Abstract
About 30% of protein-coding genes in the human genome are related through two whole genome duplication (WGD) events. Although WGD is often credited with great evolutionary importance, the processes governing the retention of these genes and their biological significance remain unclear. One increasingly popular hypothesis is that dosage balance constraints are a major determinant of duplicate gene retention. We test this hypothesis and show that WGD-duplicated genes (ohnologs) have rarely experienced subsequent small-scale duplication (SSD) and are also refractory to copy number variation (CNV) in human populations and are thus likely to be sensitive to relative quantities (i.e., they are dosage-balanced). By contrast, genes that have experienced SSD in the vertebrate lineage are more likely to also display CNV. This supports the hypothesis of biased retention of dosage-balanced genes after WGD. We also show that ohnologs have a strong association with human disease. In particular, Down Syndrome (DS) caused by trisomy 21 is widely assumed to be caused by dosage effects, and 75% of previously reported candidate genes for this syndrome are ohnologs that experienced no other copy number changes. We propose the remaining dosage-balanced ohnologs on chromosome 21 as candidate DS genes. These observations clearly show a persistent resistance to dose changes in genes duplicated by WGD. Dosage balance constraints simultaneously explain duplicate gene retention and essentiality after WGD.
Keywords: whole genome duplication, copy number variation, Down Syndrome, trisomy 21
Early in the vertebrate lineage the genome of our simple ancestor experienced radical upheaval from two rounds of whole genome duplication (WGD) and the subsequent chromosomal rearrangement and loss of many of the duplicate copies (“ohnologs”) (1–3). Although only about 20–30% of the protein-coding genes in the human genome can be traced back to these events (ref. 3 and this study), the two tetraploid episodes in vertebrate history have frequently been credited with creating the conditions for the evolution of vertebrate complexity. Understanding the patterns of ohnolog retention is crucial to develop a unified model for the evolutionary impact of WGD and many groups have uncovered significant trends such as enrichment for developmental genes (4–6) and protein complex membership (7).
Recently it was shown that mammalian ohnologs are more essential (i.e., knockout of one copy is more likely to lead to sterility or inviability) than paralogs generated by small-scale duplication (SSD) and are equally as essential as singleton genes (7). A prevalence of dosage-balanced genes among ohnologs was proposed to explain this contradiction of the theoretical, expected backup role of duplicated genes, which should buffer against such effects. Dosage balance may exist between two or more genes whose products interact or participate in the same pathway or process (8–10). According to the dosage balance hypothesis, changes in the relative dosage of gene product, such as would occur through duplication of some but not all of the balanced gene set, should be deleterious (11). WGD creates a unique opportunity for the duplication of dosage-balanced genes because it guarantees the simultaneous duplication of all components of a balanced gene set (10, 12). Furthermore, once the genes have been duplicated by WGD, subsequent loss of individual genes would result in a dosage imbalance due to insufficient gene product, thus leading to biased retention of dosage-balanced ohnologs. In fact, evidence for preferential retention of dosage-balanced genes after WGD is accumulating (4, 7, 11–20). Copy number variation [copy number polymorphism (CNV)] describes population level polymorphism of small segmental duplications and is known to directly correlate with gene expression levels (21–24). Thus, CNV of dosage-balanced genes is also expected to be deleterious. This model predicts that retained ohnologs should be enriched for dosage-balanced genes that are resistant to subsequent SSD and to CNV in human populations.
We track SSD events in vertebrate ohnologs after WGD and in sister lineages that did not experience WGD (Fig. 1 and SI Materials and Methods) in order to test the dosage-balance hypothesis and show the first large-scale evidence that ohnologs are resistant to fluctuations in relative quantities by SSD and CNV. We propose that ohnologs that have experienced neither SSD nor CNV are dosage-balanced and find that, consistent with this, they are strongly associated with disease. In particular, Down Syndrome (DS) caused by trisomy 21 appears to be caused in large part by the deleterious effects of the 1.5-fold increase in dosage of ohnologs on that chromosome.
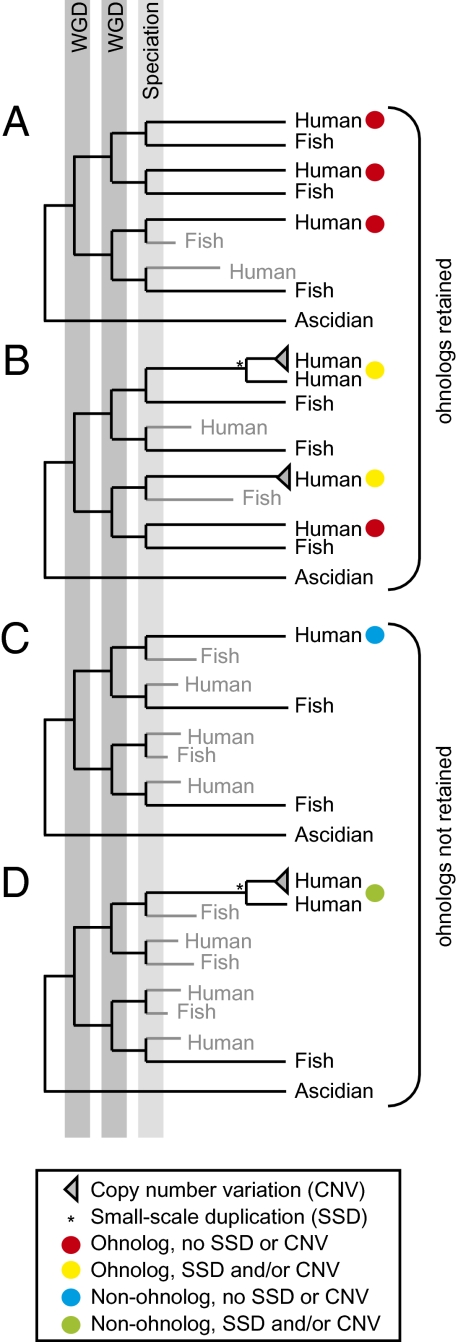
Fig. 1.
Tetrapod gene families. Each panel shows a hypothetical vertebrate gene family where members of each family have been generated by whole genome duplication (WGD) and/or small-scale duplication (SSD). Genes are labeled with the organism name. Many genes duplicated by WGD (ohnologs) are subsequently lost, and these are indicated in gray. (A and B) Ohnologs have been retained. (C and D) No ohnologs were retained. In order to compare the frequency of SSD of ohnologs and nonohnologs across a similar time-frame, vertebrate gene families are further broken down into tetrapod gene families (indicated by colored circles) and are classified into ohnologs without SSD and CNV (red), ohnologs that have experienced SSD or CNV (yellow), nonohnologs without SSD and CNV (blue) or nonohnologs that have experienced SSD or CNV (green).
Results and Discussion
To compare the frequency of SSD of different genes over a comparable period of time, we inferred the set of genes present just after the fish-tetrapod divergence and clustered all paralogs generated by subsequent duplications into “tetrapod gene families” (Fig. 1 and SI Materials and Methods). Only 6.7% of ancient ohnologs have experienced SSD in this time frame (449/6,742; blastp hit with E-value < 10−7 and alignable region > 30%), compared to 10.1% (1,109/10,976) of ancient nonohnologs (P = 4.8 × 10−15, χ2 test). This observation demonstrates that ohnologs experienced SSD less frequently than other genes in the human genome. Furthermore, when we examine genes in the ascidian (Ciona intestinalis) genome, a lineage that did not experience WGD, we find that genes that have not experienced lineage-specific SSD in ascidian are more likely to be orthologs of human ohnologs (30.1%; 1,804/5,998) than ascidian genes that did experience lineage-specific SSD (20.6%; 649/3,147; P < 2.2 × 10−16, χ2 test). We observe the same trend for fly (31.6% vs. 20.0%; P < 2.2 × 10−16), worm (31.6% vs. 21.1%; P < 2.2 × 10−16) and sea anemone (24.6% vs. 14.6%; P < 2.2 × 10−16). The resistance of retained ohnologs to the otherwise prevalent process of SSD, even in distantly-related lineages that did not experience WGD, strongly supports the inference that these genes are ancient dosage-balanced genes.
Within human populations, we expect that CNV of dosage-balanced genes should be deleterious. We compare the proportion of genes displaying CNV (PCNV) for ohnologs with that for all human protein coding genes. Any gene whose entire coding sequence is found within a CNV region is considered to have CNV. We find that the PCNV of ohnologs (22.6%, 1,648/7,294) is significantly lower than the human genome average PCNV (29.3%, 6,136/20,907; P < 2.2 × 10−16, χ2 test). By contrast, the PCNV of duplicated genes generated by SSD is significantly higher than the genome average (36.6%, 3,306/9,027; P < 2.2 × 10−16, χ2 test). This observation is true of copy loss variants (CLV) and copy gain variants (CGV) independently. The proportions of CLVs (13.1%, 957/7,294) and of CGVs for ohnologs (9.9%, 722/7,294) are significantly lower than the genome average (18.4%, 3,843/20,907 and 14.6%, 3,055/20,907, respectively; P < 2.2 × 10−16 and P < 2.2 × 10−16, respectively, χ2 test). By contrast, the proportions of CLVs (23.7%, 2,142/9,027) and of CGVs for SSD duplicates (20.6%, 1,858/9,027) are significantly higher than the genome average (P < 2.2 × 10−16 and P < 2.2 × 10−16, respectively, χ2 test).
We consider the potential impact of the gene length bias of ohnologs because the average length of ohnologs (87,287 bp) is longer than that of all genes (55,970 bp). The longer the length of a gene, the less likely that the whole coding-sequence of the gene is within CNVs. When we repeat the analysis with an extremely loose definition of CNV genes that required only 1-bp overlap, the PCNV of ohnologs (41.2%, 3,005/7,294) is still significantly lower than the genome average (42.8%, 8,945/20,907; P = 0.0073, χ2 test).
This indicates that the propensity for individual gene duplication over evolutionary time in the vertebrate lineage is closely linked to the propensity for duplication/loss within human populations and suggests a persistent deleterious effect of dosage changes for a subset of human genes. Whereas genes that have experienced recent SSD in the human lineage continue to be subject to dosage changes through CNV in human populations, ohnologs without subsequent SSD are also resistant to CNV. Over 60% of ohnologs (63.6%; 4,638/7,294) are free of SSD and CNV, compared to 32.4% (4,412/13,613) of nonohnologs in the genome, and the difference is statistically significant (P < 2.2 × 10−16, χ2 test). These results indicate that retained ohnologs in the human genome are enriched for dosage-balanced genes. We propose that these 4,638 genes are dosage-balanced ohnologs (DBOs).
This method of detecting dosage-balanced genes is indirect and we note that some dosage-balanced genes will not be detected by this method, and conversely that some genes that appear to be dosage-balanced by our measure may be dosage-insensitive genes that have not experienced duplications due to chance rather than dosage constraints. We examined some of the properties of DBOs with respect to expected characteristics of dosage-balanced genes. It has previously been shown that developmental genes, transcription factors, and protein complex members are likely to be dosage-balanced (8, 11, 18). We observe significant enrichment for protein complex membership for DBOs (14.6%, 676/4,638) compared to non-DBO ohnologs (10.5%, 280/2,656; P = 1.1 × 10−6, χ2 test) and nonohnologous genes (8.8%, 1,202/13,613; P < 2.2 × 10−16, χ2 test). Furthermore, we find that gene ontology (GO) terms “multicellular organismal development,” “cell differentiation,” “cell communication,” and “transcription regulator activity,” related to development and transcription are extensively enriched in DBOs (Table S1). On the other hand, for non-DBO ohnologs the enrichment of GO ids related to development is low and transcription regulator activity is not enriched (Table S2). These results further support that inferred DBOs in our data are genuinely dosage-balanced genes.
Several previous studies have considered the duplicability of dosage sensitive genes (both dosage-balanced and haploinsufficient). The results from these studies were somewhat contradictory and indicated both lower duplicability of genetic components of more complex proteins (more subunits) (25) and higher duplicability of genes with dominant-negative phenotypes (presumed haploinsufficient genes) (26). These observations are reconciled in the context of the special impact of whole genome duplication. As described above, protein-complex members are unlikely to be duplicated except by WGD. We find that, similarly, haploinsufficient genes are enriched within ohnologs and DBOs, and are depleted among SSD-duplicated genes (SI Materials and Methods). Thus we observe a consistent relationship between dosage constraints and duplication patterns, namely, preferential retention of ohnologs of dosage-sensitive genes and low duplicability by SSD.
CNV data from large studies of healthy individuals (such as the data used here) show that disease genes are significantly underrepresented in the lists of variable copy number genes (27) and many studies have reported a relationship between CNV and human disease (21, 28–32). The effect of duplicating a dosage-balanced gene should be deleterious and CNV of these genes is expected to lead to human disease (33). Consistent with this expectation, we find that DBOs are significantly enriched in human disease genes from Online Mendelian Inheritance in Man (34) (OMIM; 15.9%, 736/4,638) compared to other genes (11.1%, 1,812/16,269; P < 2.2 × 10−16, χ2 test), as are all ohnologs (16.5%, 1,201/7,294, of ohnologs are disease genes; P < 2.2 × 10−16). This suggests the generality of a strong relationship between ohnologs and human disorders, including several genes causing conditions that have previously been reported to be specifically due to dosage imbalance such as the genes coding for ABCA1, BMI1, CHRNB2, CHRNA4, CLOCK, NCAM1, NCAM2, NOTCH1, NOTCH2, NOTCH3, and PLP1 (35). Interestingly, the proportion of essential genes for DBOs (17.1%, 793/4,638) is significantly higher than for other ohnologs (11.7%, 311/2,656; P < 2.2 × 10−16, χ2 test) and nonohnologs (6.2%, 843/13,613; P < 2.2 × 10−16, χ2 test), which possibly reflects a higher incidence of lethal phenotypes specifically associated with perturbation of DBOs.
Trisomy is an extreme example of CNV. Trisomy 21 results in DS, which is generally considered to be due to dosage imbalance caused by the extra copy of chromosome 21 and occurs at a frequency of more than 1/1,000 in human populations (36). Most trisomies are incompatible with life and are not observed in live births. Trisomy 21 has the least severe phenotypic consequences and is thus the most commonly observed human trisomy. In keeping with this, we observe that chromosome 21 has the smallest number of DBOs of any chromosome except the Y, and that DBOs are significantly underrepresented on chromosome 21 (observation 40 vs. expectation 56.1; P = 0.010), as are all ohnologs (observation 58 vs. expectation 88; P = 4.8 × 10−5).
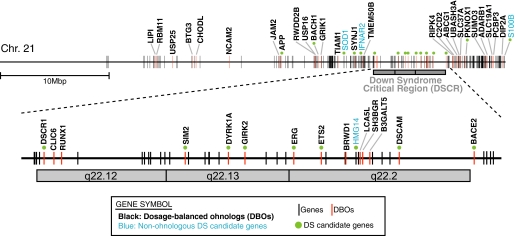
Several genes on chromosome 21 have been identified as DS-related genes (36, 37). For example, a 1.5-fold increase in dosage of DSCR1 and DYRK1A has been shown experimentally to lead to features of the DS phenotype (38). Table 1 lists all 40 DBOs from chromosome 21 and 16 candidate DS genes from the literature (36, 37). Strikingly, 75% (12/16) of reported DS candidates are also DBOs, whereas under a hypothesis of no association we would expect only two of the candidate genes to also be DBOs; this is a highly significant difference (P = 5.9 × 10−8, Fisher's exact test; Table 1). This result indicates that our results from a computational approach are consistent with previous reports based on experimental analysis. Only one previously reported DS candidate gene, S100B, displays CNV (gene gains: variation IDs 3,235 and 8,897). Interestingly, S100B is also a candidate gene for bipolar disorder where mutations in the promoter region leading to increased expression are linked to the disorder (39). In particular, duplication of a region on chromosome 21 known as the Down Syndrome critical region (DSCR) is thought to be a major determinant of the features of DS (38, 40–42), although it is still controversial (35, 43). We find significant overrepresentation of DBOs in the DSCR (P = 0.0012; Fig. 2). We propose that the contribution of the DSCR to the features of DS is determined by the enrichment of DBOs in the region (Fig. 2). A major goal of DS research is the identification of the particular genes on chromosome 21 and also genes on other chromosomes that contribute to the syndrome in order to advance detection and therapeutic strategies (36). We suggest that the DBOs on chromosome 21 are candidate DS genes worthy of further investigation. Furthermore, it is likely that ohnolog pairs of chromosome 21 DS candidates and DBOs (Table S3) are likely to participate in the same molecular processes and thus are candidate nonchromosome-21 genes involved in the DS phenotype.
Table 1.
Dosage-balanced ohnologs and Down Syndrome-related genes on chromosome 21
| Ensembl id | Gene symbol | Full name | Reference |
| ENSG00000188992 | LIPI | Lipase, member I | |
| ENSG00000185272 | RBM11 | RNA binding motif protein 11 | |
| ENSG00000155313 | USP25 | Ubiquitin specific peptidase 25 | |
| ENSG00000154640 | BTG3 | BTG family, member 3 | |
| ENSG00000154645 | CHODL | Chondrolectin | |
| ENSG00000154654 | NCAM2 | Neural cell adhesion molecule 2 | |
| ENSG00000154721 | JAM2 | Junctional adhesion molecule 2 | |
| ENSG00000142192 | APP | Amyloid β (A4) precursor protein | 37 |
| ENSG00000156253 | RWDD2B | RWD domain containing 2B | |
| ENSG00000156256 | USP16 | Ubiquitin specific peptidase 16 | |
| ENSG00000156273 | BACH1 | BTB and CNC homology 1 | 37 |
| ENSG00000171189 | GRIK1 | Glutamate receptor, ionotropic, kainate 1 | |
| ENSG00000156299 | TIAM1 | T-cell lymphoma invasion and metastasis 1 | |
| ENSG00000142168 | SOD1 | Superoxide dismutase 1 | 37 |
| ENSG00000159082 | SYNJ1 | Synaptojanin 1 | 37 |
| ENSG00000159110 | IFNAR2 | Interferon receptor 2 | 37 |
| ENSG00000142188 | TMEM50B | Transmembrane protein 50B | |
| ENSG00000159200 | DSCR1 | Down syndrome critical region gene 1 | 36, 37 |
| ENSG00000159212 | CLIC6 | Chloride intracellular channel 6 | |
| ENSG00000159216 | RUNX1 | Runt-related transcription factor 1 | |
| ENSG00000159263 | SIM2 | Single-minded homolog 2 | 36 |
| ENSG00000157540 | DYRK1A | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 36, 37 |
| ENSG00000157542 | GIRK2 | Potassium inwardly-rectifying channel, subfamily J, member 6 | 36 |
| ENSG00000157554 | ERG | V-ets erythroblastosis virus E26 oncogene homolog | 37 |
| ENSG00000157557 | ETS2 | V-ets erythroblastosis virus E26 oncogene homolog 2 | 37 |
| ENSG00000185658 | BRWD1 | Bromodomain and WD repeat domain containing 1 | |
| ENSG00000205581 | HMG14 | High-mobility group nucleosome binding domain 1 | 37 |
| ENSG00000157578 | LCA5L | Leber congenital amaurosis 5-like | |
| ENSG00000185437 | SH3BGR | SH3 domain binding glutamic acid-rich protein | |
| ENSG00000183778 | B3GALT5 | β-1,3-galactosyltransferase 5 | |
| ENSG00000171587 | DSCAM | Down syndrome cell adhesion molecule | 37 |
| ENSG00000182240 | BACE2 | β-site APP-cleaving enzyme 2 | 37 |
| ENSG00000183421 | RIPK4 | Receptor-interacting serine-threonine kinase 4 | |
| ENSG00000157617 | C2CD2 | C2 calcium-dependent domain containing 2 | |
| ENSG00000160179 | ABCG1 | ATP-binding cassette, sub-family G (WHITE), member 1 | |
| ENSG00000160185 | UBASH3A | Ubiquitin associated and SH3 domain containing, A | |
| ENSG00000160190 | SLC37A1 | Solute carrier family 37, member 1 | |
| ENSG00000160199 | PKNOX1 | PBX/knotted 1 homeobox 1 | 37 |
| ENSG00000184900 | SUMO3 | SMT3 suppressor of mif two 3 homolog 3 | |
| ENSG00000197381 | ADARB1 | Adenosine deaminase, RNA-specific, B | |
| ENSG00000173638 | SLC19A1 | Solute carrier family 19, member 1 | |
| ENSG00000183570 | PCBP3 | Poly(rC) binding protein 3 | |
| ENSG00000160305 | DIP2A | DIP2 disco-interacting protein 2 homolog A | |
| ENSG00000160307 | S100B | S100 calcium binding protein B | 37 |
Where a reference is provided, those genes were previously reported as candidate DS genes. Genes in bold are not dosage-balanced ohnologs.
Fig. 2.
Ohnologs and Down Syndrome (DS)-related genes on chromosome 21. Red and black vertical lines are ohnologs and other protein coding genes, respectively. Green dots mark reported DS candidate genes (Table 1). Gene symbols labeled in black and blue show dosage-balanced ohnologs (DBOs) and nonohnolog DS candidate genes, respectively. A gray rectangle indicates the Down Syndrome critical region covering 21q22.12, 21q22.13, and 21q22.2, which is shown in more detail below.
As previously mentioned, a clear relationship has been demonstrated between gene copy number and expression level (e.g., ref. 21). However, it has been shown that a substantial proportion of triplicated genes in DS patients or DS model mice are automatically dosage-compensated (i.e., expressed at diploid levels (44–54); in Table S4) a phenomenon that would alleviate copy number constraints on dosage-balanced genes. However, their expression patterns are not consistent between studies or tissues (55). For example, the expression level of a DS gene DYRK1A (38) is increased 1.5-fold in DS brains but not increased in DS infants (56). Other experimentally verified, robust DS candidates have 1.5-fold dosage in some tissues, but their dosages are compensated automatically in other tissues (Table S4). This expression variability may be at least partly responsible for variability in the DS phenotype (44). Overexpressed genes are considered to be likely DS candidate genes (44); however, measures of overexpression are hampered by the difficulty in comparing “like-with-like” caused by some global changes in the DS phenotype (55), and DBOs are not significantly overrepresented among reported overexpressed genes (Table S5).
We present evidence for dosage-balance constraints acting on retained ohnologs based on their patterns of small-scale duplication over the vertebrate lineage and duplication/loss within human populations. Our results support the hypothesis that ohnologs are enriched for dosage-balanced genes (4, 7, 11–20) and shed light on duplicate gene retention and essentiality for vertebrate genomes (7). We have further shown that ohnologs are frequently associated with disease including conditions known to be caused by dosage-imbalance, and in particular we propose a significant role for DBOs on chromosome 21 in determining the features of DS and propose novel DS candidate genes based on their evolutionary patterns. Application of this methodology to other human diseases caused by dosage imbalance may be effective in identifying candidate disease genes.
Materials and Methods
Gene with Copy Number Variants.
There are 20,907 protein-coding genes that have known genomic locations and that were not on alternative sequences such as chr6_COX in Ensembl release 52 were used in this study (57). We downloaded CNVs in the human genome from Database of Genomic Variants version 7 (http://projects.tcag.ca/variation/). When the entire coding-sequence of a gene is within one of the copy number variants, we defined the gene as a CNV gene. We used 6,136 CNV genes and 14,771 non-CNV genes in this study. Out of 6,136 CNV genes, 3,843 and 3,055 genes displayed copy loss and copy gain variants, respectively.
Ohnologs and SSD Duplicated Genes.
A detailed description of the identification of ohnologs (Tables S6 and S7) and SSD duplicated genes can be found in SI Materials and Methods.
GO.
GO ids and GO “slim” annotations for biological process and molecular function of human were downloaded from ftp://ftp.geneontology.org/pub/go/gene-associations/ and ftp://ftp.geneontology.org/pub/go/GO_slims, respectively. We excluded the GO ids GO:0008150 (biological process unknown) and GO:0003674 (molecular function unknown). The frequency of each GO id assigned to DBOs or non-DBO ohnologs was counted. We calculated the P value for each GO id by comparison of the observed frequency in the dataset with expectations based on a hypergeometric distribution using all genes with at least one GO id. The estimated P values were adjusted by Bonferroni correction. Significantly under- or overrepresented GO ids for DBOs and non-DBO ohnologs are shown in Table S1 and S2, respectively.
Members of Protein Complex.
We obtained a list of members of human protein complex from Human Protein Reference Database (HPRD; http://www.hprd.org). We examined the enrichment for protein complex membership for DBOs.
Haploinsufficient Genes.
As per Kondrashov and Koonin (26), we inferred haploinsufficient genes from genes with dominant-negative phenotypes (SI Materials and Methods). Disease gene lists were obtained from Lopez-Bigas et al. (58).
Underrepresentation of Dosage-Balanced Genes on Chromosome 21.
We conducted simulations to investigate whether the number of DBOs on chromosome 21 was smaller than expected. We randomly shuffled gene locations of all protein coding genes on the human genome 1,000 times, and counted the number of DBOs on chromosome 21.
Disease Genes.
We obtained 2,548 disease genes from the “Morbidmap” database produced by OMIM (ftp://ftp.ncbi.nih.gov/repository/OMIM/morbidmap).
Essential Genes.
Mouse essential genes are determined by phenotype data from Mouse Genome Informatics (MGI; http://www.informatics.jax.org/). Full details of the identification of mouse essential genes are given in Makino et al. (7). We infer human essential genes through one to one orthology relationships with the mouse genes as defined by Ensembl release 52. Finally, we defined 1,947 genes with lethal or infertile phenotypes as essential genes in human.
Supplementary Material
Acknowledgments
We thank Laurent Duret for helpful comments and Science Gallery, Trinity College Dublin, for stimulating interactions. This work is supported by Science Foundation Ireland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914697107/-/DCSupplemental.
References
- 1.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- 3.Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomme T, et al. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet FG, et al. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- 6.Hufton AL, et al. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res. 2008;18:1582–1591. doi: 10.1101/gr.080119.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino T, Hokamp K, McLysaght A. The complex relationship of gene duplication and essentiality. Trends Genet. 2009;25:152–155. doi: 10.1016/j.tig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Veitia RA. Exploring the etiology of haploinsufficiency. Bioessays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- 9.Veitia RA. Nonlinear effects in macromolecular assembly and dosage sensitivity. J Theor Biol. 2003;220:19–25. doi: 10.1006/jtbi.2003.3105. [DOI] [PubMed] [Google Scholar]
- 10.Veitia RA. Gene dosage balance in cellular pathways: Implications for dominance and gene duplicability. Genetics. 2004;168:569–574. doi: 10.1534/genetics.104.029785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 12.Veitia RA. Paralogs in polyploids: One for all and all for one? Plant Cell. 2005;17:4–11. doi: 10.1105/tpc.104.170130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- 14.Hakes L, Pinney JW, Lovell SC, Oliver SG, Robertson DL. All duplicates are not equal: The difference between small-scale and genome duplication. Genome Biol. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–464. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 19.Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 20.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: Genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Hurles ME, Dermitzakis ET, Tyler-Smith C. The functional impact of structural variation in humans. Trends Genet. 2008;24:238–245. doi: 10.1016/j.tig.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarroll SA, et al. International HapMap Consortium. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 23.Scherer SW, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39(Suppl 7):S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Lusk R, Li WH. Organismal complexity, protein complexity, and gene duplicability. Proc Natl Acad Sci USA. 2003;100:15661–15665. doi: 10.1073/pnas.2536672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–290. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen DQ, Webber C, Ponting CP. Bias of selection on human copy-number variants. PLoS Genet. 2006;2:e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 29.Estivill X, Armengol L. Copy number variants and common disorders: Filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007;3:1787–1799. doi: 10.1371/journal.pgen.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93:22–26. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conrad B, Antonarakis SE. Gene duplication: A drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- 34.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33(Database Issue):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: From genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 36.Wiseman FK, Alford KA, Tybulewicz VL, Fisher EM. Down syndrome—recent progress and future prospects. Hum Mol Genet. 2009;18(R1, R1):R75–R83. doi: 10.1093/hmg/ddp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommer CA, Henrique-Silva F. Trisomy 21 and Down syndrome: A short review. Braz J Biol. 2008;68:447–452. doi: 10.1590/s1519-69842008000200031. [DOI] [PubMed] [Google Scholar]
- 38.Arron JR, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 39.Roche S, et al. Candidate gene analysis of 21q22: Support for S100B as a susceptibility gene for bipolar affective disorder with psychosis. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:1094–1096. doi: 10.1002/ajmg.b.30556. [DOI] [PubMed] [Google Scholar]
- 40.Kisling E. Cranial Morphology in Down's syndrome: A Comparative Roentgencephalometric Study in Adult Males. Copenhagen: Munksgaard; 1966. [Google Scholar]
- 41.Richtsmeier JT, Baxter LL, Reeves RH. Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev Dyn. 2000;217:137–145. doi: 10.1002/(SICI)1097-0177(200002)217:2<137::AID-DVDY1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 42.Delabar JM, et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 43.Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aït Yahya-Graison E, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: Impact on disease phenotypes. Am J Hum Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kahlem P, et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 2004;14:1258–1267. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S, Antonarakis SE. Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res. 2004;14:1268–1274. doi: 10.1101/gr.2090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao R, et al. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol. 2005;6:R107. doi: 10.1186/gb-2005-6-13-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Y, et al. Blood expression profiles for tuberous sclerosis complex 2, neurofibromatosis type 1, and Down's syndrome. Ann Neurol. 2004;56:808–814. doi: 10.1002/ana.20291. [DOI] [PubMed] [Google Scholar]
- 49.Li CM, et al. Cell type-specific over-expression of chromosome 21 genes in fibroblasts and fetal hearts with trisomy 21. BMC Med Genet. 2006;7:24. doi: 10.1186/1471-2350-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prandini P, et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am J Hum Genet. 2007;81:252–263. doi: 10.1086/519248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Doherty A, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saran NG, Pletcher MT, Natale JE, Cheng Y, Reeves RH. Global disruption of the cerebellar transcriptome in a Down syndrome mouse model. Hum Mol Genet. 2003;12:2013–2019. doi: 10.1093/hmg/ddg217. [DOI] [PubMed] [Google Scholar]
- 53.Amano K, et al. Dosage-dependent over-expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum Mol Genet. 2004;13:1333–1340. doi: 10.1093/hmg/ddh154. [DOI] [PubMed] [Google Scholar]
- 54.Dauphinot L, et al. The cerebellar transcriptome during postnatal development of the Ts1Cje mouse, a segmental trisomy model for Down syndrome. Hum Mol Genet. 2005;14:373–384. doi: 10.1093/hmg/ddi033. [DOI] [PubMed] [Google Scholar]
- 55.FitzPatrick DR. Transcriptional consequences of autosomal trisomy: Primary gene dosage with complex downstream effects. Trends Genet. 2005;21:249–253. doi: 10.1016/j.tig.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Dowjat WK, et al. Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007;413:77–81. doi: 10.1016/j.neulet.2006.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35(Database issue):D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.López-Bigas N, Blencowe BJ, Ouzounis CA. Highly consistent patterns for inherited human diseases at the molecular level. Bioinformatics. 2006;22:269–277. doi: 10.1093/bioinformatics/bti781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.