Abstract
Approximately 100 genes undergo genomic imprinting. Mutations in fewer than 10 imprinted genetic loci, including GNAS, are associated with complex human diseases that differ phenotypically based on the parent transmitting the mutation. Besides the ubiquitously expressed Gsα, which is of broad biological importance, GNAS gives rise to an antisense transcript and to several Gsα variants that are transcribed from the nonmethylated parental allele. We previously identified two almost identical GNAS microdeletions extending from exon NESP55 to antisense (AS) exon 3 (delNESP55/delAS3-4). When inherited maternally, both deletions are associated with erasure of all maternal GNAS methylation imprints and autosomal-dominant pseudohypoparathyroidism type Ib, a disorder characterized by parathyroid hormone–resistant hypocalcemia and hyperphosphatemia. As for other imprinting disorders, the mechanisms resulting in abnormal GNAS methylation are largely unknown, in part because of a paucity of suitable animal models. We now showed in mice that deletion of the region equivalent to delNESP55/delAS3-4 on the paternal allele (ΔNesp55p) leads to healthy animals without Gnas methylation changes. In contrast, mice carrying the deletion on the maternal allele (ΔNesp55m) showed loss of all maternal Gnas methylation imprints, leading in kidney to increased 1A transcription and decreased Gsα mRNA levels, and to associated hypocalcemia, hyperphosphatemia, and secondary hyperparathyroidism. Besides representing a murine autosomal-dominant pseudohypoparathyroidism type Ib model and one of only few animal models for imprinted human disorders, our findings suggest that the Nesp55 differentially methylated region is an additional principal imprinting control region, which directs Gnas methylation and thereby affects expression of all maternal Gnas-derived transcripts.
Keywords: genomic imprinting, Gsα, pseudohypoparathyroidism, parathyroid hormone, hormonal resistance
Fewer than 100 genetic loci in mammals undergo methylation on the maternal or paternal allele, thereby limiting their expression to only one parental allele (1, 2). Mutations in fewer than 10 of these imprinted loci cause human disorders, which are associated with abnormal DNA methylation and differ phenotypically based on the parent transmitting the genetic defect. The mechanisms leading to changes in DNA methylation are unknown, partly because of a paucity of suitable animal models mimicking the human disorder.
The complex GNAS locus (chromosome 20q13.3; mouse distal chromosome 2) is one of the few differentially methylated regions of the genome that is associated with human disorders (1, 3). GNAS encodes the α-subunit of the stimulatory G protein (Gsα), which is important for cAMP-dependent signaling events downstream of a large variety of G protein–coupled receptors (3–6). By splicing three distinct first exons (A/B, XL, or NESP55; mouse exons 1A, Gnasxl, or Nesp55, respectively) onto GNAS exons 2 through 13 (mouse Gnas exons 2–12), several alternative transcripts are generated, including the paternally expressed sense transcripts XLαs, XXLαs, and A/B (in the mouse Xlαs, Xxlαs, and 1A, respectively) and the maternally expressed transcript NESP55 (in the mouse Nesp55) (7–9); furthermore, an antisense transcript AS [in the mouse, Nespas (10)] is expressed from the paternal allele (11). Allele-specific expression of the different transcripts is dictated by differential methylation of their promoters and first exons, which restrict transcription to the nonmethylated parental allele (12) (Fig. 1A).
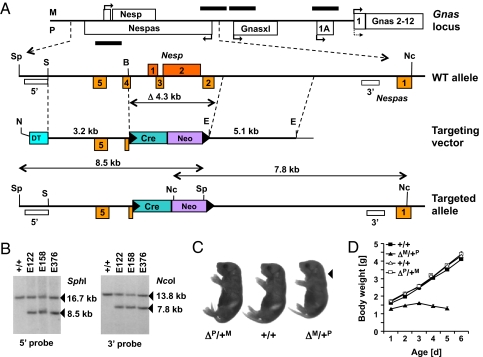
Fig. 1.
Targeted deletion of the Nesp55 DMR region and its effect on postnatal growth. (A) Schematic overview of the mouse Gnas locus and strategy to delete the Nesp55 DMR (not drawn to scale). Exons or genes transcribed in sense and antisense orientation are shown above and below (maternally and paternally expressed transcripts, respectively); promoters and direction of transcription are indicated by arrows. Although Gsα is biallelically expressed in most tissues, it is expressed in some cells/tissues predominantly from the maternal allele (solid arrow) with only limited or no expression from the paternal allele (dotted arrow). DMRs are indicated by black bars. The enlarged Nesp55/Nespas region below shows the position of selected restriction sites (B, BamHI; E, EcoRI; Nc, NcoI; S, SacI; Sp, SphI), of the 5′ and 3′ external probes (open rectangles), and of the locations of recombinogenic vector arms (dotted lines). The targeting vector furthermore contains a self-splicing floxed (filled triangles, loxP sites) pACN cassette (Cre, neo), which replaces Nesp55 exons 1 and 2, Nespas exons 2 and 3, and portions of Nespas exon 4, as well as a diphtheria cassette (DT); this vector was linearized with NotI (N; angular line). The scheme of the targeted allele indicates how the 8.5-kb and 7.8-kb targeted bands are generated. (B) Homologous recombination in three ES cell clones was verified by digestion of genomic DNA with SphI and NcoI, followed by blotting and hybridization with either the 5′ or the 3′ probe, respectively; +/+, WT ES cells. (C) Mice with the ΔNesp55 deletion on the paternal (ΔP/+M) or maternal (ΔM/+P) allele and a WT littermate from a mother of the genotype ΔNesp55p (+/+) at birth. The arrow shows s.c. edema, which was present only in ΔM/+P mice. (D) Weight curves demonstrating postnatal growth retardation of ΔNesp55m pups (ΔM/+P) compared with ΔNesp55p pups (ΔP/+M) and WT littermates from fathers (open symbols) or mothers (closed symbols) of the genotype ΔNesp55p. All data, including those of three mice that survived until postnatal d 5, are expressed as mean ± SEM.
The human disorders that are caused by heterozygous muta-tions within the GNAS locus include pseudohypoparathyroidism type Ia (PHP-Ia), which is caused by maternally inherited inactivating mutations affecting the exons encoding Gsα; pseudopseu-dohypoparathyroidism (PPHP) and progressive osseous heteroplasia (POH), which are both caused by paternally inherited inactivating mutations in these exons; and the McCune–Albright syndrome, which is caused by mutations that lead to constitutive Gsα activity (3–6). Furthermore, the autosomal-dominant (AD) form of PHP type Ib (AD-PHP-Ib) is caused by microdeletions within or upstream of GNAS or by uniparental isodisomy of chromosome 20q, and both are associated with loss of one or several methylation imprints on the maternal GNAS allele (3). PHP-Ia is associated with multiple hormone resistance, including toward parathyroid hormone (PTH) and thyroid stimulating hormone (TSH), and Albright hereditary osteodystrophy (AHO) (3–6). Like with PHP-Ia, patients affected by PHP-Ib develop resistance toward PTH leading to hypocalcemia and hyperphosphatemia, which can sometimes be associated with mild resistance to TSH; unlike PHP-Ia, PHP-Ib appears to be only rarely associated with AHO features, such as shortening of metacarpals (13–15).
AD-PHP-Ib is caused by microdeletions within STX16 (16, 17) or within GNAS (delNESP55/ASdel3-4) (18), which are associated with loss of maternal exon A/B methylation alone or with loss of methylation at all maternal GNAS differentially methylated regions (DMRs), respectively. Either of these epigenetic changes is associated, as determined in peripheral blood cells, with increased A/B transcription, which is thought to reduce Gsα expression in the proximal renal tubules, where this ubiquitous signaling protein appears to be derived predominantly from the maternal allele (6, 15); as a result, PTH resistance develops in this tissue. Mice lacking the equivalent of one of the STX16 deletions failed to reproduce human AD-PHP-Ib, suggesting that the region important for establishing or maintaining exon A/B methylation is located at a different location in mice (19). We now generated mice with targeted deletion of the Gnas region that is equivalent to delNESP55/ASdel3-4 identified in two AD-PHP-Ib families and show that maternal inheritance of this deletion leads to loss of all maternal methylation imprints and to PTH resistance.
Results and Discussion
To generate mice carrying a deletion similar to delNESP55/ASdel3-4 (18) (ΔNesp55), a targeting vector was constructed that removes a 4.3-kb genomic DNA fragment that includes most of the CpG island extending from Nespas exon 4 to Nespas intron 1 (20), thus deleting both murine Nesp55 exons, as well as Nespas exons 2 and 3 and portions of Nespas exon 4 (Fig. 1 A and B). Mice that inherited the ΔNesp55 allele from their father, i.e., ΔNesp55p offspring, revealed no obvious developmental defects or growth deficiency. In contrast, ΔNesp55m mice had lower birth weights than their WT littermates (Fig. 1 C and D) and failed to gain weight during postnatal development; no survival beyond d 5 was observed. Furthermore, some pups had narrow bodies, were hyperactive, and showed s.c. edema, i.e., abnormalities that are similar to those observed in mice with paternal uniparental disomy for distal chromosome 2 (pDp2) (21).
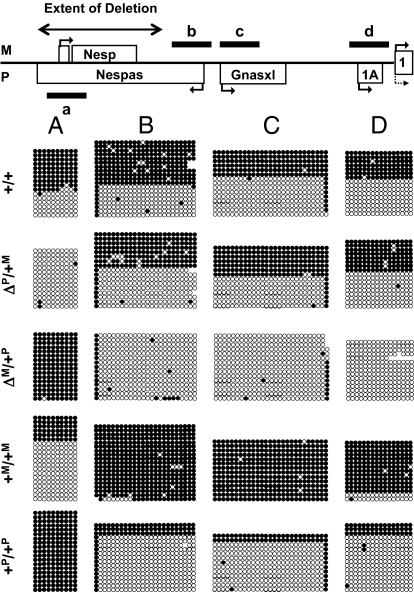
Nucleotide sequence analysis of individual PCR clones derived from bisulfite-treated genomic DNA of ΔNesp55m and pDp2 mice showed no evidence for an unmethylated Nesp55 DMR and a complete loss or close to complete loss, respectively, of all maternal methylation imprints (Fig. 2). These included the exon 1A DMR, which represents, besides the Nespas/Gnasxl DMR, another subordinated ICR within the Gnas cluster that is established in the germline (22). Loss of methylation at the corresponding human A/B DMR is invariably observed in all patients with AD-PHP-Ib and in most patients with sporadic PHP-Ib (16, 18, 23–25), and is associated with an increase in A/B mRNA transcripts (1A mRNA transcripts in the mouse). In contrast, ΔNesp55p mice showed no evidence for a methylated Nesp55 DMR, which is consistent with the extent of the introduced deletion. Compared with WT animals, there was no obvious change in the methylation pattern at the other DMRs; data were confirmed by Southern blot analyses using methylation-sensitive restriction enzymes (Fig. S1). Overall these molecular findings are indistinguishable from those of families with delNESP55/ASdel3-4 (18) and similar to those previously reported for human paternal uniparental isodisomy comprising the GNAS locus (26), thus establishing the Nesp55 DMR as another primary ICR of the Gnas locus.
Fig. 2.
Schematic representation of the mouse Gnas locus (Top) depicting the location of the analyzed DMRs (black bars) and the extent of the deletion (two-sided arrow) (Note: not drawn to scale). Analysis of the DMRs of the Gnas locus: (A) Nesp55; (B) Nespas exon 1 (Nespas/Gnasxl DMR); (C) Gnasxl exon1 (Nespas/Gnasxl DMR); and (D) exon 1A. Exons transcribed in the sense and antisense orientation are shown above and below (maternally and paternally expressed transcripts, respectively); promoters and direction of transcription are indicated by arrows. Bisulfite-treated genomic DNA from livers of 2-d-old WT littermates (+/+) or animals with paternally or maternally inherited ∆Nesp55 (ΔP/+M or ΔM/+P, respectively) was PCR-amplified and cloned for nucleotide sequence analysis. Maternal (+M/+M) and paternal (+P/+P) uniparental disomic mice with respect to distal chromosome 2 were included in the analysis. Each row of circles represents a clone and each circle corresponds to a separate CpG (filled circles, methylated CpG; open circles, nonmethylated CpG). Each block of circles represents the data from an individual mouse.
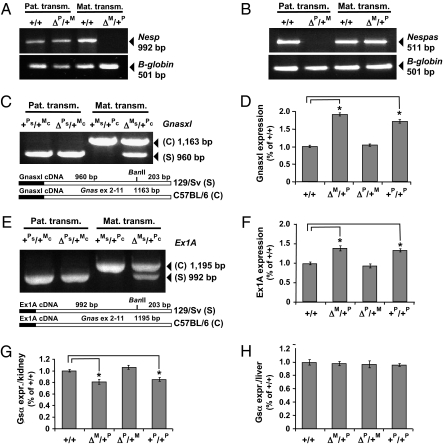
Analysis of total brain RNA by RT-PCR (Fig. 3A) and Northern blot (Fig. S2) revealed no evidence for Nesp55 transcripts in ΔNesp55m mice, but normal Nesp55 expression in brains of ΔNesp55p mice. Similar to the findings in patients with maternally inherited delNESP55/ASdel3-4, Nespas expression occurred from the paternal allele in ΔNesp55m animals, as determined by analysis of total embryo RNA. However, unlike in healthy carriers with paternally inherited delNESP55/ASdel3-4, no Nespas transcript was detected in ΔNesp55p embryos (Fig. 3B and data not shown). This discrepancy is most likely related to the fact that the ablated mouse genomic region included both Nesp exons, Nespas exons 2 and 3, as well as portions of Nespas exon 4, possibly leading to an unstable message. Consistent with the observed methylation changes at the Nespas/Gnasxl DMR, Xlαs transcription occurred in ΔNesp55m animals from both parental alleles leading to an approximate 1.9-fold increase of its message; in contrast, analysis of the mRNA encoding Xlαs from ΔNesp55p animals revealed no evidence for biallelic expression and thus no increased expression (Fig. 3 C and D). Likewise, because of the loss of methylation at the maternal exon 1A DMR, 1A transcription occurred biallelically, resulting in an approximate 1.4-fold increase in mRNA, as judged by quantitative real-time RT-PCR (qRT-PCR; Fig. 3 E and F); both changes were similar to those observed in pDp2 mice.
Fig. 3.
RT-PCR analysis of mRNA transcripts derived from the imprinted Gnas promoters. (A) Analysis of Nesp55 expression using total RNA from brain of 2-d-old WT mice (+/+) or animals with paternally or maternally inherited ΔNesp55 (ΔP/+M or ΔM/+P, respectively); β-globin, amplification control. (B) analysis of Nespas expression using total RNA from 15.5 d postcoitus embryos; β-globin, amplification control; analysis of Gnasxl (C) and 1A expression (E) using total RNA from brain and kidney, respectively, of 2-d-old pups; reciprocal crosses between mice with the ΔNesp55 allele in the 129/SvJ background (Δs, recombinant allele; +s, WT allele) and C57BL/6 mice (+c, WT allele); to determine whether Xlαs or 1A transcripts were derived from the maternal or the paternal allele, PCR products were incubated with BanII, which cuts cDNA derived from 129/SvJ (s) RNA, but not from C57BL/6J (c) RNA, due to a SNP located in Gnas exon 10, as described previously (19). qRT-PCR using total RNA from WT mice (+/+), animals with paternally or maternally inherited ΔNesp55 (ΔP/+M or ΔM/+P, respectively), as well as pDp2 animals (+P/+P); all expression levels were normalized to Actb mRNA and are shown relative to the expression levels in WT mice (normalized to 100%); mean ± SEM of six independent experiments with genetically manipulated mice and of 12 independent experiments with WT mice; asterisk, P < 0.001 vs. +/+. (D) Expression of Xlαs in brain. (F) Expression of 1A in kidney. (G) Expression of Gsα in kidney. (H) Expression of Gsα in liver.
Loss of exon 1A methylation and the resulting biallelic 1A mRNA transcription had previously been predicted to suppress biallelic Gsα transcription in the renal proximal tubules (and presumably in other tissues/cells in which Gsα expression is thought to occur only from the maternal allele) to an extent that is sufficient to induce PTH-resistance and the resulting changes in mineral ion homeostasis (16, 23–25). Indeed, a readily detectable decrease in Gsα mRNA transcripts was observed in total RNA from whole kidneys of ΔNesp55m mice (Fig. 3G), although monoallelic Gsα expression is thought to be confined to the proximal tubules (27, 28); in contrast, no obvious change in Gsα expression was detected in kidneys from ΔNesp55p mice or in liver from ΔNesp55m mice (Fig. 3H). Reduced Gsα expression, presumably in the proximal renal tubules, was associated with increased serum levels of PTH and phosphorous, as well as reduced ionized calcium (Table 1). These biochemical changes on postnatal d 2 were consistent with PTH resistance, which occurred earlier than observed in humans with AD or sporadic PHP-Ib, who usually do not develop symptomatic hypocalcemia early in life (16, 18, 24, 25). It is conceivable that a rapid further decline in blood calcium levels occurred over the next 2 to 3 d, which could have contributed to the invariable demise of ΔNesp55m mice by postnatal d 5. pDp2 mice also die perinatally and show a similar phenotype as ΔNesp55m mice, and both animals have biallelic Xlαs expression (Fig. 3D). Despite these similarities, however, it appears unlikely that increased Xlαs levels contribute to the postnatal death of ΔNesp55m mice, as patients with PHP-Ib with broad methylation GNAS changes resulting from delNESP55/ASdel3-4 (18), patUPD20q (26), or yet unknown molecular defects (25) show no early lethality.
Table 1.
PTH, ionized calcium, and phosphorous concentrations in blood from WT and ΔNesp55 mice at day 2 after birth
| Genotype of offspring | PTH (pg/mL) | Ca (mmol/L) | Pi (mmol/L) |
| Parental genotype: mother ΔP/+M, father +/+ | |||
| +/+ | 21.9 ± 0.51 | 1.42 ± 0.02 | 8.86 ± 0.85 |
| ΔM/+P | 55.2 ± 5.10* | 1.24 ± 0.06† | 12.81 ± 0.68‡ |
| Parental genotype: mother +/+, father ΔP/+M | |||
| +/+ | 22.4 ± 1.71 | 1.38 ± 0.04 | 8.91 ± 0.90 |
| ΔP/+M | 22.8 ± 1.18 | 1.36 ± 0.04 | 9.00 ± 0.39 |
Results are mean ± SEM (n = 8 in each group). Statistical comparisons were made using the two-tailed Student t test within paternal or maternal offspring groups. ΔM/+P mice were significantly different from WT littermates (+/+), while +/+ and ΔP/+M mice had indistinguishable findings.
*P = 0.0003, serum PTH.
†P = 0.019, ionized calcium.
‡P = 0.009, inorganic serum phosphorous.
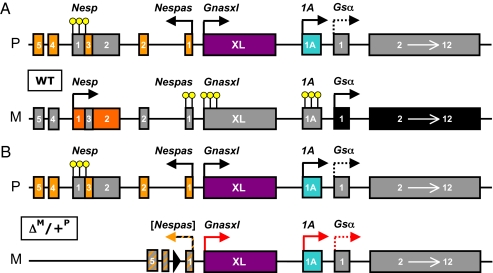
It has proven difficult to generate mouse models of imprinted human diseases (1, 19, 29). Different from the previously generated mice lacking three Stx16 exons, which failed to develop PTH resistance (19), ΔNesp55m mice recapitulate the epigenetic changes observed in patients with maternally inherited delNESP55/ASdel3-4 and develop hypocalcemia and hyperphosphatemia despite elevated PTH levels, thus providing a model of AD-PHP-Ib. Our findings furthermore indicate that the Nesp55 DMR represents another principal ICR within the complex Gnas cluster, which regulates in cis methylation of Nespas/Gnasxl and 1A. Both the Nesp55 DMR and the hierarchically equal Nespas/Gnasxl DMR (30) appear to regulate methylation of the subordinated exon 1A DMR (30, 31) through mechanisms involving active Nesp55 transcription in the oocyte (32). Derepression of 1A transcription through loss of methylation limits Gsα expression in imprinted tissues such as the proximal renal tubules, thus leading to the development of hormonal resistance (Fig. 4). The role of Nespas RNA in this regulatory process may be similarly important as that of other noncoding RNAs, which appear to be involved in silencing several autosomal and X-chromosomal genes (33).
Fig. 4.
Schematic presentation of the changes induced by the 4.3-kb deletion from the Nesp55 DMR. (A) Methylation status of the Gnas DMR and parent-specific expression of transcripts derived from the Gnas locus in imprinted tissues. Paternal (P) and maternal (M) alleles are depicted as lines with exons (boxes), methylated promoters (yellow circles), and transcriptional arrows (black arrows) showing the start site and direction of transcription of Gnas-derived coding and noncoding/nontranslated mRNAs. Exons filled in gray indicate silenced, or in the case of Gsα, poorly expressed transcripts. Although Gsα is biallelically expressed in most tissues, it is predominantly expressed from the maternal allele in some tissues (bold arrow) with little or no expression from the paternal allele (dotted arrow). (B) Model of the mechanism through which the loss of the Nesp55 DMR might lead to PTH resistance. Lack of the Nesp55 DMR on the maternal allele results in a loss of methylation at the two downstream DMRs, Nespas/Gnasxl and 1A, and consequent expression of Xlαs and 1A transcripts (red arrows), which are normally expressed only from the paternal allele. This suggests that the maternal unmethylated allele of Nesp55 could serve to repress expression of these transcripts from the paternal chromosome. As a consequence of biallelic 1A expression, Gsα expression is reduced in some tissues, including the proximal renal tubules, thus leading to PTH resistance. Because of the extent of the introduced deletion, its effect on the expression of the Nespas transcript could not be investigated (gray/red-shaded box and black/red-labeled arrow). The filled triangle represents the single loxP site that remains at the site of the introduced deletion. No effect on the imprinted expression of Gsα was observed after paternal transmission of ΔNesp55.
Methods
Construction of the Targeting Vector.
The pACN targeting vector was designed to delete the entire Nesp55 DMR of Gnas (nucleotides 70,086–74,480) encompassing Nesp55 exons 1 and 2, and the Nespas exons 2 and 3 as well as part of Nespas exon 4 (Fig. 1A and SI Methods). The vector contained the diphtheria toxin cassette driven by the thymidine kinase promoter and the neomycin gene driven by the RNA polymerase II promoter for negative and positive selection, respectively, of transfected embryonic stem (ES) cells; this selection cassette was flanked by loxP sites (34). The Cre recombinase gene (Cre) was driven by the testis-specific promoter (tACE) of the gene encoding angiotensin-converting enzyme, thus allowing self-excision of the selection cassette upon germline transmission.
Targeting of ES Cells and Mouse Breedings/Analyses.
The pACN vector was linearized with NotI before transfection of male J1 ES cells from mouse strain 129/SvJ (34). Colonies surviving G418 selection were screened by Southern blot analysis using SphI-digested genomic DNA, which was probed with a 32P-labeled 1.2-kb DNA fragment (nucleotides 77,987–79,227). Correct targeting at the 3′ end was confirmed by probing NcoI digests with a 289-bp fragment (nucleotides 62,540–62,829; Fig. 1 A and B). Three independently targeted ES cell clones were injected into C57BL/6J blastocysts, which were transferred into uteri of 2.5-d postcoitus pseudopregnant CD1 mice. Agouti-marked male chimeric mice were mated with 129/SvJ or C57BL/6J females to generate ΔNesp55p mice in either background. Female and male ΔNesp55p mice were then mated to generate ΔNesp55m and ΔNesp55p animals; both lines were maintained through ΔNesp55p males. RNA and methylation analyses and quantification of Gnas-derived transcripts by qRT-PCR was performed using standard techniques (SI Methods). Animal studies were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care (protocol number 2001N000183/2) and by the University of Veterinary Medicine Vienna institutional ethics committee (GZ BMBWK-68.205/0247-BrGT/2005).
Measurement of Ionized Calcium, Phosphorus, and PTH.
Ionized blood calcium concentration was measured using a 9180 Electrolyte Analyzer (AVL Medical Instruments), inorganic phosphorous was measured using the Stanbio Phosphorus Liqui-UV Procedure (Stanbio), and PTH concentrations were measured using a two-side enzyme-linked immunoassay specific for intact mouse PTH (Immutopics).
Statistical Analyses.
Data are presented as means ± SEM. Differences between WT and ΔNesp55p or ΔNesp55m mice were evaluated using the two-tailed Student t test.
GenBank Accession Numbers.
Nucleotide numbers cited in this publication refer to sequence accession number AL929537; pACN, AF169416.
Supplementary Material
Acknowledgments
We thank Dr. En Li (Massachusetts General Hospital) for J1 ES cells; G. Bounoutas for help with the electroporation of ES cells; T. Doetschman for the 129Sv library; K. R. Thomas for the pACN vector; M. Stuerzl for the pAL41 plasmid; and I. H. Maxwell for the DT-A cassette. This work was supported by grants from National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health Grants R01DK46718 (to H.J.) and R01DK073911 (to M.B.).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910224107/DCSupplemental.
References
- 1.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu Rev Genomics Hum Genet. 2009;10:241–262. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- 3.Bastepe M, Jüppner H. Pseudohypoparathyroidism, Gs {alpha}, and the GNAS locus. IBMS BoneKEy. 2005;2:20–32. [Google Scholar]
- 4.Spiegel AM, Weinstein LS. Inherited diseases involving g proteins and g protein-coupled receptors. Annu Rev Med. 2004;55:27–39. doi: 10.1146/annurev.med.55.091902.103843. [DOI] [PubMed] [Google Scholar]
- 5.Weinstein LS, Xie T, Zhang QH, Chen M. Studies of the regulation and function of the Gs alpha gene Gnas using gene targeting technology. Pharmacol Ther. 2007;115:271–291. doi: 10.1016/j.pharmthera.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plagge A, Kelsey G, Germain-Lee EL. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLalpha(s) in human and mouse. J Endocrinol. 2008;196:193–214. doi: 10.1677/JOE-07-0544. [DOI] [PubMed] [Google Scholar]
- 7.Hayward BE, et al. The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci USA. 1998;95:10038–10043. doi: 10.1073/pnas.95.17.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward BE, Moran V, Strain L, Bonthron DT. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA. 1998;95:15475–15480. doi: 10.1073/pnas.95.26.15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters J, et al. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wroe SF, et al. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc Natl Acad Sci USA. 2000;97:3342–3346. doi: 10.1073/pnas.050015397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayward BE, Bonthron DT. An imprinted antisense transcript at the human GNAS1 locus. Hum Mol Genet. 2000;9:835–841. doi: 10.1093/hmg/9.5.835. [DOI] [PubMed] [Google Scholar]
- 12.Peters J, et al. Imprinting control within the compact Gnas locus. Cytogenet Genome Res. 2006;113:194–201. doi: 10.1159/000090832. [DOI] [PubMed] [Google Scholar]
- 13.de Nanclares GP, et al. Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright's hereditary osteodystrophy. J Clin Endocrinol Metab. 2007;92:2370–2373. doi: 10.1210/jc.2006-2287. [DOI] [PubMed] [Google Scholar]
- 14.Mariot V, Maupetit-Méhouas S, Sinding C, Kottler ML, Linglart A. A maternal epimutation of GNAS leads to Albright osteodystrophy and parathyroid hormone resistance. J Clin Endocrinol Metab. 2008;93:661–665. doi: 10.1210/jc.2007-0927. [DOI] [PubMed] [Google Scholar]
- 15.Bastepe M, Jüppner H. GNAS locus and pseudohypoparathyroidism. Horm Res. 2005;63:65–74. doi: 10.1159/000083895. [DOI] [PubMed] [Google Scholar]
- 16.Bastepe M, et al. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest. 2003;112:1255–1263. doi: 10.1172/JCI19159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linglart A, Gensure RC, Olney RC, Jüppner H, Bastepe M. A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS. Am J Hum Genet. 2005;76:804–814. doi: 10.1086/429932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastepe M, et al. Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat Genet. 2005;37:25–27. doi: 10.1038/ng1487. [DOI] [PubMed] [Google Scholar]
- 19.Fröhlich LF, Bastepe M, Ozturk D, Abu-Zahra H, Jüppner H. Lack of Gnas epigenetic changes and pseudohypoparathyroidism type Ib in mice with targeted disruption of syntaxin-16. Endocrinology. 2007;148:2925–2935. doi: 10.1210/en.2006-1298. [DOI] [PubMed] [Google Scholar]
- 20.Coombes C, et al. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol Cell Biol. 2003;23:5475–5488. doi: 10.1128/MCB.23.16.5475-5488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20:5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, et al. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest. 2000;106:1167–1174. doi: 10.1172/JCI10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastepe M, et al. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet. 2001;10:1231–1241. doi: 10.1093/hmg/10.12.1231. [DOI] [PubMed] [Google Scholar]
- 25.Linglart A, Bastepe M, Jüppner H. Similar clinical and laboratory findings in patients with symptomatic autosomal dominant and sporadic pseudohypoparathyroidism type Ib despite different epigenetic changes at the GNAS locus. Clin Endocrinol (Oxf) 2007;67:822–831. doi: 10.1111/j.1365-2265.2007.02969.x. [DOI] [PubMed] [Google Scholar]
- 26.Bastepe M, Lane AH, Jüppner H. Paternal uniparental isodisomy of chromo-some 20q—and the resulting changes in GNAS1 methylation—as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet. 2001;68:1283–1289. doi: 10.1086/320117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu S, et al. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc Natl Acad Sci USA. 1998;95:8715–8720. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstein LS, Yu S, Ecelbarger CA. Variable imprinting of the heterotrimeric G protein G(s) alpha-subunit within different segments of the nephron. Am J Physiol Renal Physiol. 2000;278:F507–F514. doi: 10.1152/ajprenal.2000.278.4.F507. [DOI] [PubMed] [Google Scholar]
- 29.Peery EG, Elmore MD, Resnick JL, Brannan CI, Johnstone KA. A targeted deletion upstream of Snrpn does not result in an imprinting defect. Mamm Genome. 2007;18:255–262. doi: 10.1007/s00335-007-9019-3. [DOI] [PubMed] [Google Scholar]
- 30.Williamson CM, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- 31.Williamson CM, et al. A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat Genet. 2004;36:894–899. doi: 10.1038/ng1398. [DOI] [PubMed] [Google Scholar]
- 32.Chotalia M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.