Abstract
Poly(ADP-ribosyl)ation by poly(ADP-ribose) polymerases regulates the interaction of many DNA damage and repair factors with sites of DNA strand lesions. The interaction of these factors with poly(ADP-ribose) (PAR) is mediated by specific domains, including the recently identified PAR-binding zinc finger (PBZ) domain. However, the mechanism governing these interactions is unclear. To better understand the PBZ-PAR interaction, we performed a detailed examination of the representative PBZ-containing protein involved in the DNA damage response, aprataxin polynucleotide-kinase-like factor (APLF), which possesses two tandem PBZ domains. Here we present structural and biochemical studies that identify Y381/Y386 and Y423/Y428 residues in the conserved C(M/P)Y and CYR motifs within each APLF PBZ domain that are critical for the interaction with the adenine ring of ADP-ribose. Basic residues (R387 and R429 in the first and second PBZ domains, respectively) coordinate additional interactions with the phosphate backbone of ADP-ribose, suggesting that APLF binds to multiple ADP-ribose residues along PAR polymers. These C(M/P)Y and CYR motifs form a basic/hydrophobic pocket within a variant zinc finger structure and are required for APLF recruitment to sites of DNA damage in vivo.
Keywords: DNA damage signaling, high affinity
The DNA damage response (DDR) and maintenance of chromosomal stability is regulated in part by posttranslational modifications, including poly(ADP-ribosyl)ation by poly(ADP-ribose) polymerases (PARPs), which direct the recruitment of proteins involved in the signaling and repair of DNA damage. In addition, PARP enzymes are important regulators of chromatin remodeling, apoptosis, and transcription (1). In the early response to DNA damage, PARP1 is the predominant PARP activated by DNA strand lesions and catalyzes the attachment of multiple ADP-ribose (ADPr) units from nicotinamide adenine dinucleotide (NAD+) onto target proteins, including PARP1 itself (2). Poly(ADP-ribose) (PAR) accumulated at DNA breaks is subsequently metabolized to ADPr by PAR glycohydrolase (3). To date, PAR-binding motifs have been described in some macrodomains, which also bind to ADPr (4, 5), in a basic residue-rich motif interspersed with hydrophobic amino acids (6), and in the more recently described PAR-binding zinc finger (PBZ) domains (7). PBZ domains, present in proteins either as single or two tandem motifs, are limited to multicellular eukaryotes, and the majority of PBZ-containing proteins have putative roles in PAR metabolism, DNA repair, or DNA damage signaling (7–9).
APLF (aprataxin polynucleotide kinase (PNK)-like factor, also known as PALF and Xip1) is a newly identified protein involved in the DDR possessing a forkhead-associated (FHA) domain and two tandem PBZ motifs (7–11) (Fig. 1A). APLF was originally identified based on the similarity of its FHA domain to those of PNK and aprataxin (12), which share functional similarities and direct FHA- and phosphothreonine-dependent interactions with the DNA repair proteins XRCC1 and XRCC4 (8–16). APLF has been shown to participate in DNA repair, and human cells depleted of APLF by siRNA demonstrate cellular sensitivity to a number of DNA damaging agents that resembles the cellular sensitivities of PARP1-deficient cells (8–11). The two tandem PBZ domains are essential for the recruitment of APLF to sites of DNA damage and may serve to modulate PAR metabolism in vivo (10, 11, 17).
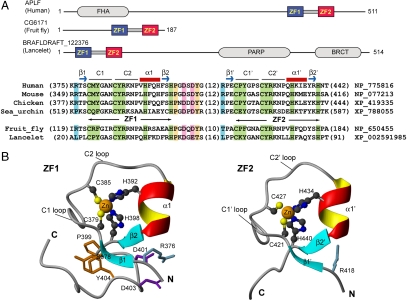
Fig. 1.
APLF PBZ domains comprise unique zinc finger folds. (A) Schematic representation of APLF and all other known tandem PBZ-containing proteins with domain organization, and alignment of PBZ amino acid sequences. (B) NMR-derived structure of the APLF PBZ domains (ZF1 and ZF2) and linker region.
Given that the PBZ- and PAR-dependent recruitment of APLF to sites of DNA damage is likely important for its function, we sought to better understand the mechanism of the APLF-PAR interaction. Structural studies presented here demonstrate that the tandem APLF PBZ domains each contain C(M/P)Y and CYR motifs that form a basic/hydrophobic pocket within a variant zinc finger (ZF)-structure that are essential for the interaction with the adenine ring of ADP-ribose. The tandem PBZ domains dramatically enhanced the binding to PAR compared to the isolated individual PBZ domains, suggesting the existence of an allosteric interaction between the two APLF PBZ domains as discussed below. Importantly, the C(M/P)Y and CYR motifs were also found to be critical for the interaction of APLF with PAR associated with sites of DNA damage in vivo. We propose that the evolutionarily conserved motifs, C(M/P)Y and CYR, are a key signature of PAR recognition.
Results
Structural Characterization of the APLF PBZ Domains
To understand the mechanism by which the APLF PBZ domains (ZF1 and ZF2) recognize PAR, we determined the structure of the tandem PBZ-containing peptide (TZF, APLF residues 360–448) by NMR (Fig. 1 B, Fig. S1A, and Table S1). ZF1 and ZF2 are well-structured, nearly identical in their folds (rmsd = 1.1 Å), and are connected via a 22-residue linker (residues 399–420), which appears to be largely flexible thus causing some degree of freedom in the domain orientation (Fig. S1). The polypeptide backbone of each domain consists of two short β-strands (β1 and β2 in ZF1 and β1′ and β2′ in ZF2) and a short α-helix (α1 in ZF1 and α1′ in ZF2) with a long loop connecting β1 and α1 in ZF1, and β1′ and α1′ in ZF2. This loop contains two conserved cysteine residues (C379 and C385 in ZF1, and C421 and C427 in ZF2) responsible for Zn2+ coordination. The other Zn2+ coordinating residues (H392 and H398 in ZF1, and H434 and H440 in ZF2) reside within α1 and after β1, respectively (Fig. 1 B and C), and all use the imidazole ring NE2 atom for Zn2+ ligation (Fig. S2A). One notable difference between ZF1 and ZF2 was found in the domain-linker interaction. In ZF1, Y404 covers an otherwise exposed hydrophobic patch formed by residues S378, C379, G382, A383, S397, and P399 that is further augmented by electrostatic interactions between D401/D403 and R376. In ZF2, these interactions are absent.
PBZ Domain Possesses a Distinct Structural Topology.
Structural folds homologous to the APLF PBZ structure were not detected using DALI (http://ekhidna.biocenter.helsinki.fi/dali_server/) and iCOPS (http://cops.services.came.sbg.ac.at/). We then performed a manual comparison of the PBZ structure with representative zinc-binding domain structures. In this manual analysis, we followed the fold classification of the ZF protein family as previously proposed (18), in which eight fold subclasses are defined. We compared those eight representative zinc-binding domain structures (including those represented in Fig. S2B) to the PBZ structure of APLF, and identified some degree of similarity to the RNA binding ZFs of TIS11d (19), which has a polypeptide chain trace resembling that of PBZ (rmsd ∼ 1.7 Å). Despite this similarity, the TIS11d ZF and PBZ domain possess distinct motifs for zinc binding (CX5CX6HX5H in PBZ and CX8CX5CX3H in TIS11d), and differ in their structural topologies. These results suggest that the PBZ structure is a variant of the TIS11d ZF fold.
Adenine Ring of ADPr Interacts with the APLF PBZ Domains.
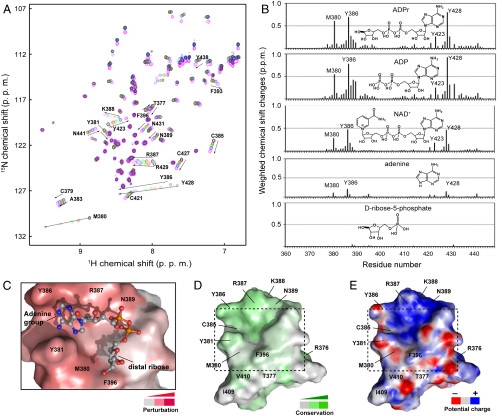
In order to investigate the interaction of the APLF TZF with PAR, we used ADPr in our NMR titration experiments (Fig. 2A). We found that Y386 in ZF1 and Y428 in ZF2 displayed dramatic upfield chemical shift changes of 0.7 and 0.8 ppm in the 1H dimension, respectively. A similar magnitude of chemical shift perturbation (∼0.6 ppm in 1H) was observed for a less conserved residue, M380 in ZF1. The corresponding position in ZF2 is a proline (P422), which does not have a backbone amide and therefore no data were available for this residue. Nevertheless, the preceding and following residues, C421 and Y423, respectively, showed marked perturbations (0.1–0.3 ppm in 1H) akin to the changes observed for the corresponding residues in ZF1 (C379 and Y381). Our NMR titration data using several ADPr-related chemical compounds (Fig. 2B) coupled with the observation of intermolecular NOEs between TZF and ADPr (Fig. S3C) suggested that the large changes in chemical shift observed for Y386 in ZF1 and Y428 in ZF2 may be due to the ring current effect from the adenine ring of ADPr. Taken together, these data strongly argue that the C(M/P)Y and CYR motifs containing the first and second zinc coordinating cysteine residues are both involved in the interaction with ADPr.
Fig. 2.
NMR characterization of the ADPr-binding C(M/P)Y and CYR motifs. (A) Titration of TZF with ADPr monitored by 1H-15N heteronuclear single quantum coherence (HSQC) spectra. (B) Chemical shift perturbation profiles of TZF by ADPr, ADP, NAD+, adenine, and D-ribose-5-phosphate. (C) A representative HADDOCK model of ADPr at the binding site of ZF1. (D) Conservation and (E) electrostatic potential surface representation of ZF1 with the ADPr-binding site as indicated.
Nucleotide Binding Specificity.
In addition to the four ADPr-related molecules described in Fig. 2B, we examined the binding of GDP with the APLF TZF using NMR spectroscopy (Fig. S4). The addition of GDP induced qualitatively similar but somewhat reduced chemical shift perturbations in both PBZ domains of TZF when compared with ADPr, ADP, or NAD+. Y386, R387, Y428, and R429 all displayed chemical shift changes in smaller magnitude upon binding to GDP. The chemical shift change observed for M380 within the CMY motif with GDP (0.02 ppm in 1H and 0.03 ppm in 15N) was drastically reduced when compared with the change seen with ADP (0.53 ppm in 1H and 0.91 ppm in 15N). However, we could not detect a significant difference in the dissociation constants between GDP and ADPr or ADP (Table S3). These data indicate that the PBZ nucleotide binding pocket can also accommodate the guanine ring of GDP at a low affinity equivalent to that observed with ADPr or ADP (Kd ∼ 10-3 M). It is possible that higher nucleotide binding specificity is achieved only when multiple adenine rings are presented in the form of PAR polymer chains.
APLF PBZ Domains Primarily Recognize ADPr Independently.
To further investigate the role of the C(M/P)Y and CYR motifs in ADPr recognition, we employed site-directed mutagenesis to generate the mutant peptides Y381A/Y386A (ZFY1), Y423A/Y428A (ZFY2), and Y381A/Y386A/Y423A/Y428A (ZFY4) in the context of TZF. ZFY1 dramatically reduced chemical shift perturbations within ZF1 but had little impact on the ADPr-induced chemical shift changes in ZF2. Similarly, ZFY2 only affected ZF2 and did not affect ZF1. On the other hand, ZFY4 affected both ZF1 and ZF2 domains (Fig. S5). These data support the notion that the PBZ domains are ADPr-binding motifs, and further indicate that ZF1 and ZF2 are primarily independent in ADPr recognition.
C(M/P)Y and CYR Motifs are Critical Elements of the ADPr-Binding Pocket.
We then mapped the ADPr-binding sites on our NMR structure of TZF and identified a well-defined pocket consisting of the first CMY/CYR motif along with N389 and F396 (Fig. 2 C–E), and the second CPY/CYR motif along with N431 and Y438 (Fig. S2C). These residues are highly conserved in APLF homologs and in other PBZ domains, except for M380, which is often P, R, K, or Q. Although this pocket is positively charged with R387 in ZF1 and R429 in ZF2, it is also hydrophobic due to the presence of Y381 and M380 in the C1 loop and Y386 in the C2 loop (Fig. 1). Therefore, the C(M/P)Y and CYR motifs are key elements of this ADPr-binding basic/hydrophobic pocket, and are most likely important for the interaction with PAR. Interestingly, this basic/hydrophobic pocket is adjacent to a large positively charged area formed by residues R387, K388, and N389 in ZF1 (Fig. 2E) and R429, K430, and N431 in ZF2 (Fig. S2C), which may provide additional interaction sites for the negatively charged phosphate groups of PAR chains.
Similar Mechanisms of ADPr Recognition are Utilized by PBZ and Macrodomains.
In order to gain insight into how ADPr interacts with our PBZ structure, we used HADDOCK (20) to build a structural model of an APLF ZF1 and ADPr complex using the experimental constraints described in the SI Text. The resulting model (Fig. 2C and Fig. S3A) showed that the adenine ring is sandwiched between the aromatic rings of Y386 and Y381, and the phosphate groups are close to the side chains of R387 and N389. Interestingly, this ADPr-binding site resembles that of the macrodomain of archaebacterial Af1521 (4) whereby the conserved Y176 residue stacks with the adenine ring of the ADPr molecule (Fig. S3B). A similar mode of interaction between ADPr and the macrodomain of histone macroH2A1.1 has also been recently reported (5). This structural similarity in the nucleotide binding site strongly argues that these conserved tyrosine residues are critical for nucleotide recognition.
Tandem APLF PBZ Domains Enhance PAR-Binding.
The APLF ZFs appear to primarily recognize ADPr independently. Furthermore, we found that TZF, ZF1, and ZF2 all bound to ADPr with similar affinities (in the range of Kd 10-3 M) as determined from our NMR titration experiments (Table S3). However, in the context of interacting with longer PAR polymers, we questioned whether the tethered tandem ZF domains would exhibit different binding properties. To examine this further, we performed surface plasmon resonance (SPR) and dot blot analyses. Remarkably, TZF displayed over 1,000-fold higher binding affinity (Kd 9.5 × 10-10 M) compared to ZF1 or ZF2 (Fig. 3B and Table S2). Interestingly, the PAR-binding affinity of the isolated ZF1 peptide (Kd 5.2 × 10-7 M) was almost 20-fold greater than ZF2 (Kd 8.3 × 10-6 M). Consistent with these observations, the mutant ZFY1 disrupted PAR binding to a greater extent than ZFY2, whereas ZFY4 completely abolished PAR binding in vitro (Fig. 3). These data indicate that substitutions within ZF1 were generally less tolerated in terms of in vitro PAR binding than those in ZF2 (Fig. 3). Furthermore, the higher binding affinities for PAR over ADPr, even with the isolated ZF domains, suggests that the covalent linkage between multiple ADPr subunits promotes a significant shift to the bound state in the binding equilibrium, which may be further augmented by higher local concentrations of ADPr in the context of PAR polymer.
Fig. 3.
The tandem APLF PBZ domains enhance interactions with PAR. (A) APLF peptides as indicated were spotted onto nitrocellulose, incubated with 32P-radio-labeled PAR polymers and visualized using autoradiography. (B) SPR-derived association constants (Ka) of APLF peptides with PAR. (Inset) TZF-PAR binding by SPR. FL, full-length APLF; RU, response units.
C(M/P)Y and CYR Motifs are Critical for PAR-Binding in Vivo.
Our structural and biochemical data suggested that the C(M/P)Y and CYR motifs were critical for ADPr and PAR binding in vitro. To further examine this in vivo, we employed real-time live cell imaging analyses following laser microirradiation of a defined nuclear subvolume of U2OS cells stably expressing full-length EGFP-tagged APLF proteins, and monitored EGFP-APLF recruitment kinetics to sites of laser-induced DNA damage (Fig. S6). As was the case in vitro, we found that substitutions within ZF1 had more deleterious consequences on EGFP-APLF recruitment kinetics than those in ZF2. For example, the Y386A, R376A, and M380P more severely impaired the recruitment of EGFP-APLF to sites of laser-induced DNA damage than the equivalent substitutions within ZF2 (Y428A, R418A, and P422M, respectively) (Fig. 4). Moreover, the mutants ZFY1 and ZFY2 severely impaired EGFP-APLF recruitment and were as deleterious as the ZF1m and ZF2m mutants, which substitute both zinc-binding cysteine residues to glycine in ZF1 and ZF2, respectively (Fig. 4). Therefore, the C(M/P)Y and CYR motifs are important not only for ADPr and PAR binding in vitro but also for PAR binding in vivo.
Fig. 4.
Recruitment kinetics of EGFP-APLF and PBZ mutants following laser-induced DNA damage. (A) Live cell imaging analyses of U2OS cells stably expressing EGFP-APLF proteins (full-length) as indicated following laser microirradiation and time-lapse imaging over 5 min. (B) Quantitation of the recruitment kinetics in A. The average relative fluorescence (± SE) of EGFP-APLF from at least 10 cells from three independent experiments was plotted over the 5-min time course.
Discussion
APLF PBZ Structure Represents a Variant Among the Zinc Finger Superfamily.
ZFs are an extremely abundant structural motif mediating diverse biological functions (21). ZFs were originally identified as specific DNA-binding motifs, and are now also known to be involved in the recognition of proteins, RNA and PAR. We compared our PBZ structure with various structures of ZF proteins and we found that the PBZ domain is distinct from other representative Zn2+ binding structures in terms of structural topology and zinc coordination (Fig. S2B). The transcription factor SP1 contains the classic C2H2 ZF motif (Fig. S2B), which recognizes double-stranded DNA (22). Although the C2H2 motif found in APLF resembles this classic DNA-binding ZF motif, a low amino acid conservation (10% sequence identity between the SP1 ZF motif and APLF ZF1) and the different spacing of the zinc coordinating residues result in two distinct structural topologies, as illustrated in Fig. S2B. PARP1, which plays a key role in the DNA damage response, contains three ZFs, two of which use the treble clef type CCHC finger motif composed of an antiparallel β-sheet and an α-helix, which is markedly different from our PBZ structure (Fig. S2B). Interestingly, the RNA binding CCCH zinc fingers of TIS11d (19) are similar to the PBZ domain in its global backbone trace, despite the marked differences in secondary structure topology and zinc coordination. In TIS11d, RNA binds to conserved aromatic residues (Y170/Y208 and F176/F214). The structural configuration of these aromatic residues on the solvent exposed surface resembles that of the corresponding residues in APLF (Y381/Y423 and Y386/Y428), suggesting a functionally conserved role of this Tyr/Phe-rich surface in nucleotide binding.
Functional Significance of Tandem PBZ Domains
There are at least 30 known PBZ-containing proteins, the majority of which contain only a single PBZ domain. In contrast, there are only three known tandem PBZ-containing proteins, including APLF (Fig. 1A). Therefore, we posed the question of whether or not the presence of two adjacent PBZ domains may be of functional significance. Our NMR and SPR data demonstrated that the isolated ZF1 and ZF2 of APLF can independently bind ADPr and PAR. However, we found that the TZF peptide bound to PAR polymer with a remarkably higher affinity (Kd 9.5 × 10-10 M) than the individual ZF domains (Table S2). In contrast, TZF did not demonstrate enhancement in ADPr binding, as TZF, ZF1, and ZF2 bound to ADPr with similar affinities (in the range of Kd 10-3 M; Table S3). We also found that the isolated ZF1 peptide had a higher affinity for PAR (Kd 5.2 × 10-7 M) compared to ZF2 (Kd 8.3 × 10-6 M), and that mutations in ZF1 had more deleterious effects on in vitro and in vivo PAR binding compared to those in ZF2 (Figs. 3 and 4). Consistent with our results, a more severe impairment in in vitro PAR binding was also observed when the zinc-binding cysteine residues were substituted to alanine in the first compared to the second APLF ZF domain as judged by dot blot analyses (7). Collectively, these observations suggest that the first APLF ZF may serve as the primary high affinity “anchoring” site for PAR, and that the close proximity between the first and second ZF domains generates synergy in PAR binding as discussed below.
To further understand the nature of the difference in PAR binding between ZF1 and ZF2, we carried out a more detailed structural comparison of the two ZFs. Despite the similarity in zinc ligation and topological fold, we found a significant difference in the hydrophobic core structure between ZF1 and ZF2. In ZF1, the first seven residues (P399GDSDYG405) of the interdomain linker interact with both strands of the β-sheet, and together contribute to an extensive hydrophobic core of this ZF domain (Fig. 1 B and Fig. S1A). R376 in β1 and D401/D403 from the linker are involved in electrostatic interactions that appear to further stabilize the ZF1-linker interaction. The {1H}-15N NOE measurement showed that the first seven residues of the linker possessed the same mobility as the core structure of ZF1 (average NOE value at ∼0.6; Fig. S1C). Four out of the seven residues in the linker are invariant in APLF homologs (P399, D401, D403, and Y404) (9) and are conserved among the other tandem PBZ-containing proteins (Fig. 1A). In contrast, the corresponding residues in single PBZ-containing proteins are not conserved. We have defined the ZF1 domain from residue C379 to H398 based on the function of these conserved residues in zinc coordination. However, based on our structural data, it is possible that these seven “linker” residues in fact comprise part of the ZF1 fold. In either scenario, these differences in the protein hydrophobic core structure between ZF1 and ZF2 may contribute to the structural stability and to the property of PAR binding. It should be noted that our structural data were collected on a truncated protein and we cannot rule out the possibility that other portion(s) of the protein might contribute to the structural stability and/or PAR-binding properties of ZF2. Nevertheless, when we compared the substitutions R376A and R418A in the context of the full-length EGFP-APLF protein in vivo, we found that R376A was far more deleterious to EGFP-APLF recruitment kinetics than R418A (Fig. 4). Although it is unclear exactly how R376 contributes to PAR binding in vivo, we speculate that electrostatic interactions between R376 and the conserved residues D401/D403 serve to stabilize ZF1–PAR interactions.
Flexible Linker Region.
In the present structure of APLF TZF, the relative orientation of ZF1 and ZF2 could not be determined due to the lack of sufficient distance restraints between the two ZFs. A low value of {1H}-15N heteronuclear NOEs was observed for residues G406–D416, suggesting that this portion of the domain linker is flexible (average NOE ∼ 0.2), which is consistent with our finding of a “disordered” linker region in the ensemble of TZF structures (Fig. S1C). To further examine the nature of the domain linker and the domain orientation, we employed residual dipolar coupling (RDC) measurements on TZF. We found that the experimental RDC data for each domain fit well with the individual structures of ZF1 and ZF2. However, the same dataset of RDC did not fit well with any of the ensemble structures of TZF (Fig. S1D). Furthermore, we found that the NMR spectra of the isolated ZF1 and ZF2 peptides were virtually superimposable with the spectrum of TZF (Fig. S1E). Collectively, our structure and NMR data show that APLF TZF is made up with two relatively independent domains connected by a largely flexible linker due to the high degree of internal mobility of residues G406–D416. It has previously been demonstrated that disordered linker regions of other ZF proteins may become ordered upon ligand binding. Indeed, the disorder-to-order transition of the TFIIIA ZF linker induced upon DNA binding is known to contribute to DNA binding (23). Furthermore, it has been suggested that linker length is a critical determinant of binding affinity, and that induced folding of relatively longer linkers (such as that of APLF) could occur upon binding to ligand (24). Therefore, it is tempting to speculate that the increased affinity of TZF for PAR is due to enhanced ordering of the linker region upon PAR binding. It is also possible that the ZF1 and ZF2 core regions make new interdomain contacts only when complexed with PAR. Such a ligand-induced conformational allostery in APLF could provide a structural explanation for the observed dramatic enhancement in PAR binding (over 1,000-fold) with the tandem PBZ domains of TZF relative to the single domains, ZF1 or ZF2. Further studies are needed to elucidate how exactly the APLF TZF domain displays this marked enhancement in binding to the polymer chain of PAR. Unfortunately, homogenous PAR polymer or oligomer in a well-defined length, which is required for NMR or X-ray crystallography, is currently unavailable.
Comparison Between PBZ Domains and PAR Binding Macrodomains.
Certain macrodomains interact with both ADPr and PAR, including that of histone macroH2A1.1. The crystal structure of the ADPr-bound macrodomain of macroH2A1.1 identified specific interactions with the 2′ OH and 3′ OH groups of the proximal ribose in a deep pocket of the binding site, therefore making the macrodomain incompatible with binding continuously along PAR polymers. This suggests an important functional distinction between the proposed PAR-binding mechanisms of the APLF PBZ domains and the macrodomain of macroH2A1.1. The histone macroH2A1.1 macrodomain does not likely bind along PAR polymers, but specifically recognizes terminal ADP-ribose rings, hence functioning to cap the PAR chain (5). In contrast, the basic/hydrophobic ADPr-binding site of PBZ domains is largely exposed. By virtue of its tandem PBZ motifs and the molecular mobility, APLF would specifically and strongly bind multiple ADP-ribose residues along PAR polymers in vivo. We speculate that these different types of PAR-binding activities contribute to functionally distinct mechanisms in vivo.
Materials and Methods
Cloning and Plasmid Constructions.
To generate APLF TZF (pGEX4T3-APLF360–448), pGEX4T3-APLF (9) was digested with BamHI and SphI, blunt-ended, religated in-frame, and a stop codon was inserted by site-directed mutagenesis to truncate the GST-fusion protein after APLF amino acid residue 448. ZF1 and ZF2 (pGEX4T1-APLF368–410 and pGEX4T1-APLF411–448, respectively) were PCR-amplified from pGEX4T3-APLF360–448, digested with BamHI and XhoI, and these fragments were ligated in-frame into BamHI/XhoI digested pGEX4T1. PEGFPC2-APLF was generated by HindIII/ApaI digestion of pcDNA3.1/V5-His-APLF (9) and ligating in-frame into HindIII/ApaI digested pEGFP-C2 (Stratagene). All mutations were generated by QuikChange site-directed mutagenesis (Stratagene), using either pEGFPC2-APLF or pGEX4T3-APLF360–448 APLF TZF as templates. Plasmid constructions were verified by sequence analysis.
NMR Spectroscopy.
NMR samples were prepared in a buffer containing 20 mM Bis-Tris pH 6.0 (22 °C), 50 mM NaCl, 50 μM ZnCl2, and 1 mM NaN3 in 93% H2O/7% D2O, or 99% D2O, except for the RDC samples, which required an elevated salt concentration of 500 mM NaCl. NMR spectra were acquired at 25 °C on Bruker Avance 600 and 800 MHz spectrometers, both equipped with cryoprobes. Triple resonance spectra were collected on 13C/15N-labeled APLF TZF at a concentration of 0.5–0.8 mM. Complete protein backbone resonance assignments for 1H, 13C, and 15N nuclei were obtained using standard procedures as previously described (25). Further experimental details are described in SI Text.
Structure Calculations and Molecular Docking.
The three-dimensional structure of APLF TZF domain was calculated using CYANA (26), and further water-refinement of structures was performed using the RECOORD scripts (27) in Crystallography and NMR System (28). The lowest energy structure of ZF1 in the APLF TZF domain was used as a template for HADDOCK calculations (20), which provided a molecular model for the ZF1–ADPr interaction. Specific details of these computational procedures are provided in the SI Text.
Supplementary Material
Acknowledgments.
We thank the Advanced Optical Microscopy Facility at the Ontario Cancer Institute for support with live-cell imaging analyses. This work was funded by the Canadian Institute of Health Research (C.A.K.). NMR instruments were supported by the Canadian Foundation of Innovation. M.I. holds a Canada Research Chair. A.L.F. is supported by the Excellence in Radiation Research for the 21st Century Program.
Note added in proof.
While our manuscript was under review, similar results were published by Eustermann et al. (29).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB code 2KUO). Assignments of 1H, 13C, and 15N chemical shifts for APLF TZF (360–448) have been deposited in the BioMagResBank (BMRB code 16744).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000556107/-/DCSupplemental.
References
- 1.Rouleau M, Aubin RA, Poirier GG. Poly(ADP-ribosyl)ated chromatin domains: Access granted. J Cell Sci. 2004;117(Pt 6):815–825. doi: 10.1242/jcs.01080. [DOI] [PubMed] [Google Scholar]
- 2.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 3.Desnoyers S, et al. Biochemical properties and function of poly(ADP-ribose) glycohydrolase. Biochimie. 1995;77(6):433–438. doi: 10.1016/0300-9084(96)88156-9. [DOI] [PubMed] [Google Scholar]
- 4.Karras GI, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24(11):1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16(9):923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 6.Gagne JP, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36(22):6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451(7174):81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 8.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol. 2007;27(10):3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair. 2008;7(2):292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Kanno S, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 2007;26(8):2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bekker-Jensen S, et al. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J Biol Chem. 2007;282(27):19638–19643. doi: 10.1074/jbc.C700060200. [DOI] [PubMed] [Google Scholar]
- 12.Koch CA, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23(19):3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loizou JI, et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117(1):17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 14.Clements PM, et al. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair. 2004;3(11):1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Luo H, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol Cell Biol. 2004;24(19):8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein NK, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17(5):657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol. 2008;28(14):4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003;31(2):532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11(3):257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 20.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 21.Laity JH, Lee BM, Wright PE. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 22.Narayan VA, Kriwacki RW, Caradonna JP. Structures of zinc finger domains from transcription factor Sp1. Insights into sequence-specific protein-DNA recognition. J Biol Chem. 1997;272(12):7801–7809. doi: 10.1074/jbc.272.12.7801. [DOI] [PubMed] [Google Scholar]
- 23.Laity JH, Dyson HJ, Wright PE. DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000;295(4):719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- 24.Shamoo Y, Abdul-Manan N, Williams KR. Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res. 1995;23(5):725–728. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plevin MJ, Zhang J, Guo C, Roeder RG, Ikura M. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proc Natl Acad Sci USA. 2006;103(27):10242–10247. doi: 10.1073/pnas.0603463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 27.Nederveen AJ, et al. RECOORD: A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59(4):662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Eustermann S, et al. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat Struct Mol Biol. 2010;17(2):241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.