Abstract
Retroviral Gag polyproteins coopt host factors to traffic from cytosolic ribosomes to the plasma membrane, where virions are released. Before membrane transport, the multidomain Gag protein of Rous sarcoma virus (RSV) undergoes importin-mediated nuclear import and CRM1-dependent nuclear export, an intrinsic step in the assembly pathway. Transient nuclear trafficking of Gag is required for efficient viral RNA (vRNA) encapsidation, suggesting that Gag:vRNA binding might occur in the nucleus. Here, we show that Gag is imported into the nucleus through direct interactions of the Gag NC domain with importin-α (imp-α) and the MA domain with importin-11 (imp-11). The vRNA packaging signal, known as ψ, inhibited imp-α binding to Gag, indicating that the NC domain does not bind to imp-α and vRNA simultaneously. Unexpectedly, vRNA binding also prevented the association of imp-11 with both the MA domain alone and with Gag, suggesting that the MA domain may bind to the vRNA genome. In contrast, direct binding of Gag to the nuclear export factor CRM1, via the CRM1-RanGTP heterodimer, was stimulated by ψRNA. These findings suggest a model whereby the genomic vRNA serves as a switch to regulate the ordered association of host import/export factors that mediate Gag nucleocytoplasmic trafficking for virion assembly. The Gag:vRNA interaction appears to serve multiple critical roles in assembly: specific selection of the vRNA genome for packaging, stimulating the formation of Gag dimers, and triggering export of viral ribonucleoprotein complexes from the nucleus.
Keywords: protein-RNA binding, retrovirus assembly, importin-α/β, importin-11, CRM1
Translocation of macromolecules across the nuclear membrane is a tightly regulated process that maintains order in the intracellular environment. Proteins gain entry into the nucleus only if they possess the appropriate signals recognized by nuclear import receptors (importins) or if they interact directly with the nuclear pore complex. For reentry into the cytoplasm, proteins in the nucleus must contain sequences that interact with export factors (exportins) to exit through the nuclear pore. Collectively, importins and exportins are known as karyopherins, members of the structurally related importin-β superfamily of proteins that control nucleocytoplasmic trafficking of proteins and RNA molecules (1, 2). Some proteins that undergo nucleocytoplasmic shuttling are subsequently transported to the plasma membrane, where they perform their biological roles (3–5). The mechanisms that regulate these complex trafficking signals to coordinate spatiotemporal control of intracellular protein movement are the subject of this report.
The Rous sarcoma virus (RSV) Gag polyprotein, the major structural component of the virus particle, orchestrates virion assembly. RSV Gag is synthesized on cytosolic ribosomes, transiently traffics through the nucleus, and then travels to the plasma membrane, where complexes of Gag bound to the viral RNA (vRNA) genome form virus particles that emerge from the cell via budding (6, 7). Nuclear trafficking of RSV Gag is necessary for efficient incorporation of vRNA into nascent particles, suggesting that Gag might bind its genome in the nucleus (8). This model represents a unique paradigm for how retroviruses package their genomes. Whether other retroviruses follow a similar pathway for genome encapsidation remains unknown, although the Gag proteins of HIV type 1, murine leukemia virus, human and simian foamy viruses, and retrotransposon Tf1 have been reported to gain access to the nucleus (9–12). Because genome packaging is essential for viral infectivity, and thus serves as a potential target for antiviral therapy, understanding the mechanisms underlying Gag:vRNA interactions and intracellular transport is of critical importance.
RSV Gag possesses two independent nuclear localization signals (NLSs), a classic NLS in NC and a noncanonical NLS in MA (13), and a CRM1-dependent nuclear export signal (NES) in p10 (14) (Fig. S1). Genetic analysis using Saccharomyces cerevisiae revealed that nuclear import of NC depends on the importin-α/β (imp-α/β) pathway, whereas Kap120p and Mtr10p [transportin-SR and importin-11 (imp-11), respectively, in higher eukaryotes] are import receptors for MA (13). Toxicity of the Gag protein in yeast prevented determination of whether it interacts with the same karyopherins as the isolated MA and NC domains. Dissecting activities of the nucleocytoplasmic targeting signals in Gag is further complicated by overlap of the NLSs and NES with sequences required for additional steps in the assembly process (Fig. S1). The NLS in MA coincides with the plasma membrane-binding signal (15), the NC NLS overlaps the Gag:vRNA interaction domain (16), and the NES in p10 corresponds to the Gag multimerization interface (17, 18).
Virus assembly depends on recognition of appropriate Gag subcellular trafficking signals and assembly motifs at the proper time and location. A crucial question is how the accessibilities of Gag NLSs and NES are controlled to permit ordered interactions with host import/export machinery. Here, we present data suggesting a mechanistic role for Gag-nucleic acid binding in regulating the ordered events required for nucleocytoplasmic trafficking and virus particle assembly. Importins and vRNA compete for binding of Gag, ensuring that Gag enters the nucleus before Gag-Gag dimerization. In the nucleus, Gag-vRNA binding serves as a signal for nuclear export by stimulating association of Gag with the CRM1:RanGTP complex, delivering Gag:vRNA complexes back into the cytoplasm for particle assembly.
Results
Imp-β and Imp-11 Mediate Nuclear Entry of the Gag Polyprotein in Avian Cells.
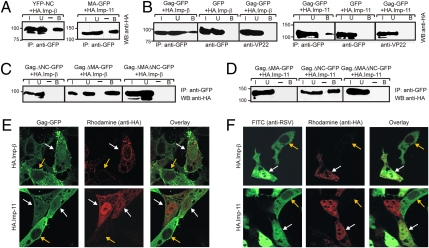
Our earlier studies identified the nuclear import receptors for the isolated MA and NC domains in yeast (13). To test whether the mammalian homologues of these karyopherins were associated with MA and NC in avian cells, the natural host for RSV, MA-GFP and YFP-NC (Fig. S1) were coexpressed with HA-tagged imp-11 and imp-β. YFP-NC interacted with imp-β, and MA-GFP was pulled down with imp-11 (Fig. 1A); however, YFP-NC did not pull down with imp-11, and MA-GFP did not pull down with imp-β (Fig. S2A).
Fig. 1.
Imp-11 and imp-β drive import of Gag-GFP and interact with Gag in vivo. Coimmunoprecipitation of importins with YFP-NC and MA-GFP (A), Gag-GFP (B), ΔNC.Gag-GFP and ΔMAΔNC.Gag-GFP (C), and ΔMA.Gag-GFP (D). I (input, 3% of total lysate), U (unbound), and B (bound) proteins detected by Western blotting (WB) and antibodies used for immunoprecipitation (IP) are indicated. Molecular weight standards in kilodaltons (kDa) are indicated to the left. The experiments were performed three times, and representative blots are shown. (E) QT6 cells were transiently cotransfected with Gag-GFP derivatives and HA.imp-β or HA.imp-11. HA-tagged importins were detected by rhodamine-conjugated anti-HA, and cells were imaged through the nuclear plane using confocal microscopy. White arrows indicate cells expressing both HA.imp-11 and Gag-GFP, and yellow arrows indicate cells expressing only Gag-GFP. (F) RSV-infected QT6 cells transiently expressed either HA.imp-β or HA.imp-11. RSV Gag was detected with anti-RSV serum and a mouse-anti-rabbit FITC-conjugated secondary antibody. White arrows indicate cells expressing HA-tagged importins and Gag, and yellow arrows point to infected but untransfected cells. The fluorescence intensity of the nucleus was measured in the presence and absence of overexpressed imp-β and imp-11 and divided by the total cellular fluorescence to calculate percent nuclear fluorescence; five cells were measured to calculate the mean.
To establish whether the MA and NC domains mediate association of the Gag polyprotein with imp-11 and imp-β, immunoprecipitation of Gag-GFP followed by immunoblotting with anti-HA was performed, revealing that Gag was pulled down in complex with HA-tagged imp-β and imp-11 (Fig. 1B). Neither HA.imp-β nor HA.imp-11 associated with GFP, nor was Gag-GFP immunoprecipitated with an irrelevant antibody (anti-VP22). Removal of the NC sequence in ΔNC.Gag-GFP and ΔMA.ΔNC.Gag-GFP abrogated interaction with imp-β, whereas deletion of MA (Gag.ΔMA-GFP) did not (Fig. 1C and Fig. S1). Removal of MA (ΔMA.Gag-GFP and ΔMA.ΔNC.Gag-GFP) eliminated interaction with imp-11, although deletion of NC (Gag.ΔNC-GFP) had no effect (Fig. 1D and Fig. S1). Importantly, Gag-GFP and all mutant derivatives were expressed at similar levels, indicating that the failure of Gag deletion constructs to interact with importins was not attributable to low expression levels (Fig. S2B). Reciprocal pull-downs using anti-HA antibody to immunoprecipitate Gag-GFP were not informative because of nonspecific interaction of Gag with the antiserum.
To examine the functional activity of the importins in Gag nuclear entry, imp-β and imp-11 were overexpressed in avian cells and subcellular localization of Gag-GFP was examined using confocal microscopy. Increasing the intracellular levels of imp-11 and imp-β will enhance the nuclear accumulation of their respective cargoes because concentrations of these importins are limiting in cells (19–21). Overexpression of imp-β or imp-11 resulted in accumulation of Gag-GFP in the nucleus (Fig. 1E, white arrows). In cells expressing HA.imp-11, the mean nuclear fluorescence of Gag-GFP was 28.3% compared with 11.8% for cells containing only endogenous levels of imp-11 (yellow arrows; P = 0.001), whereas the mean nuclear level of Gag-GFP in cells expressing HA.imp-β was 31.8% (P = 0.002). Together, these results demonstrated a functional role for imp-11 and imp-β in the nuclear import of Gag in avian cells (13).
To determine whether the same import machinery was involved in Gag nuclear entry during RSV infection, imp-β and imp-11 were overexpressed in cells expressing stably integrated proviral genomes (Fig. 1F). Expression of HA.imp-11 or HA.imp-β demonstrated a marked increase in Gag nuclear localization (Fig. 1F, white arrows), with mean nuclear fluorescence increasing to 58.8% for imp-11 and 66.2% for imp-β overexpression in contrast to cells expressing endogenous levels of the import factors (Fig. 1F, yellow arrows, 11.6%; P = 0.003 and P = 0.005, respectively). In addition, Gag:imp-β and Gag:imp-11 complexes were specifically coimmunoprecipitated from RSV-infected cells (Fig. S2). Together, these data confirmed that imp-β and imp-11 are constituents of the Gag nuclear import machinery in RSV infection.
Gag Interacts Directly with Imp-α, the Adaptor Protein for Imp-β.
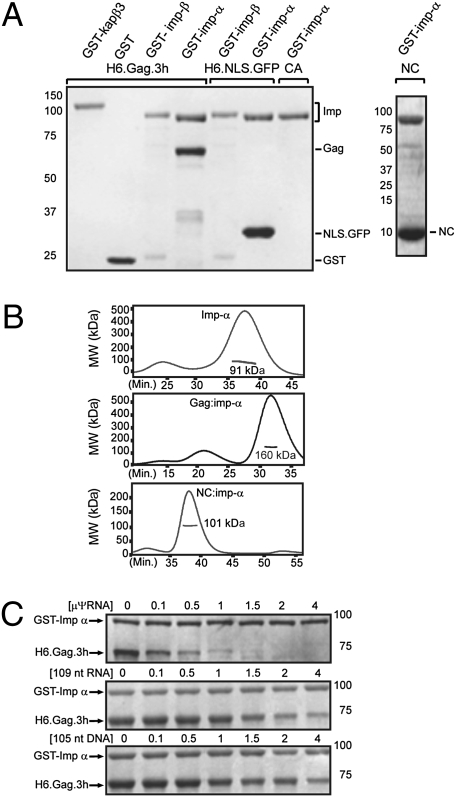
Our previous work indicated that Gag uses imp-α/β for nuclear import; thus, we considered it likely that Gag binds directly to the imp-α subunit to bridge the association with imp-β. To test this idea, we performed in vitro binding experiments using recombinant GST-tagged importins and H6.Gag.3h (Fig. S1). Incubation of Gag with GST-tagged karyopherins demonstrated binding to GST-imp-α but not to GST-kapβ3, GST-imp-β, or GST alone (Fig. 2A). NC also bound imp-α, but the isolated CA domain did not. Imp-α associated with the SV40 NLS (H6.NLS.GFP), recapitulating a biologically relevant interaction (22). Multiangle laser light scattering using recombinant Gag, imp-α, and NC revealed that Gag:imp-α and NC:imp-α each formed a complex with 1:1 stoichiometry (Fig. 2B), suggesting that the NC domain of Gag binds as a monomer to imp-α in the cytoplasm before nuclear translocation.
Fig. 2.
vRNA potently inhibits direct binding of Gag to imp-α. (A) Recombinant GST-tagged karyopherins were incubated with equimolar ratios of H6.Gag.3h and NC or control proteins H6.NLS.GFP and CA. After incubation with GST beads, bound proteins were stained with Coomassie blue. (B) Multiangle light scattering-size exclusion chromatography was performed for imp-α and mixtures of NC:imp-α and Gag:imp-α. The apparent molecular weight was determined, with that of Gag being 62 kDa, that of imp-α being 92 kDa, and that of NC being 9.5 kDa. The major peak for Gag:imp-α complexes was 160 kDa, with a predicted molecular weight of 154 kDa for a complex with 1:1 stoichiometry. For NC:imp-α, the experimentally determined molecular weight was 101 kDa, consistent with an expected mass of 101.5 kDa for 1:1 stoichiometry. (C) Binding reactions were performed as in A with preincubation of μψ-RNA, 105-nt DNA, or nonviral 109-nt RNA.
Nucleic Acids Compete with Imp-α for Binding to Gag.
Although the NC region of Gag binds directly to imp-α, NC also forms a high-affinity complex with the ψ packaging signal to select the viral genome for encapsidation (7). To address whether the interactions of Gag:vRNA and Gag:imp-α are mutually exclusive, we examined whether the nucleic acids μψ-RNA (the minimal vRNA packaging signal composed of 82 nt plus 32 nonviral bases introduced by cloning) (23), a non-vRNA (109 nt), or an oligodeoxynucleotide (105 nt) would interfere with Gag:imp-α binding. Each nucleic acid inhibited the Gag:imp-α interaction, with μψ-RNA being the most efficient competitor (Fig. 2C). Quantification of Gag band intensities from three experiments illustrated the differences in affinities for μψ-RNA, non-vRNA, and DNA (Fig. S3A). Even at the lowest molar ratio (0.1), competition was readily apparent for μψ-RNA, whereas equivalent degrees of competition required approximately a 4-fold higher molar ratio for DNA and non-vRNA (Fig. 2C and Fig. S3A). Nucleic acids did not affect binding of imp-α to SV40 T-antigen NLS (H6.NLS.GFP; Fig. S3B).
We considered two main possibilities to explain the competition results: Either μψ-RNA blocks imp-α binding to Gag through direct competition for a common binding site in NC or RNA binding induces a conformational change in Gag that prevents recognition of imp-α by the NC domain. Analysis of the structure of RSV NC bound to μψ-RNA (24) compared with the structure of imp-α bound to SV40 T-antigen NLS (22) suggests overlapping binding sites within NC for imp-α and RNA (Fig. S4). Superimposition of backbone atoms of K36-R38 in NC (a portion of the predicted NLS) onto the equivalent sequence of T-antigen NLS shows that these sequences could bind imp-α in an analogous manner. The backbone of μψ-RNA colocalizes with imp-α, suggesting that μψ-RNA and imp-α cannot simultaneously bind to the NC domain because of overlap of the binding sites; alternatively, NC or imp-α would have to undergo a significant conformational change for all components to bind.
Far Western Analysis of Gag:Imp-11 Interaction.
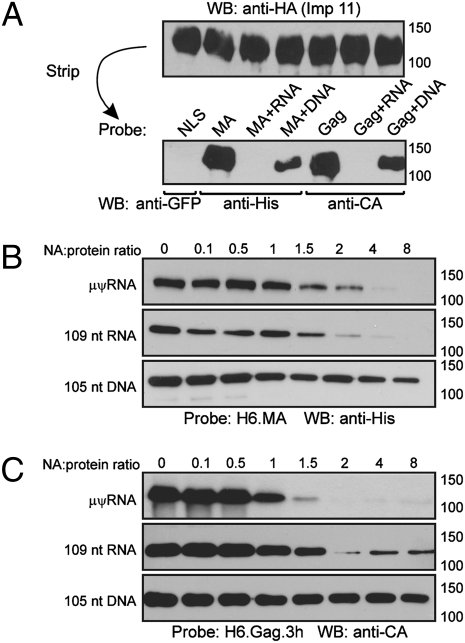
To probe the nature of the interaction between imp-11 and the MA domain in Gag, we used a far Western approach. HA-tagged imp-11 was immunoprecipitated from transfected QT6 cells and detected via immunoblotting, showing equivalent expression (Fig. 3A). Using H6.MA (Fig. S1), H6.Gag.3h, or H6.NLS.GFP as a protein probe, we found that MA and Gag bound directly to imp-11. The canonical NLS (H6.NLS.GFP) did not interact with imp-11, suggesting that imp-11 binding to H6.MA and H6.Gag was not attributable to the H6 tag itself.
Fig. 3.
Direct binding of Gag to imp-11 is inhibited by viral and non-vRNA. (A) HA-tagged imp-11 was immunoprecipitated from transfected QT6 cells using anti-HA, followed by Western blotting. The membrane was stripped and reprobed with the indicated proteins, which were detected via Western blotting using the antibodies below. (B) Recombinant H6.MA protein was preincubated with ratios of nucleic acid (NA)/protein increasing from 0.1 to 8 before use as a probe against imp-11 bound to the membrane. H6.MA bound to immobilized imp-11 was detected with anti-HA antibody. (C) Recombinant H6.Gag.3h protein mixed with increasing concentrations of NAs was used as the probe against immobilized imp-11, and anti-RSV antibody was used for detection. For A–C, each set of experiments was performed three times with representative blots shown.
RSV MA possesses nonspecific nucleic acid binding activity (25–27), so we tested whether nucleic acids would alter MA:imp-11 or Gag:imp-11 interactions. H6.MA was incubated before far Western analysis with the DNA and RNA preparations used previously (Fig. 3B). μψ-RNA and non-vRNA were equally effective competitors of MA:imp-11 binding, although both RNA species were more efficacious than DNA. For Gag, μψ-RNA was the most efficient competitor, with complete inhibition at a 2:1 RNA/protein molar ratio (Fig. 3C). The enhanced activity of μψ-RNA compared with non-vRNA in competing for Gag raises the possibility that selective binding of ψ to the NC region of Gag induces a conformational change that prevents MA from associating with imp-11. Alternatively, although RSV MA is not known to play a role in genomic RNA packaging, it is possible that the MA region of Gag makes direct contact with ψ within the assembling virus particle.
Nucleic Acids Stimulate Cooperative Binding of Gag to the Nuclear Export Factor CRM1.
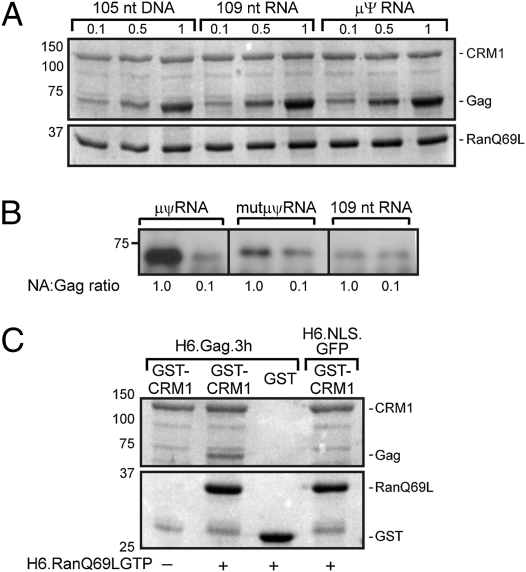
Nuclear trafficking of Gag is required for efficient incorporation of vRNA into virus particles (8); thus, Gag might bind vRNA in the nucleus. To determine whether the Gag:RNA complex may be the actual substrate for CRM1, H6.Gag.3h and GST-CRM1 were incubated with increasing concentrations of nucleic acids before GST pull-down to assess the effect of nucleic acids on the interaction between Gag and CRM1. Because CRM1 binding to NESs on cargoes depends on the formation of a ternary complex with RanGTP (28), we performed Gag:CRM1 in vitro binding experiments in the presence of the constitutively active mutant H6.RanQ69LGTP (29). At a high concentration of Gag (5 μM), each nucleic acid tested enhanced Gag binding to CRM1:Ran, although there was a statistically significant preference for μψ-RNA compared with non-vRNA (P = 0.0071) or DNA (P = 0.0004; Fig. 4A).
Fig. 4.
Gag binding to CRM1:RanGTP complexes is stabilized by nucleic acids. (A) Increasing concentrations of each indicated nucleic acid were added to H6.Gag.3h at the specified molar ratios before incubation with CRM1 and the nonhydrolyzable mutant RanQ69LGST before GST pull-down and bound proteins were detected by Coomassie blue. (B) RNA (μψ, mutant-μψ, or nonviral 109mer) was added to a mixture of proteins (GST.CRM1, RanQ69LGTP, and H6.Gag.3h) at a 0.1 or 1.0 molar ratio. Complexes were pulled down, and Gag was detected by Western blotting. (C) GST.CRM1 was incubated with H6.Gag.3h or H6.NLS.GFP, and proteins bound to glutathione beads were visualized using Coomassie blue. The presence (+) or absence (−) of RanQ69LGST is indicated. For A–C, the data are representative of three independent experiments.
To determine more quantitatively whether this effect was selective for μψ-RNA, we performed the pull-downs using a lower concentration of Gag (20 nM) and μψ-RNA, a non-vRNA, or a nonfunctional mutant of μψ (mut-μψ-RNA) (23) (Fig. 4B). At a 0.1 molar ratio of nucleic acids to Gag, there was no major difference in the degree of Gag:CRM1 association, but at a higher molar ratio (1.0), significantly more Gag was pulled down with CRM1 using μψ-RNA compared with non-vRNA (372-fold increase; P = 0.0001) or mut-μψ-RNA (9.78-fold increase; P = 0.0002) (Fig. S5). In the absence of nucleic acids, the small amount of Gag that bound to GST-CRM1 did so in a Ran-dependent manner (Fig. 4C). Specificity was demonstrated by the absence of Gag:GST interaction and by the lack of H6.NLS binding to CRM1. Together, these results suggest that Gag binds directly to CRM1 and that Gag:CRM1:RanGTP ternary complexes form cooperatively in the presence of vRNA, promoting nuclear egress of viral ribonucleoprotein complexes bound for the plasma membrane for particle assembly.
Discussion
RSV serves as an informative model for studying the regulation of complex subcellular transport pathways. The Gag polyprotein undergoes an ordered series of steps that lead from synthesis on cytosolic ribosomes, into and out of the nucleus, and, ultimately, to the plasma membrane, where nascent virus particles emerge. Our studies demonstrate that Gag binds directly to karyopherins imp-α and imp-11 but that binding is inhibited by nucleic acids, most notably by the vRNA packaging signal ψ. Binding to the exportin CRM1 is stimulated by Gag-nucleic acid interactions, suggesting that Gag-vRNA binding serves as a switch to signal nuclear export of the viral ribonucleoprotein complex, the earliest building block of the assembling virus particle.
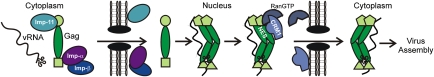
Based on our data, we present a working model (Fig. 5) in which the MA and NC NLSs in newly synthesized Gag proteins engage imp-11 and imp-α/β. Gag and imp-α form stable 1:1 complexes in vitro, suggesting that Gag binds to the import receptors as a monomer. Once in the nucleus, Gag is released from the import factors and binds to genomic vRNA, primarily through interaction of the NC domain with ψ. MA might also bind to vRNA, likely at a different site. On vRNA binding, Gag forms a dimer or oligomer (30) that exposes the p10 NES or increases its affinity for CRM1-RanGTP, leading to export of the viral ribonucleoprotein complex through the nuclear pore. Although ψ-RNA selectively promotes Gag:CRM1 binding, considering that only two copies of vRNA are incorporated into each virion, it is possible that other RNAs in the nucleus [e.g., U6 and other small RNAs enriched in retroviral particles (31–34)] might also bind Gag to stimulate nuclear export. Once in the cytoplasm, the MA and NC regions of Gag do not reassociate with importins because they are engaged in higher affinity interactions with RNA. We speculate that the membrane-binding signal in Gag becomes active, via either a structural change in MA or interaction with a cellular partner, to direct Gag:vRNA complexes toward the plasma membrane, where they associate with additional Gag proteins to complete particle assembly.
Fig. 5.
Model of vRNA-regulated nucleocytoplasmic trafficking of Gag. After synthesis in the cytoplasm, monomeric Gag binds to host import receptors, preventing Gag from associating with nucleic acids. The Gag:importin complex translocates into the nucleus and disassociates, whereupon Gag binds to vRNA and dimerizes, resulting in a conformational change that exposes the p10 NES and facilitates binding to CRM1:RanGTP. The Gag:vRNA complex is exported through the nuclear pore, releasing CRM1 and RanGTP. Gag, bound to its vRNA genome, is unable to reassociate with the importins and is instead transported to the plasma membrane for virus particle assembly and release.
Although the RanGTP/GDP gradient across the nuclear envelope provides directionality for nucleocytoplasmic trafficking (2), we present compelling evidence that vRNA binding serves as an additional mechanism to ensure efficient transport of the Gag protein out of the nucleus. Cellular proteins adopt related strategies, including the RNA-editing enzyme ADAR-1, for which binding of double-stranded RNA inhibits nuclear import and facilitates export (35). Thus, Gag-vRNA binding serves multiple critical roles, such as the molecular switch to trigger nuclear export of Gag, a platform for Gag dimerization (36), and the mechanism for specific encapsidation of the viral genome.
The results of this study provide an explanation for how competing subcellular targeting signals in Gag are regulated; however, a number of intriguing questions remain. Why does Gag contain two independent NLSs that interact with different importins? It is possible that the NLSs in MA and NC are redundant to enhance efficiency of nuclear localization (37). Alternatively, the NLSs might not be equivalent; their interactions with importins may be hierarchical, allowing intranuclear levels of Gag to be “fine-tuned” (1, 38). Thus, the amount of Gag in the nucleus might vary if import occurs through the NC:imp-α/β pathway vs. the MA:imp-11 pathway (13, 38). This mechanism might maintain low concentrations of Gag in the nucleus to prevent premature particle assembly. Additionally, it is plausible that each importin directs Gag to a specific subnuclear site that plays a role in virus assembly (39). Finally, it is possible that imp-11 and imp-α/β interactions are involved in nuclear import of mature MA and NC proteins during early infection to facilitate nuclear localization of the incoming proviral genome.
Finding that the NC:imp-α interaction was inhibited by nucleic acids was unexpected, because other protein-protein interactions mediated by retroviral NC sequences use nucleic acids as a bridge (40, 41). The observation that ψ-RNA and imp-α compete for binding to NC suggests that the binding sites for imp-α and nucleic acids overlap. It will be of interest to determine the structural basis of NC-imp-α binding to elucidate how the same region of NC adapts to participate in protein-RNA binding (with vRNA) or protein-protein interactions (with imp-α).
In contrast to Gag-importin interactions, Gag:CRM1:RanGTP complex formation was stabilized by ψ-RNA. CRM1 likely associates with a dimer of Gag, based on studies showing that nucleic acid binding promotes Gag dimerization (36, 42) and Gag dimers form in the nucleus (30). However, the crystal structure of the p10-CA Gag dimer shows that the NES is buried in hydrophobic interactions with CA, and is therefore unavailable to bind to CRM1 (17). One possible explanation for this apparent discrepancy is that the structure was determined for a p10-CA fragment lacking NC, such that the contribution of nucleic acids to the structure could not be evaluated. We propose that NC-RNA binding induces a conformational change in Gag, exposing the p10 NES to promote CRM1 binding. Further structural studies of the full-length Gag protein bound to nucleic acids and in complex with CRM1 are needed to answer this intriguing question.
Collectively, our findings illustrate a unique mechanism that links regulation of nucleocytoplasmic trafficking to retroviral genome packaging, demonstrating the intricate interplay that has evolved between karyopherins, nuclear transport pathways, and the intracellular pathogen RSV. Furthermore, these results promote understanding of the spatiotemporal regulation of subcellular trafficking in proteins that transit the nucleus en route to their final destination at the plasma membrane. Future experiments will test the predictions of our proposed model using structural biochemistry to define the conformational changes induced by sequential binding of karyopherins and vRNA to regulate the dual localization of the Gag protein.
Methods
Cell Lines, Viruses, and Plasmids.
QT6 quail fibroblasts were maintained as described and transfected using calcium phosphate (43). pRC.V8 (44), pGag-GFP (45), pMA-GFP, pΔMA.Gag-GFP, pYFP-NC (13), pΔNC.Gag-GFP (46), pKH3.importin.11, pKH3.importin.β (20), GST.importin-α, GST.importin-β, GST.H6.RanQ69L (47), pGST.CRM1 (48), pH6.NLS-GFP (49), GST-tagged karyopherins β2 and β3 (50), RSV NC (51), and RSV CA protein (52) were described. pΔMA.Gag-GFP was PCR-amplified and inserted into pΔNC.Gag-GFP using SstI-SdaI to make pΔMA.ΔNC.Gag-GFP. A KpnI-SstI fragment from pRC.V8 was transferred into pQE30 to make pH6.MA. The Gag.3h (53) sequence was PCR-amplified using a variant of pRC.V8 that differed from the published pAT.V8 (44) sequence [T1327 to C, resulting in a valine to alanine substitution that does not affect infectivity, RNA packaging, or budding (8)] and inserted into pET-28TEV (54). The RSV minimal packaging signal μψ (23) was amplified from pGEM7Zf+RSV−LTR.MA (43) and transferred into pGEM7Zf+ (Promega). A 109-nt non-vRNA was synthesized from pGEM72f+ after digestion with NsiI. RNAs were synthesized using Ribomax (Promega).
Recombinant Protein Expression and Purification.
Protein constructs were expressed in Escherichia coli (BL21 DE3 pRIL; Stratagene) in ZYP-5052 autoinduction media (55). For H6.Gag.3h, the pellet was lysed in BugBuster lysis buffer (Novagen), centrifuged at 20,000 × g, resuspended in buffer A [10 mM Hepes (pH 7.5), 0.5 M NaCl, and 50 mM imidazole], sonicated, and loaded onto a NiNTA column (GE Healthcare). Proteins were eluted with Buffer B [10 mM Hepes (pH 7.5), 0.5 M NaCl, and 0.5 M imidazole]. Fractions containing H6.Gag.3h were purified on a Superdex200 column (GE Healthcare) or by phenyl sepharose chromatography using 0.1 M NaCl in 25 mM Tris-HCl (pH 7.5) and 0.5 mM DTT. Pellets from H6.NLS-GFP and H6.MA cultures were not sonicated; NiNTA column fractions were pooled and loaded onto a Superdex75 column (Pharmacia) in 25 mM Tris-HCl (pH 7.5), 0.5 M NaCl, and 0.5 mM DTT. H6.RanQ69LGTP was purified as described elsewhere (56). Purified proteins were concentrated to ∼5 mg/mL. The absorbance ratio at wavelengths 260:280 for each protein was less than 0.62, indicating negligible nucleic acid contamination. For GST-tagged proteins, bacterial pellets were resuspended in 20 mM Tris (pH 7.5), 0.1% Triton X-100, and benzonase (15 U/mL); sonicated; and purified using a glutathione column, followed by Superdex 200 (or Superdex 75 for GST) in 25 mM Tris-HCl (pH 7.5) and 0.5 M NaCl.
Laser-Scanning Confocal Microscopy.
QT6 cells were fixed and permeabilized with 2% (wt/vol) paraformaldehyde and methanol. HA-tagged importins were visualized with anti-HA rhodamine-conjugated antibody (Roche), and Gag was visualized with anti-RSV rabbit antibody and FITC-conjugated goat-anti-mouse antibody. Cells expressing GFP (excited at 488 nm) and double-stranded red or rhodamine (excited at 543 nm) were imaged with a Leica SP2 AOBS confocal microscope using sequential scanning. Images were equally adjusted for intensity using CorelDraw ×3 (Corel Corporation). Quantification of the nuclear fluorescence intensity of Gag proteins was calculated by dividing the fluorescence intensity of the nucleus by the fluorescence intensity of the entire cell (Leica Microsystems software) (30).
Coimmunoprecipitation.
Cells expressing HA.imp-β were lysed in 25 mM Tris, 10 mM MgCl2, 100 mM NaCl, 0.5% Nonidet P-40, and 10% (vol/vol) glycerol. Cells expressing HA.imp-11 were lysed as described elsewhere (57). Soluble fractions were incubated with anti-GFP (Abcam), anti-RSV (53), or anti-VP22 (58) and bound to protein A beads (Pierce). Western blots were probed with anti-HA (Roche), anti-RSV, or anti-GFP and with HRP-conjugated secondary antibodies (Sigma), with detection by chemiluminescence.
In Vitro Binding.
Recombinant proteins (5 μM or 20 nM) were mixed with nucleic acids at a range of molar ratios extending from 0 to 1.0 in 25 mM Tris-HCl (pH 7.5), 0.05 mM DTT, and 0.1 M NaCl at 4 °C, and glutathione beads (GE Healthcare) were added. Bound proteins were eluted and visualized using either Coomassie blue (Bio-Rad) or Western blotting for H6.Gag.3h utilizing anti-RSV, followed by mouse-anti-rabbit FITC-conjugated secondary antibody. Binding reactions were performed in triplicate, and bands were quantified using ImageJ software (59). For far Western blot analysis, KH3-importin.11 was immunoprecipitated from transfected QT6 cells using anti-HA antibody and detected by Western blotting. The blot was stripped and incubated with 0–6 M guanidinium-HCl (60, 61). Purified protein H6.MA, H6.Gag.3h, or H6.NLS-GFP was used as a probe, incubated with appropriate antibodies, detected via Western blotting, and quantified using densitometry and ImageJ software.
Multiangle Laser Light Scattering.
A total of 16 μM H6.Gag.3h or NC ± GST.imp-α at a 1:1 molar ratio was applied to a Superdex200 HR 10/300 GL column (Amersham Pharmacia); equilibrated in 20 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 1 mM DTT, and 0.5% sodium azide; and eluted at a flow rate of 0.4 mL/min at 27 °C. The molecular weights of the complexes were determined using an in-line Dawn Heleos II multiangle laser light-scattering detector (Wyatt Technology) and ASTRA software (ASTRA Software Corp.). The protein concentration was monitored by changes in refractive index using an Optilab rEX detector (Wyatt Technology Corp.).
Supplementary Material
Acknowledgments
We are grateful for the generosity of scientists who contributed reagents: Stephen Burley, Gino Cingolani, Richard Courtney, Rebecca Craven, Bryan Cullen, Robert Gorelick, Michael Malim, Junona Moroianu, and Scott Plafker. We thank members of the Parent and Flanagan laboratories and the Department of Microbiology and Immunology for critical discussions of this work. This project was supported by Grant R01CA076534 (to L.J.P.) from the National Institutes of Health and the National Cancer Institute. Funding was provided to L.J.P. and N.G. from the Pennsylvania Department of Health using Tobacco Settlement Funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000304107/-/DCSupplemental.
References
- 1.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 2.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 3.Gardiner FC, Costa R, Ayscough KR. Nucleocytoplasmic trafficking is required for functioning of the adaptor protein Sla1p in endocytosis. Traffic. 2007;8:347–358. doi: 10.1111/j.1600-0854.2007.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens J, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 5.Itoh K, Brott BK, Bae GU, Ratcliffe MJ, Sokol SY. Nuclear localization is required for Dishevelled function in Wnt/beta-catenin signaling. J Biol. 2005 doi: 10.1186/jbiol20. 10.1186/jbiol20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc Natl Acad Sci USA. 2002;99:3944–3949. doi: 10.1073/pnas.062652199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanstrom R, Wills JW. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 8.Garbitt-Hirst R, Kenney SP, Parent LJ. Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging. J Virol. 2009;83:6790–6797. doi: 10.1128/JVI.00101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont S, et al. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402:681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 10.Nash MA, Meyer MK, Decker GL, Arlinghaus RB. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J Virol. 1993;67:1350–1356. doi: 10.1128/jvi.67.3.1350-1356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasundaram D, Benedik MJ, Morphew M, Dang VD, Levin HL. Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol Cell Biol. 1999;19:5768–5784. doi: 10.1128/mcb.19.8.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schliephake AW, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ. Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J Virol. 2006;80:1798–1806. doi: 10.1128/JVI.80.4.1798-1806.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheifele LZ, Ryan EP, Parent LJ. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J Virol. 2005;79:8732–8741. doi: 10.1128/JVI.79.14.8732-8741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verderame MF, Nelle TD, Wills JW. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weldon RA, Jr, Wills JW. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandhagopal N, et al. Dimeric rous sarcoma virus capsid protein structure relevant to immature Gag assembly. J Mol Biol. 2004;335:275–282. doi: 10.1016/j.jmb.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 18.Scheifele LZ, Kenney SP, Cairns TM, Craven RC, Parent LJ. Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology. J Virol. 2007;81:10718–10728. doi: 10.1128/JVI.01061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timney BL, et al. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plafker SM, Macara IG. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 2000;19:5502–5513. doi: 10.1093/emboj/19.20.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Musser SM. Nuclear import time and transport efficiency depend on importin beta concentration. J Cell Biol. 2006;174:951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 23.Banks JD, Linial ML. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J Virol. 2000;74:456–464. doi: 10.1128/jvi.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, McAllen JK, Tailor Y, Summers MF. High affinity nucleocapsid protein binding to the muPsi RNA packaging signal of Rous sarcoma virus. J Mol Biol. 2005;349:976–988. doi: 10.1016/j.jmb.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 25.Steeg CM, Vogt VM. RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J Virol. 1990;64:847–855. doi: 10.1128/jvi.64.2.847-855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darlix JL, Spahr PF. Binding sites of viral protein P19 onto Rous sarcoma virus RNA and possible controls of viral functions. J Mol Biol. 1982;160:147–161. doi: 10.1016/0022-2836(82)90172-3. [DOI] [PubMed] [Google Scholar]
- 27.Leis JP, McGinnis J, Green RW. Rous sarcoma virus p19 binds to specific double-stranded regions of viral RNA: Effect of p19 on cleavage of viral RNA by RNase III. Virology. 1978;84:87–98. doi: 10.1016/0042-6822(78)90220-9. [DOI] [PubMed] [Google Scholar]
- 28.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: To the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenney SP, Lochmann TL, Schmid CL, Parent LJ. Intermolecular interactions between retroviral Gag proteins in the nucleus. J Virol. 2008;82:683–691. doi: 10.1128/JVI.02049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rulli SJ, Jr, et al. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giles KE, Caputi M, Beemon KL. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA. 2004;10:299–307. doi: 10.1261/rna.2150604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen PJ, Cywinski A, Taylor JM. Reverse transcription of 7S L RNA by an avian retrovirus. J Virol. 1985;54:278–284. doi: 10.1128/jvi.54.2.278-284.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia EL, et al. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J Virol. 2009;83:12526–12534. doi: 10.1128/JVI.01219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fritz J, et al. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol. 2009;29:1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma YM, Vogt VM. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J Virol. 2004;78:52–60. doi: 10.1128/JVI.78.1.52-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wodrich H, et al. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J Virol. 2006;80:9608–9618. doi: 10.1128/JVI.00850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo M, Pang CW, Gerken AE, Brock TG. Multiple nuclear localization sequences allow modulation of 5-lipoxygenase nuclear import. Traffic. 2004;5:847–854. doi: 10.1111/j.1600-0854.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- 39.Pemberton LF, Rosenblum JS, Blobel G. Nuclear import of the TATA-binding protein: Mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol. 1999;145:1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svarovskaia ES, et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 42.Ma YM, Vogt VM. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J Virol. 2002;76:5452–5462. doi: 10.1128/JVI.76.11.5452-5462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garbitt RA, Albert JA, Kessler MD, Parent LJ. trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J Virol. 2001;75:260–268. doi: 10.1128/JVI.75.1.260-268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craven RC, Leure-duPree AE, Erdie CR, Wilson CB, Wills JW. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus gag protein. J Virol. 1993;67:6246–6252. doi: 10.1128/jvi.67.10.6246-6252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callahan EM, Wills JW. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J Virol. 2000;74:11222–11229. doi: 10.1128/jvi.74.23.11222-11229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callahan EM, Wills JW. Link between genome packaging and rate of budding for Rous sarcoma virus. J Virol. 2003;77:9388–9398. doi: 10.1128/JVI.77.17.9388-9398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Cullen BR. Structural and functional analysis of the avian leukemia virus constitutive transport element. RNA. 1999;5:1645–1655. doi: 10.1017/s1355838299991616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen MH, et al. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin alpha. J Biol Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- 50.Yaseen NR, Blobel G. Cloning and characterization of human karyopherin beta3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart-Maynard KM, et al. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purdy JG, Flanagan JM, Ropson IJ, Rennoll-Bankert KE, Craven RC. Critical role of conserved hydrophobic residues within the major homology region in mature retroviral capsid assembly. J Virol. 2008;82:5951–5961. doi: 10.1128/JVI.00214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weldon RA, Jr, Erdie CR, Oliver MG, Wills JW. Incorporation of chimeric Gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonanno JB, et al. Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis. Proc Natl Acad Sci USA. 2001;98:12896–12901. doi: 10.1073/pnas.181466998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Hieda M, et al. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J Cell Biol. 1999;144:645–655. doi: 10.1083/jcb.144.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lischka P, Sorg G, Kann M, Winkler M, Stamminger T. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J Virol. 2003;77:3734–3748. doi: 10.1128/JVI.77.6.3734-3748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Regan KJ, Murphy MA, Bucks MA, Wills JW, Courtney RJ. Incorporation of the herpes simplex virus type 1 tegument protein VP22 into the virus particle is independent of interaction with VP16. Virology. 2007;369:263–280. doi: 10.1016/j.virol.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 59.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 60.Guichet A, et al. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz CJ, et al. FTZ-Factor1 and Fushi tarazu interact via conserved nuclear receptor and coactivator motifs. EMBO J. 2001;20:510–519. doi: 10.1093/emboj/20.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.