Abstract
The genome has often been called the operating system (OS) for a living organism. A computer OS is described by a regulatory control network termed the call graph, which is analogous to the transcriptional regulatory network in a cell. To apply our firsthand knowledge of the architecture of software systems to understand cellular design principles, we present a comparison between the transcriptional regulatory network of a well-studied bacterium (Escherichia coli) and the call graph of a canonical OS (Linux) in terms of topology and evolution. We show that both networks have a fundamentally hierarchical layout, but there is a key difference: The transcriptional regulatory network possesses a few global regulators at the top and many targets at the bottom; conversely, the call graph has many regulators controlling a small set of generic functions. This top-heavy organization leads to highly overlapping functional modules in the call graph, in contrast to the relatively independent modules in the regulatory network. We further develop a way to measure evolutionary rates comparably between the two networks and explain this difference in terms of network evolution. The process of biological evolution via random mutation and subsequent selection tightly constrains the evolution of regulatory network hubs. The call graph, however, exhibits rapid evolution of its highly connected generic components, made possible by designers’ continual fine-tuning. These findings stem from the design principles of the two systems: robustness for biological systems and cost effectiveness (reuse) for software systems.
Keywords: systems biology, adaptive complex systems
Complex systems are characterized by interactions among huge numbers of heterogeneous constituents. In particular, many complex systems are adaptive, meaning the interconnections are shaped progressively by a changing environment. The driving forces of adaptation are common design principles such as the reduction of cost and the enhancement of system robustness (1). Optimal solutions are determined by trade-offs between conflicting principles and therefore vary from system to system. Over the past decade, the study of networks has emerged as an interdisciplinary research field aiming to discover the underlying principles of complex systems and to develop tools or algorithms for analyzing them. By capturing the interconnections between individual components, networks not only serve as backbones to study the emergent properties of complex systems, but they also provide an abstract framework that facilitates the cross-disciplinary comparison of different adaptive complex systems, ranging from biological systems to technological ones (2). Cross-disciplinary comparison between biological systems and commonplace systems such as organization hierarchies (3, 4) and engineering devices should be of particular interest to systems biologists. Despite tremendous advancement in high-throughput experiments and computational algorithms, the study of biological systems in general still suffers from limitations in accuracy and completeness of data. Insights gained from systems in which we have direct access and thorough understanding can leverage our knowledge to biological ones.
Like biological systems, software systems such as a computer operating system (OS) are adaptive systems undergoing evolution. Whereas the evolution of biological systems is subject to natural selection, the evolution of software systems is under the constraints of hardware architecture and customer requirements. Since the pioneering work of Lehman (5), the evolutionary pressure on software has been studied among engineers. Interestingly enough, biological and software systems both execute information processing tasks. Whereas biological information processing is mediated by complex interactions between genes, proteins, and various small molecules, software systems exhibit a comparable level of complexity in the interconnections between functions. Understanding the structure and evolution of their underlying networks sheds light on the design principles of both natural and man-made information processing systems.
The master control plan of a cell is its transcriptional regulatory network. The transcriptional regulatory network coordinates gene expression in response to environmental and intracellular signals, resulting in the execution of cellular processes such as cell divisions and metabolism. Understanding how cellular control processes are orchestrated by transcription factors (TFs) is a fundamental objective of systems biology (6–9), and therefore a great deal of effort has been focused on understanding the structure and evolution of transcriptional regulatory networks. Analogous to the transcriptional regulatory network in a cell, a computer OS consists of thousands of functions organized into a so-called call graph, which is a directed network whose nodes are functions with directed edges leading from a function to each other function it calls. Whereas the genome-wide transcriptional regulatory network and the call graph are static representations of all possible regulatory relationships and calls, both transcription regulation and function activation are dynamic. Different sets of transcription factors and target genes forming so-called functional modules (10) are activated at different times and in response to different environmental conditions. In the same way, complex OSs are organized into modules consisting of functions that are executed for various tasks.
Here we perform a one-to-one comparison between the transcriptional regulatory network of Escherichia coli and the call graph of the Linux kernel, which are both canonical systems. E. coli is one of the most well-annotated model organisms. The study of its transcriptional regulatory network has a long history (11–15). On the software side, the Linux kernel is the central component of one of the most popular and well-documented OSs. Since its creation by Linus Torvalds in 1991, it has been continuously revised, and its source lines of code has increased from around 10,000 in the original version 0.01 to more than 12 million in version 2.6.33. Therefore, the two systems are ideal candidates for an in-depth cross-disciplinary comparison.
Results
Comparison of Basic Topology and Hierarchical Structure.
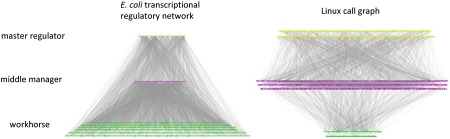
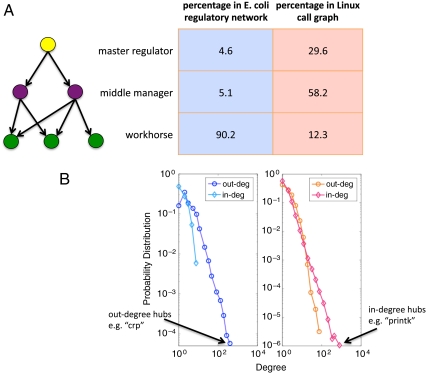
In a directed network, the in-degree and out-degree of a node refer to the number of regulators calling the node and the number of target genes or functions called by the node, respectively. The networks of interest in this study are displayed in Fig. 1 and their key attributes are listed in Table 1. As discussed in earlier studies (3, 13), transcriptional regulatory networks exhibit a characteristic pyramidal hierarchical layout, in which there are a few master TFs on the top and most TFs are at the middle, regulating a set of non-TF target genes. We refer to these non-TF targets as workhorses (16). The existence of a hierarchical organization implies the existence of a downward information flow in response to various forms of stimuli. The Linux call graph has a similar intrinsic direction, where the chain of command starts from high-level starting functions like “main” and flows to many other downstream functions following the outgoing edges. To further investigate the structure of the two networks, we divide nodes into three categories (Fig. 1): master regulators (nodes with zero in-degree), workhorses (nodes with zero out-degree), and middle managers (nodes with nonzero in- and out-degree). Fig. 2A shows the distribution of these categories. In the E. coli transcriptional regulatory network, the fraction of workhorses is large and the top two layers each comprise less than 5% of the total number of genes. In the call graph, on the contrary, over 80% of functions are located in the upper levels of the hierarchy. In other words, unlike the conventional pyramidal hierarchy exhibited by the E. coli transcriptional regulatory network, the Linux call graph exhibits a top-heavy structure.
Fig. 1.
The hierarchical layout of the E. coli transcriptional regulatory network and the Linux call graph. (Left) The transcriptional regulatory network of E. coli. (Right) The call graph of the Linux Kernel. Nodes are classified into three categories on the basis of their location in the hierarchy: master regulators (nodes with zero in-degree, Yellow), workhorses (nodes with zero out-degree, Green), and middle managers (nodes with nonzero in- and out-degree, Purple). Persistent genes and persistent functions (as defined in the main text) are shown in a larger size. The majority of persistent genes are located at the workhorse level, but persistent functions are underrepresented in the workhorse level. For easy visualization of the Linux call graph, we sampled 10% of the nodes for display. Under the sampling, the relative portion of nodes in the three levels and the ratio between persistent and nonpersistent nodes are preserved compared to the original network. The entire E. coli transcriptional regulatory network is displayed.
Table 1.
Statistics of the E. coli regulatory network and the Linux call graph
|
E. coli transcriptional regulatory network |
Linux call graph |
|
| Number of nodes | 1,378 | 12,391 |
| Number of persistent nodes | 72* (5%) | 5,120 (41%) |
| Number of edges | 2,967 | 33,553 |
| Number of modules | 64 | 3,665 |
| Number of comparative references | 200 bacterial genomes | 24 versions of kernels |
| Years of evolution | Billions | 20 |
*In the E. coli genome 72 out of 212 persistent genes could be mapped to the transcriptional regulatory network.
Fig. 2.
Comparison of the E. coli transcriptional regulatory network and Linux call graph in terms of topology and hierarchical structure. (A) The distribution of the three categories in the E. coli transcriptional regulatory network and the Linux call graph. The transcriptional regulatory network (1,378 nodes) follows a conventional hierarchical picture, with a few top regulators and many workhorse proteins. The Linux call graph (12,391 nodes), on the other hand, possesses many regulators; the number of workhorse routines is much lower in proportion. (B) Degree distributions of the E. coli transcriptional regulatory network and the Linux call graph. The regulatory network has a broad out-degree distribution but a narrow in-degree distribution. The situation is reversed in the call graph, where we can find in-degree hubs, but the out-degree distribution is rather narrow. An out-degree hub in the E. coli regulatory network and an in-degree hub in the Linux call graph are shown.
The discrepancy we find in the hierarchical organization is related to the discrepancy in-degree distribution. Like other complex networks such as social networks and the World Wide Web, both transcriptional regulatory networks and call graphs possess hubs, the highly connected nodes at the tail of the skewed degree distribution (17). The Linux call graph possesses in-degree hubs (nodes with many incoming edges) but no out-degree hubs (nodes with a high number of outgoing edges) (see Fig. 2B). The skewed in-degree distribution has been reported in software networks other than the Linux call graph (18). In particular, in-degree hubs in the Linux call graph are enriched at the bottom of the network hierarchy. They are workhorses called by a large number of regulators from the upper levels. In contrast, in the E. coli regulatory network, there are hubs with high out-degree but not high in-degree; i.e., no gene is regulated by many different transcription factors (see Fig. 2B). The out-degree hubs in the E. coli regulatory network regulate many workhorses at the bottom of the hierarchy.
Comparison of Functional Modules and Node Reuse.
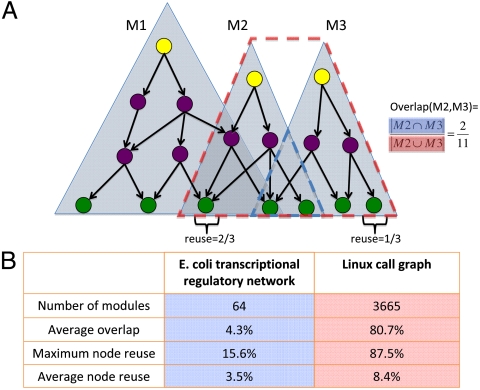
Modularity is an important concept in both biology and engineering (19). In fact, the technique of modular programming is widely employed in modern software design (20). As discussed earlier, dynamical functional modules expressed under different conditions in transcriptional regulatory networks resemble the modules of functions responsible for different computational tasks. Modules can be labeled naturally by the master regulators controlling them, because every middle manager and workhorse in the hierarchy is controlled by at least one master regulator. Modules defined in this way have been termed regulons (15) or origons (21). Specifically, we define a functional module in both call graphs and transcriptional regulatory networks as the subnetwork that consists of all the downstream nodes executed or controlled by a specific master regulator (Fig. 3A).
Fig. 3.
Modules in the E. coli transcriptional regulatory network and Linux call graph. (A) Definition of modules, reuse, and overlap. A module is characterized by a master regulator, with zero in-degree, and all of the nodes regulated directly or indirectly by the master regulator. Here there are three modules (M1, M2, and M3) represented by three triangles. Reuse of a node is defined as the fraction of modules to which the node belongs. This quantity is illustrated with the two labeled nodes. One is shared by M1 and M2 but not M3, and thus the reuse is 2/3. The other belongs to only M3; its reuse is therefore 1/3. The overlap between a pair of modules is defined by the size of their intersection normalized by their union. The overlap of M2 and M3 is thus 2/11. (B) Statistics of modules in the E. coli transcriptional regulatory network and the Linux call graph. The average overlap is given by the mean overlap between pairs of randomly chosen modules. Nodes in the call graph are in general more generic; i.e., they are reused by more modules.
Many nodes can be members of several different functional modules. To quantify this phenomenon, we define the reuse of a node on the basis of the fraction of modules in the network to which it belongs. Nodes with high reuse are called generic. Unsurprisingly, we find that the in-degree hubs are executed most often and thus are more reusable than other nodes (Pearson correlation r = 0.16, P < 10-95 for the Linux call graph, and r = 0.53, P < 10-100 for the E. coli regulatory network). The most generic function is the well known function “printk,” which is responsible for standard display and thus called by over 90% functional modules. In the E. coli regulatory network, one of the most generic nodes is the outer membrane porin “ompF” that controls the diffusion of various metabolites. It is reused by 20% of the modules. Generally speaking, nodes in the Linux call graph have on average higher reuse than those in the E. coli transcriptional regulatory network (8.4% and 3.5%, respectively, P < 10-12 in t test; see Fig. 3B). The difference is topologically attributed to the pyramidal versus top-heavy organization. The narrow base in the Linux call graph leads to a higher average reuse. Indeed, many generic functions are workhorses such as string manipulation function “strlen.”
As shown in Fig. 3B, one of the most striking differences concerning the organization of modules in the transcriptional regulatory network and the call graph is the overlap of modules. In the Linux call graph, two randomly chosen modules overlap by more than 80%. On the other hand, the average overlap in the E. coli transcriptional regulatory network is less than 5%. We shall discuss later how such differences in the overlap of modules play a key role in robustness and fragility of the two systems.
Comparison of Network Evolution and Node Persistence.
The core components of a system are usually those that survive the evolutionary process. It is instructive to study those “survivors” in both the E. coli transcriptional regulatory network and the Linux call graph. In the Linux kernel, we focus on persistent functions, defined as those that exist in every version of software development. Persistent functions in software systems are analogous to persistent genes in biological systems, which are genes that are consistently present in a large number of genomes (22). We identified persistent functions in the Linux kernel on the basis of their appearance in all versions of the Linux source code used in this study and persistent genes in the E. coli genome by examining their distribution across a group of over 200 phylogenetically diverse bacterial genomes (see Materials and Methods for details). As shown in Fig. 1, most persistent genes in the E. coli regulatory network are workhorses: 71 out of 72 compared to 1,243 out of 1,378 for all genes (P < 10-3 by permutation test). On the other hand, in the Linux call graph, persistent functions are present at all three levels but are significantly enriched only among the master regulators and middle managers (4,680 out of 5,120 persistent functions are master regulators and middle managers, compared to 10,872 out of 12,391 for all functions, P < 10-100 by permutation test).
Of particular interest is the relationship between reuse and persistence among the workhorses. Specifically, we observe opposite correlation behaviors in the two systems: Reuse and persistence are negatively correlated in the E. coli regulatory network but positively correlated in the Linux call graph [Spearman correlation r = -0.074 (P < 0.01) and r = 0.10 (P < 10-4), respectively]. In other words, specialized nodes are more likely to be preserved in the regulatory network, but generic or reusable functions are persistent in the Linux call graph.
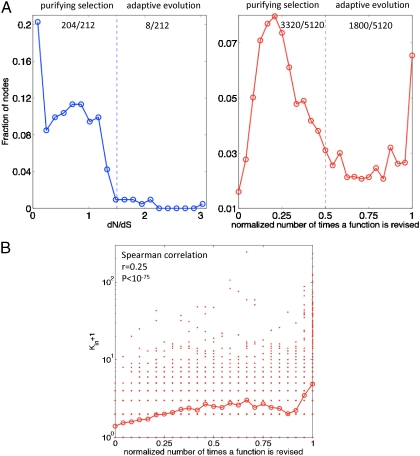
The idea of persistence is closely related to the rate of evolution. In biological systems, the fundamental components of life exist in every genome independently of environmental conditions. These persistent genes, say, ribosomal proteins and dnaA, are under high selective pressure and evolve very slowly. For example, the ratio of nonsynonymous to synonymous substitutions (dN/dS) is smaller among the persistent genes (or, rather, their corresponding proteins) than in the overall E. coli proteome (Wilcoxon rank-sum test P < 10-44), suggesting that the selection pressure on persistent genes is indeed stronger. As shown in Fig. 4A (Left), the distribution of dN/dS among persistent genes in E. coli has a gradual decreasing trend. Most of the 212 persistent genes are under strong constraints; only 51 of them are under positive selection (dN/dS > 1). Among the 51, only 8 are outliers that exhibit relatively fast evolution (right portion of the panel). The situation is remarkably different for persistent functions in the Linux kernel. Fig. 4A (Right) shows the number of times persistent functions were revised in the source code. The bimodal nature of the plot suggests that persistent functions fall into two classes. Though the majority of functions (3,320 out of 5,120, 65%) such as the string manipulation function “strlen” are not revised very often, a significant fraction of functions (1,800 out of 5,120, 35%) in the Linux call graph are undergoing adaptive evolution; i.e., they are evolving rapidly in response to external conditions. In fact, 335 (19%) of them were updated in every version, including a set of functions related to memory management such as “mempool_alloc.” The evolutionary features of persistent functions are connected to their topological features. As shown in Fig. 4B, the rate of revision of the functions is actually positively correlated with their in-degree (Spearman correlation r = 0.26, P < 10-75); i.e., highly reused functions are revised more often. In fact, the adaptive functions distinguish themselves by having higher values of reuse (12.6% versus 4.4%, Wilcoxon rank-sum test P < 10-20) than the conservative functions.
Fig. 4.
The rate of evolution of persistent genes and persistent functions. (A) Distribution of the rate of evolution. In the case of the E. coli transcriptional regulatory network (Left), the rate of evolution is quantified by dN/dS, the ratio of nonsynonymous to synonymous substitution rate. On the basis of the rate of evolution, we divide the histogram into two parts representing genes evolving in a more conservative (Left) or a more adaptive (Right) way, respectively. The overall trend of the distribution is decreasing: 204 out of 212 persistent genes are evolving under purifying selection, and only 8 out of 212 undergo some degree of adaptive evolution. The fraction of genes under positive selection, by definition dN/dS > 1, is 51 out of 212. In the case of the Linux call graph (Right), we quantify the rate of evolution by the number of revisions to the function in the source code. That number is then normalized by the total number of releases we studied—i.e., 24 (refer to Materials and Methods). The distribution is bimodal: 3,320 out of 5,120 persistent functions are revised infrequently (left portion), but there are 1,800 persistent functions that are adaptive (right portion) and 335 of them got updated in every version. (B) Correlation between the in-degree (Kin + 1) and the rate of evolution in persistent functions. In the Linux call graph, the rate of revision of persistent functions is positively correlated with their in-degrees (Spearman correlation r = 0.25). Highly used functions are revised more often. (Note that more than one persistent function may coincide at a single dot shown in the scatter plot. Each open circle represents the geometric mean in the corresponding bin.)
Discussion
We have presented a comparative analysis between the transcriptional regulatory network of E. coli and the call graph of the Linux operating system and explored their similarities and differences in hierarchical structure, modularity of organization, and persistence of nodes. A summary of the comparison can be found in Table 2. The two networks are shaped by different underlying design principles, which are deeply connected to the interplay between the systems and their environments. From a topological standpoint, it is intriguing that two distinct evolutionary processes both lead to the emergence of hierarchy in the control and regulation layouts, probably because hierarchy is a most effective way to transfer information and coordinate processes. Nevertheless, we have observed several intrinsic differences between the two hierarchical networks. To a certain extent, the presence of in-degree hub functions and the top-heavy hierarchy found in the call graph can be readily explained by common programming practices. In general, for the sake of clarity and easy debugging, programmers are encouraged to break down a code into pieces and reuse certain functions; functions that are called by many others, i.e., in-degree hubs, are therefore favored. The reuse of code leads to generic functions, which also accounts for the increase of overlap between modules in the Linux call graph. These programming practices are rooted in considerations of cost effectiveness. From an engineering point of view, the reuse of common nodes between modules is a cost-effective way to construct a complex system. However, such optimized usage of functions comes at the expense of robustness, because breakdown of a generic function causes problems in many modules. More importantly, generic functions lead to potential fragility in the sense that modifying any module may require compensating changes in a generic function. As a result, generic functions have to be updated more often (as reflected by the class of rapidly revising functions in Fig. 4A). The low overlap between modules in biological networks, on the other hand, increases robustness. Modules tend to work more independently by recruiting different sets of workhorses from the broad base of the network hierarchy.
Table 2.
One-to-one comparison between the E. coli regulatory network and the Linux call graph
|
E. coli transcriptional regulatory network |
Linux call graph |
||
| Basic properties of systems | Nodes | Genes (TFs & targets) | Functions (subroutines) |
| Edges | Transcriptional regulation | Function calls | |
| External constraints | Natural environment | Hardware architecture, customer requirements | |
| Origin of evolutionary changes | Random mutation & natural selection | Designers’ fine-tuning | |
| Hierarchical organization | Structure | Pyramidal | Top-heavy |
| Characteristic hubs | Upper-level TFs with high out-degree | Generic workhorse functions with high in-degree | |
| Organization of modules | Downstream modules as labeled by | Master TFs responsible for sensing environmental signals | High-level starting functions that initiate execution for specific tasks |
| Node reuse | Low | High | |
| Overlap between modules | Low | High | |
| Persistent nodes | Characteristics | Specialized (nongeneric) workhorses | Generic or reusable functions |
| Location in hierarchy | Mostly bottom | Mostly top | |
| Evolutionary rate | Mostly conservative (e.g., dnaA) | Conservative (e.g., strlen) & adaptive (e.g., mempool_alloc) | |
| Design principles | Building of hierarchy | Bottom up | Top down |
| Optimal solution favors | Robustness | Cost effectiveness (reuse of components) | |
The study of persistent genes in biological networks and persistent functions in call graphs offers insight into the evolution of hierarchies. Persistent genes form the core machinery of life, the so-called paleome (23). They usually are not regulators but workhorse genes that perform vital tasks. In fact, most persistent genes are enzymes. The enrichment of persistent genes at the bottom of the regulatory hierarchy in E. coli is in accordance with the view that orthologous proteins are rather similar in function whereas regulatory changes are the main driving forces of evolution (9). To a certain extent, biological evolution is building from the bottom to the top. In contrast, persistent functions in the Linux call graph are usually not bottom-level workhorses but “controllers.” This difference suggests that not only do software networks possess more regulators than workhorses, the regulators are maintained on purpose and thus the evolution goes from top to bottom.
The trade-off between robustness and cost effectiveness biological and software systems is deeply related to the nature of their evolutionary processes. Biological evolution is mediated by random mutations followed by natural selection; a hub protein in a biological network is in general hard to evolve because of the constraints imposed by its many interactions. This constrained evolution is evinced by the negative correlation between node centrality and evolutionary rate in biological networks (24, 25). The random mutation and selection process underlying biological evolution prohibits the frequent targeted changes required for nodes to become generic. The system is then forced to pay for maintaining a large set of specially designed components performing a variety of functions in response to environmental changes. In contrast, engineering systems are fundamentally different. Both in-degree and betweenness centrality (26) are positively correlated with the rate of revision in the Linux call graph (see Fig. 4B for in-degree, Spearman correlation r = 0.26, P < 10-82 for betweenness). In other words, in software engineering, a system that needs to continually adapt to new conditions is cost effective only by paying the price of constantly fine-tuning its most highly accessed functions.
Reuse is extremely common in designing man-made systems. For biological systems, to what extent they reuse their repertoires and by what means sustain robustness at the same time are questions of much interest. It was recently proposed that the repertoire of enzymes could be viewed as the toolbox of an organism (27). As the genome of an organism grows larger, it can reuse its tools more often and thus require fewer and fewer new tools for novel metabolic tasks. In other words, the number of enzymes grows slower than the number of transcription factors when the size of the genome increases. Previous studies (4) have made the related finding that as one moves towards more complex organisms, the transcriptional regulatory network has an increasingly top-heavy structure with a relatively narrow base. Thus, it may be that further analysis will demonstrate the increasing resemblance of more complex eukaryotic regulatory networks to the structure of the Linux call graph.
Materials and Methods
Network Information.
Data on the E. coli transcriptional regulatory network were obtained from RegulonDB (15). The largest connected component of the network consists of 1,378 genes with 2,967 interactions. Linux source code was downloaded from the Linux Kernel Archives (http://www.kernel.org). To address the evolution of the kernel, 24 stable versions were used, from 2.6.4 to 2.6.27, spanning from March, 2004 to October, 2008. In general, the release of a new version, say, from 2.5 to 2.6, is accompanied by major changes. We worked on the 24 releases restricted in version 2.6 and focused on the gradual evolution exhibited in these releases. For some of these releases, an additional patch was required in order to compile (SI Text). The source codes were compiled on a MacBook with a 2 GHz Intel Core 2 Duo processor and 2 GB of memory by using the compiler GCC 3.4.6, and call graphs were extracted from the compiled code by using the tool CodeViz (release 1.0.11) by Gorman (http://www.csn.ul.ie/~mel/projects/codeviz/) (see SI Text). The network analysis presented in this study was performed on the most recent version of the Linux kernel downloaded (v. 2.6.27), in which there are 12,391 functions related by 33,553 calls. The network can be downloaded from http://networks.gersteinlab.org/callgraph.
Persistent Genes.
The persistence index and the list of 212 persistent genes in E. coli K12 were obtained from ref. 22. Among them, 72 can be mapped to the largest component of the transcription regulatory network. We quantify conservation by the ratio of nonsynonymous to synonymous substitution rates (dN/dS) (28). The two rates were estimated by aligning E. coli K12 proteins with their orthologs from Salmonella typhimurium LT2. The list of orthologs was downloaded from the ATGC database (29). Alignment was done by using the tool PAL2NAL (30), and dN/dS values were estimated by the PAML package (31).
Persistent Functions.
A function is defined as persistent if it appears in all the compiled call graphs (v. 2.6.4 to v. 2.6.27). The list of persistent functions can be found at http://networks.gersteinlab.org/callgraph. In this definition, we do not take into account the precise changes in the code of the function. The frequency of revision for a particular function was estimated by parsing the patch files (see SI Text). A function is regarded as revised if there is any change in its code.
Supplementary Material
Acknowledgments.
We thank the anonymous reviewers whose valuable suggestions helped to improve the quality of the manuscript. K.-K.Y. acknowledges Lucas Lochovsky for useful discussion and critical reading of an early manuscript. K.-K.Y. acknowledges Kevin Yip for useful discussion. This work is supported by the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914771107/-/DCSupplemental.
References
- 1.Alon U. An Introduction to Systems Biology. London: Chapman & Hall/CRC; 2007. [Google Scholar]
- 2.Barabási A. LINKED: The New Science of Networks. Cambridge, MA: Perseus; 2002. [Google Scholar]
- 3.Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci USA. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhardwaj N, Yan KK, Gerstein M. Analysis of diverse regulatory networks in a hierarchical context shows consistent tendencies for collaboration in the middle levels. Proc Natl Acad Sci USA. 2010;107:6841–6846. doi: 10.1073/pnas.0910867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehman MM. Programs, life cycles, and laws of software evolution. Proc IEEE. 1980;68:1060–1076. [Google Scholar]
- 6.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 7.Bolouri H, Davidson EH. Modeling transcriptional regulatory networks. Bioessays. 2002;24:1118–1129. doi: 10.1002/bies.10189. [DOI] [PubMed] [Google Scholar]
- 8.Barabási A, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 9.Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Luscombe NM, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 11.Thieffry D, Huerta AM, Perez-Rueda E, Collado-Vides J. From specific gene regulation to genomic networks: A global analysis of transcriptional regulation in Escherichia coli. Bioessays. 1998;20:433–440. doi: 10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 13.Ma H, et al. An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs. Nucleic Acids Res. 2004;32:6643–6649. doi: 10.1093/nar/gkh1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seshasayee AS, Fraser GM, Babu MM, Luscombe NM. Principles of transcriptional regulation and evolution of the metabolic system in E. coli. Genome Res. 2009;19:79–91. doi: 10.1101/gr.079715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gama-Castro S, et al. RegulonDB (version 6.0): Gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maslov S, Sneppen K. Computational architecture of the yeast regulatory network. Phys Biol. 2005;2:S94–100. doi: 10.1088/1478-3975/2/4/S03. [DOI] [PubMed] [Google Scholar]
- 17.Barabasi AL, Albert R. Emergence scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 18.Myers CR. Software systems as complex networks: Structure, function, and evolvability of software collaboration graphs. Phys Rev E. 2003;68:046116. doi: 10.1103/PhysRevE.68.046116. [DOI] [PubMed] [Google Scholar]
- 19.Alon U. Biological networks: The tinkerer as an engineer. Science. 2003;301:1866–1867. doi: 10.1126/science.1089072. [DOI] [PubMed] [Google Scholar]
- 20.Parnas DL. On the criteria to be used in decomposing systems into modules. Commun ACM. 1972;15:1053–1058. [Google Scholar]
- 21.Balazsi G, Barabasi A, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang G, Rocha EPC, Danchin A. Persistence drives gene clustering in bacterial genomes. BMC Genomics. 2008;9:4. doi: 10.1186/1471-2164-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danchin A. Bacteria as computers making computers. FEMS Microbiol Rev. 2009;33:3–26. doi: 10.1111/j.1574-6976.2008.00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 25.Kim PM, Korbel JO, Gerstein MB. Positive selection at the protein network periphery: Evaluation in terms of structural constraints and cellular context. Proc Natl Acad Sci USA. 2007;104:20274–20279. doi: 10.1073/pnas.0710183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: Correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maslov S, Krishna S, Pang TY, Sneppen K. Toolbox model of evolution of prokaryotic metabolic networks and their regulation. Proc Natl Acad Sci USA. 2009;106:9743–9748. doi: 10.1073/pnas.0903206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novichkov PS, Ratnere I, Wolf YI, Koonin EV, Dubchak I. ATGC: A database of orthologous genes from closely related prokaryotic genomes and a research platform for microevolution of prokaryotes. Nucleic Acids Res. 2009;37:D448–454. doi: 10.1093/nar/gkn684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.