assay. Binding of the M34-Y fusion protein was eliminated upon formation of the residue-19 cross-link in FliN (Fig. 4A).
Fig. 4.
Effect of cross-linking through the N-terminal segment of FliN on the binding of FliN to the FliM1–34–CheY construct. Residue Trp-19 of FliN (in the GST–FliN fusion) was replaced with Cys. In the samples indicated, Cu-Tris[1,10-phenanthroline] was added to induce disulfide formation.
Discussion
Switch bias mutants were known to occur in FliN but at a low frequency relative to FliM or FliG (35, 36). This has been taken as evidence that FliN has a relatively small role in switching, but is also consistent with the protein having a crucial role—though one that involves comparatively few amino acid residues. The present findings show that FliN has a critical role in switching, interacting with CheYP to trigger or enable the switch to CW rotation. The interaction involves a conserved surface region on FliN where mutations were shown previously to cause CCW phenotype, and requires that CheYP be bound to the N-terminal segment of FliM. Accordingly, we propose that the N-terminal segment of FliM serves to capture CheYP, and that switching is triggered by the subsequent interaction of this FliM-bound CheYP with FliN.
Mutations in the N-terminal segment of FliM had effects suggesting that the segment contributes directly to the interaction with FliN; replacements on the exterior-facing part of the FliM segment permitted normal binding of the segment to CheYP (Fig. 3D) but weakened binding of the M34-Y fusion protein to FliN (Fig. 3B). In the context of full-length FliM, the E10W replacement eliminated chemotactic migration in soft-agar plates (Fig. 3C), and tethered cells displayed the fully CCW bias expected of motors unable to respond to CheYP. The retention of some chemotactic function in the A9W mutant might appear at odds with the pronounced defect in FliN binding, but note that the pull-down experiment involves binding between two separate proteins, whereas motor switching in the present scenario depends only on interaction between the FliM-tethered CheY and the FliN units nearby in the motor. Given the constrained nature of the CheY–FliN interaction, even a partial defect such as that in the A9W mutant might be taken to indicate a substantial weakening of the binding. In addition to Ala-9, the FliM residues Ala-13 and Leu-14 also lie on the exterior of the segment, and together with two nearby residues of CheY (Ile-96 and Ala-99) form a surface hydrophobic patch that might interact with the hydrophobic patch on FliN. In this scheme, the FliM segment would become sandwiched between CheYP and its target on the flagellar switch, a possibility first noted in the structural study of the CheY–FliM peptide complex (23). Further structural studies will be needed to define the FliN–CheY interaction in fuller detail.
Because the N-terminal segment of FliN has been shown to lie near the hydrophobic patch, it might be expected to occlude the CheY-binding site (19) (Fig. 4B). Although this segment of FliN was present in the constructs used in pull-down experiments and did not prevent the CheY–FliN interaction, when the N-terminal segments were stably associated through a disulfide crosslink, the binding of CheYP was decreased. We suggest, therefore, that the N-terminal segment could function to modulate the binding of CheYP to FliN, possibly blocking the binding of CheYP to any FliN–FliM complexes not yet installed in motors. Once FliM-FliN4 complexes are installed in the motor, the segment could be displaced to expose the site for interaction with CheYP.
The CheYP-binding site identified here overlaps closely with the binding site identified previously for FliH, a protein that functions in the export process needed for assembly of the exterior parts of the flagellum (28). The N-terminal segments of FliN might therefore serve also to prevent premature interactions with FliH. Further, if FliH and CheYP compete for overlapping binding sites on FliN, we might expect some interplay between flagellar assembly and switching.
The capture of CheYP through the initial FliMN–CheY interaction would, besides producing the determinants for binding FliN, also ensure that CheYP is present at a high “local concentration” in the vicinity of FliN. In a system such as the flagellar switch that involves the cooperative action of many CheYP molecules, it might be particularly important to hold many copies of CheYP poised nearby. A local concentration effect appeared important in the CheYP–FliMM interaction reported in a recent NMR study (27). This interaction was seen most clearly when the N-terminal and middle parts of FliM were present in the same polypeptide, so that CheYP was tethered in the vicinity of the FliMM domain through its interaction with FliMN. An interaction between CheY and FliMM appears compatible with the CheY–FliN interaction described here, in the sense that both types of interaction could occur within a single motor (and both can be accommodated in the current working model for switch-complex organization). Further work will determine whether the CheYP–FliMM interaction is, like the FliN–CheY interaction, essential for the switch to CW rotation.
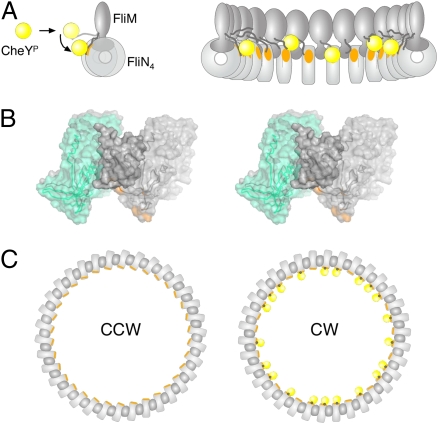
Cross-linking experiments showed that the lower portion of the C-ring is formed from an alternating array of FliN tetramers and FliM C-terminal domains, and that the switch from CCW to CW rotation is accompanied by relative movement at one of the FliN–FliMC interfaces (15). The CheYP-binding site identified here on FliN is predicted to lie near the adjacent FliMC domains and to become somewhat more accessible upon switching to the CW state (Fig. 5). Accordingly, we propose that the interaction of CheYP with FliN directly induces the relative movement of FliN and FliMC, and that this is the initiating event in the conformational switch to the CW state. Given their close proximity in the emerging structural model, a direct interaction between CheY and FliMC also appears possible and might also contribute to the energetics of switching. We note, in this context, that a previous study of the binding interactions of various FliM deletion constructs gave evidence for a direct interaction between CheY and FliMC (33). Because switching will presumably alter the relationship between rotor and stator, the movements initiated near the bottom of the C-ring must be transmitted “up” through FliMM to affect FliGC. The nature of the movements in FliMM and FliGC are presently unknown. The origins of cooperativity are likewise not yet clear, although important factors are likely to be the substantial number of FliMC and FliN4 units (and CheY-binding sites) present and the close association among these elements within the C-ring.
Fig. 5.
Model of CheYP-induced flagellar motor switching. (A Left) CheYP (yellow) interacts initially with the N-terminal segment of FliM, which is flexible enough to allow subsequent binding to a site on FliN (orange) in the vicinity of the hydrophobic patch. Only the FliM and FliN proteins of the switch are shown; FliG would be at the top (Fig. 1). (Right) View showing multiple FliM–FliN units in the lower part of the C-ring, and the binding of multiple CheYP molecules. (B) Relationship of FliN4 and FliMC units in the bottom of the C-ring, as determined from cross-linking and mutational analysis (15). One FliN4–FliMC–FliN4 unit is shown, in stereoview. Altered yields of certain cross-links upon switching (15) indicated that motor reversal is accompanied by a movement along one of the FliN4–FliMC interfaces, shown here by the two locations for the left-hand FliN4 unit (CW state, gray; CCW state, cyan). The CheYP-binding site on FliN is colored orange. (C) Hypothesis for switching in all of the FliN4–FliMC units, shown in top view. CheYP molecules are yellow and binding sites are orange; the black dot signifies the N-terminal segment of FliM sandwiched between CheYP and FliN. These segments would attach to the FliM middle domain, by a flexible linker (A).
In summary, the present findings show that flagellar motor reversal depends on an interaction between the CW signal CheYP and the rotor protein FliN. The interaction with FliN should occur subsequently to CheYP capture by the N-terminal domain of FliM. The proximity of the CheYP-binding site to the FliN–FliMC interface suggests a simple mechanism by which CheYP binding could induce relative subunit movements that initiate the process of switching.
Experimental Procedures
Plasmids and Strains.
The E. coli strains and plasmids used are listed in Table 1. Procedures for DNA manipulation were as described previously (37). The gene encoding the FliM1–34–CheY fusion was cloned into the IPTG-inducible expression plasmid pTBM30 (ApR) (38), yielding pKP235. The FliM1–34–CheY(D57A) mutant protein was obtained by site-directed mutagenesis using the QuikChange (Qiagen) procedure, yielding pKP453. The GST–FliN mutant variants were cloned in plasmid pHT96 (34), also under an IPTG-inducible promoter. All binding experiments were carried out in the ΔflhDC strain (RP3098), a gift from J.S. Parkinson (Salt Lake City, UT). The flhDC genes are required for expression of all other flagellar operons (39, 40), and so this strain expresses no flagellar genes from the chromosome. Swimming motility in soft agar and in liquid were assayed as described previously (37), using the fliM-deletion strain DFB228 transformed with wild-type or mutant plasmids. To monitor the direction of motor rotation in mutant strains, cells were tethered to coverslips using anti-flagellin antibody using procedures described previously (37). Swarm plates contained TB, 0.27% agar, appropriate antibiotic(s), and IPTG at concentrations of 0, 40, and 100 μM, for assay of FliM function at different levels of expression.
Table 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or property | Source |
| DFB228 | fliM-null strain | (30) |
| RP3098 | flhDC deletion strain; expresses no chromosomal flagellar genes | Gift from J.S. Parkinson |
| pTBM30 | Ptac expression vector; Apr | (31) |
| pHT100 | GST-only expression vector; Kmr | (30) |
| pHT96 | GST-FliN expression vector; Kmr | (30) |
| pKP235 | FliM1–34CheY fusion in pTBM30 expression vector; Apr | This study |
| pKP453 | FliM1–34CheY(D57A) in pTBM30 expression vector; Apr | This study |
GST Fusion Coprecipitation Procedure.
Coprecipitation experiments were carried out essentially as described (33, 41) with minor modifications. In experiments to probe interactions of FliM1–34–CheY fusion with FliN, the RP3098 strain was transformed with a plasmid expressing the GST–FliN fusion or its mutant variants, or a plasmid expressing the FliM1–34–CheY fusion. Negative-control experiments used GST, expressed from plasmid pHT100 (41).
Cells were cultured overnight at 32 °C in 40 mL LB containing appropriate antibiotics and 400 μM IPTG. Cells expressing GST–FliN and its mutant variants were mixed with cells expressing FliM1–34–CheY, pelleted, and resuspended in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) containing 100 μL of lysozyme (5 mg/mL in 50% glycerol), 10 μL APMSF (4-amidinophenylmethanesulfonyl fluoride), 60 μL of 1 M MgCl2, and 100 μL of either water (nonphosphorylating conditions) or 0.5 M acetyl phosphate (final concentration, 40 mM). After 1 h on ice, cells were disrupted by sonication. Debris was pelleted (16,000 × g, 30 min, 4 °C), and 50 μL of the supernatant was saved for use in estimating the amount of FliN present before addition of affinity beads. The rest (∼1 mL) was transferred to a clean tube, mixed with 150 μL of a 50% slurry of glutathione-Sepharose 4B (Pharmacia) prepared according to the manufacturer's directions, and incubated for 1 h at 4 °C with gentle rotation to allow binding. The beads were pelleted (14,000 × g, 1 min), washed with 1 mL of PBS, and repelleted. Supernatant was removed, and GST–FliN and associated proteins were released from the beads by addition of 50 μL elution buffer [50 mM reduced glutathione in 50 mM Tris-HCl (pH 8)] for 10 min at room temperature with gentle rotation. Beads were pelleted and the supernatant was collected for analysis by SDS/PAGE and immunoblotting using anti-FliM antibody, which showed high sensitivity toward the FliM segment present in the fusion protein (and not toward CheY alone). Experiments with mutant variants were done in the same way, except always under phosphorylating conditions and with wild-type proteins as control. In the experiment with Cys introduced at position 19 of FliN in the GST–FliN fusion, disulfide cross-linking was induced using Cu-phenanthroline, prepared as described (19). Pull-down experiments with the disulfide cross-linked GST–FliN used the same protocol as above.
SDS/PAGE and Immunoblotting.
Protein samples were separated on 8.5% SDS/PAGE minigels (Bio-Rad Mini-PROTEAN system) and transferred to nitrocellulose using a semidry apparatus (Bio-Rad). Rabbit polyclonal antibody against FliM was prepared as described (37) and was used at 2,000-fold dilution. Bands were visualized using the SuperSignal West Pico Luminol system (Pierce) and X-ray film (Kodak).
Acknowledgments
This work was supported by the National Institute of General Medical Sciences Grant R01 GM64664.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown DA, Berg HC. Temporal stimulation of chemotaxis in Escherichia coli. Proc Natl Acad Sci USA. 1974;71:1388–1392. doi: 10.1073/pnas.71.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi S, et al. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones CJ, Macnab RM, Okino H, Aizawa S-I. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 5.Sosinsky GE, et al. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc Natl Acad Sci USA. 1992;89:4801–4805. doi: 10.1073/pnas.89.11.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Yonekura K, Namba K. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol. 2004;337:105–113. doi: 10.1016/j.jmb.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 8.Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi S, Fujita H, Ishihara A, Aizawa S-I, Macnab RM. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 11.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowder BJ, Duyvesteyn MD, Blair DF. FliG subunit arrangement in the flagellar rotor probed by targeted cross-linking. J Bacteriol. 2005;187:5640–5647. doi: 10.1128/JB.187.16.5640-5647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown PN, Terrazas M, Paul K, Blair DF. Mutational analysis of the flagellar protein FliG: Sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol. 2007;189:305–312. doi: 10.1128/JB.01281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc Natl Acad Sci USA. 2006;103:11886–11891. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar MK, Paul K, Blair DF. Subunit organization and reversal-associated movements in the flagellar switch of Escherichia coli. J Biol Chem. 2010;285:675–684. doi: 10.1074/jbc.M109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown PN, Hill CP, Blair DF. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown PN, Mathews MAA, Joss LA, Hill CP, Blair DF. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol. 2005;187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul K, Blair DF. Organization of FliN subunits in the flagellar motor of E. coli. J Bacteriol. 2006;288:2502–2511. doi: 10.1128/JB.188.7.2502-2511.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess JF, Oosawa K, Kaplan N, Simon MI. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 21.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bren A, Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 23.Lee S-Y, et al. Crystal structure of an activated response regulator bound to its target. Nat Struct Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 24.McEvoy MM, Bren A, Eisenbach M, Dahlquist FW. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein, FliM. J Mol Biol. 1999;189:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 25.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry. 1994;33:10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- 26.Halkides CJ, et al. The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY. Biochemistry. 2000;39:5280–5286. doi: 10.1021/bi9925524. [DOI] [PubMed] [Google Scholar]
- 27.Dyer CM, Vartanian AS, Zhou H, Dahlquist FW. A molecular mechanism of bacterial flagellar motor switching. J Mol Biol. 2009;388:71–84. doi: 10.1016/j.jmb.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul K, Harmon JG, Blair DF. Mutational analysis of the flagellar rotor protein FliN: Identification of surfaces important for flagellar assembly and switching. J Bacteriol. 2006;188:5240–5248. doi: 10.1128/JB.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakushi T, Yang J, Fukuoka H, Homma M, Blair DF. Roles of charged residues of rotor and stator in flagellar rotation: Comparative study using H+-driven and Na+-driven motors in Escherichia coli. J Bacteriol. 2006;188:1466–1472. doi: 10.1128/JB.188.4.1466-1472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Way SM, Millas SG, Lee AH, Manson MD. Rusty, jammed, and well-oiled hinges: Mutations affecting the interdomain region of FliG, a rotor element of the Escherichia coli flagellar motor. J Bacteriol. 2004;186:3173–3181. doi: 10.1128/JB.186.10.3173-3181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 32.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews MAA, Tang HL, Blair DF. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H, Billings S, Wang X, Sharp L, Blair DF. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol. 1995;177:3496–3503. doi: 10.1128/jb.177.12.3496-3503.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irikura VM, Kihara M, Yamaguchi S, Sockett H, Macnab RM. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Blair DF. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison TB, Parkinson JS. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 41.Tang H, Braun TF, Blair DF. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]