Abstract
The Na+,K+-ATPase pump achieves thermodynamically uphill exchange of cytoplasmic Na+ ions for extracellular K+ ions by using ATP-mediated phosphorylation, followed by autodephosphorylation, to power conformational changes that allow ion access to the pump's binding sites from only one side of the membrane at a time. Formally, the pump behaves like an ion channel with two tightly coupled gates that are constrained to open and close alternately. The marine agent palytoxin disrupts this coupling, allowing both gates to sometimes be open, so temporarily transforming a pump into an ion channel. We made a cysteine scan of Na+,K+-ATPase transmembrane (TM) segments TM1 to TM6, and used recordings of Na+ current flow through palytoxin-bound pump-channels to monitor accessibility of introduced cysteine residues via their reaction with hydrophilic methanethiosulfonate (MTS) reagents. To visualize the open-channel pathway, the reactive positions were mapped onto a homology model of Na+,K+-ATPase based on the structure of the related sarcoplasmic- and endoplasmic-reticulum (SERCA) Ca2+-ATPase in a BeF3−-trapped state1,2, in which the extra-cytoplasmic gate is wide open (although the cytoplasmic access pathway is firmly shut). The results revealed a single unbroken chain of reactive positions that traverses the pump from the extracellular surface to the cytoplasm, comprises residues from TM1, TM2, TM4, and TM6, and passes through the equivalent of cation binding site II in SERCA, but not through site I. Cavity search analysis of the homology model validated its use for mapping the data by yielding a calculated extra-cytoplasmic pathway surrounded by MTS-reactive residues. As predicted by previous experimental results, that calculated extra-cytoplasmic pathway abruptly broadens above residue T806, at the outermost end of TM6 which forms the floor of the extracellular-facing vestibule. These findings provide a structural basis for further understanding cation translocation by the Na+,K+-ATPase and by other P-type pumps like the Ca2+- and H+,K+-ATPases.
The Na+,K+ pump, a P-type ATPase (named for the phosphorylated intermediate), generates the steep transmembrane concentration gradients of Na+ and K+ that are essential for cell life. Each pump exchanges three intracellular Na+ ions for two external K+ ions per ATP molecule hydrolyzed, up to one hundred times a second. This cation exchange is accomplished by conformational changes linked to phosphorylation and dephosphorylation of the pump3,4 that permit strictly alternating access to its ion binding sites: from the extracellular side in the phosphorylated state, called E2P, but from the cytoplasm in the dephosphorylated state, E1. The marine toxin palytoxin somehow disrupts this strict coupling between the Na+,K+ pump's two gates, so allowing both to sometimes be open, whereupon the pump is temporarily transformed into an ion channel5-7. Palytoxin thus provides an unprecedented opportunity to use direct recordings of ion flow through the Na+,K+ pump to monitor accessibility to small water-soluble probes of target cysteine residues introduced along the ion pathway8, and thereby characterize the routes to and from the ion binding sites9-13.
Mapping the resulting outline of the pathway onto a 3-dimensional structure is not straightforward, however, as the two available X-ray crystal structures of Na+,K+-ATPase14-15 both captured the same occluded-ion state with the two access routes closed. In contrast, current recordings in palytoxin-bound Na+,K+ pump-channels evidently monitor a conformation with both access routes open. Palytoxin is believed to stabilize an E2P-related conformation of the Na+,K+-ATPase because it binds most strongly to phosphorylated Na+,K+ pumps in the presence of Na+ ions7,16. The crystal structure of a BeF3−-trapped E2P-like state (PDB code 3B9B) of the sarcoplasmic- and endoplasmic-reticulum Ca2+-ATPase (SERCA), a related P-type pump, that captured an open extra-cytoplasmic pathway1,2, therefore provides a starting basis for structural evaluation of the palytoxin-bound Na+,K+ pump-channel pathway, despite the fact that the cytoplasmic-side access pathway of the SERCA pump in that structure is firmly shut.
In previous work, we used charged, or neutral, hydrophilic methanethiosulfonate (MTS) reagents to probe accessibility of strategically located cysteines, and we monitored consequences of their reactions for Na+ ion flow through palytoxin-bound pump-channels12,13. We found that residues in transmembrane (TM) segments TM4 and TM6 (cf. refs. 10,11) contribute to an ion pathway characterized by a wide vestibule that opens to the extracellular surface12 and penetrates deep into the Na+,K+-ATPase where it narrows and leads to a cation-selectivity filter13 comprising conserved acidic residues (E336 and D813) that help coordinate cations in the Na+,K+ pump equivalent of cation binding site II (and, partly, site I; ref. 15) in SERCA.
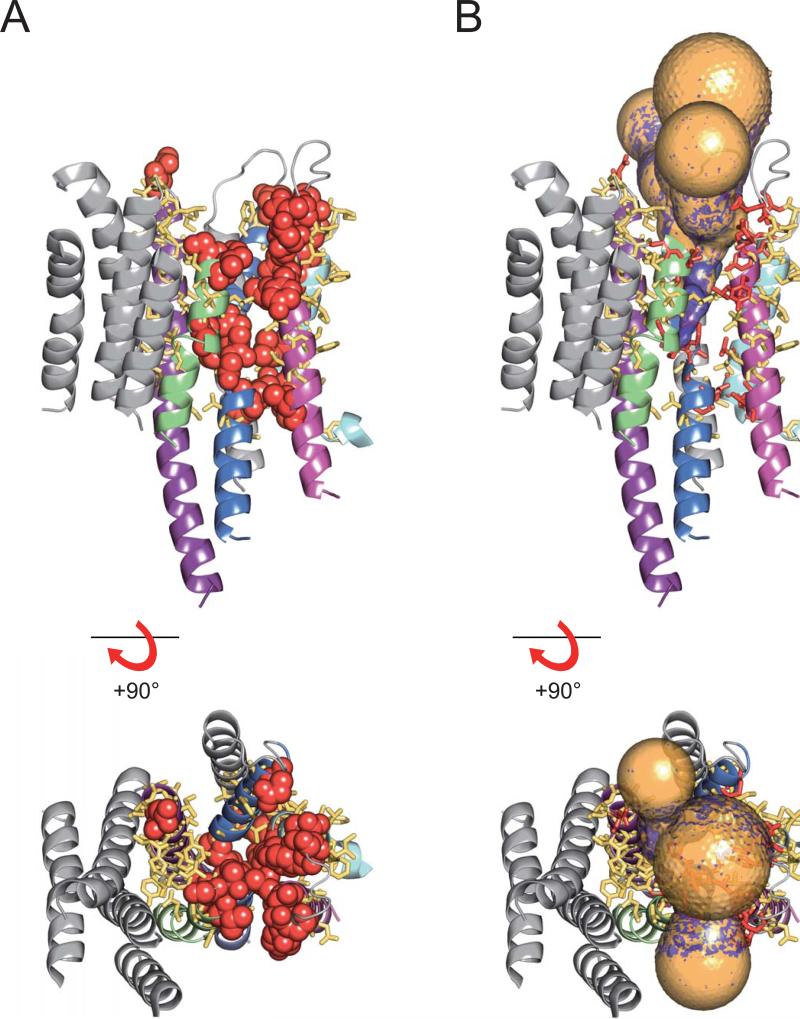
Very recently, to obtain a snapshot of the entire ion pathway, we17 used positively charged MTSET+ (2-trimethylammonium-ethyl-methanethiosulfonate) to extensively scan TM1, the TM1-2 linker, TM2, TM5, and the TM5-6 linker, as well as strategic positions in TM3. We combined these results with those from our previous representative scans of TM4 and TM6, and we displayed them (Fig. 1A) on a homology model of the Na+,K+ pump based on the SERCA E2-BeF3− structure. Reactive positions (where the ~6 Å × 8 Å MTSET+ adduct altered current ≥10%), with atoms shown as red spheres (Fig. 1A), enclose a continuous pathway through the Na+,K+-ATPase from one side of the membrane to the other, comprising residues from TM1, TM2, TM4, and TM6, and passing through cation binding site II, but were not found in TM5 or in binding site I. Non-responsive positions, shown as yellow sticks in Fig. 1, surround the reactive positions, confirming that the scan was complete. And near abolition of Na+ current by modification of a single cysteine at several positions along the pathway argue that there is no other route for Na+ ion flow through palytoxin-bound Na+,K+ pump-channels17 (for review, see ref. 18).
Figure 1.
Snapshot of the ion pathway through the Na+,K+ pump. (A) Cysteine-scanning results mapped onto a homology model of the Na+,K+ pump transmembrane domain, based on the SERCA E2-BeF3− structure, viewed from side (upper) and extracellular surface (bottom). Helices are colored grey except TM1 (pale blue), TM2 (magenta), TM4 (blue), TM5 (purple), and TM6 (green). Residues at MTSET+-responsive and non-responsive positions are shown as red balls and yellow sticks, respectively. B. MOLE analysis of the homology model shown in the same orientations as in A. Gold: tunnels starting from S784, T785 and E788, equivalent to cation-binding site I in SERCA. Purple: tunnels starting from E336 and D813, equivalent to cation-binding site II. Red and yellow sticks mark MTSET+-responsive and non-responsive positions.
To further evaluate the scanning results, we used MOLE (ref. 19; http://troll.chemi.muni.cz/whitezone/development/mole/online/moleonline1.3/) to examine the tunnels that connect binding sites I (gold tunnels, starting between positions S784, N785 and E788) and II (purple tunnels, starting between positions E336 and D813) with the extracellular space in the Na+,K+ pump model based on the SERCA BeF3− structure.
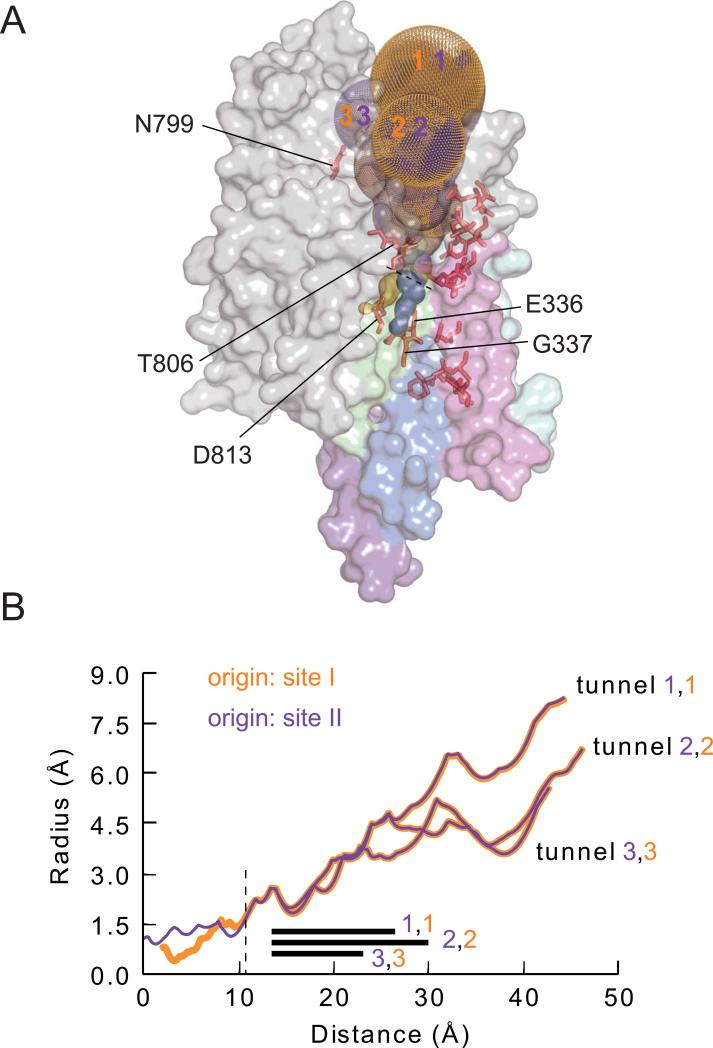
Each starting point yields three principal tunnels (Figs. 1B, 2A). The three tunnels that arise from a given binding site are initially narrow and almost identical to each other (Fig. 2). Close to their origins, the three near identical site I tunnels are distinct from the three near identical site II tunnels (Fig. 2A), but they merge with the site II tunnels a few Å above site II (Fig. 2A,B). All tunnels then run together until, ~20 Å from binding site II, they relatively abruptly broaden and diverge somewhat, to form the irregular wide vestibule that opens to the exterior (Figs. 1B, 2). Once they broaden, each of the three site I tunnels is essentially identical to one of the three site II tunnels (Figs. 1B, 2). Notably, the site II tunnels and post-merger site I tunnels are surrounded by MTSET+-reactive positions (Figs. 1B, 2A), and a narrow extension of the site II tunnel reaches towards the deep reactive TM4 position, G337. Even MTSET+-responsive position N799 in the TM5-TM6 external loop, distant from most reactive positions in the homology model (Fig. 1A), contacts a pair of tunnels, supporting applicability of the model.
Figure 2.
Characteristics of tunnels in the Na+,K+ pump homology model. (A) Gold and purple dots show tunnels starting from cation-binding sites I and II. Key reactive positions along those tunnels are labeled. Transmembrane helices are colored as in Fig. 1. (B) Radii of tunnels against distance from the level of binding site II. The three tunnels originating from site I are shown as gold lines, and the three from site II as purple lines. The dashed line at ~11Å marks the position of the dashed line in A, after which site I- and site II-originating tunnels superimpose. The black bars indicate the distance over which T806 is associated with the tunnels.
The tunnel branch leading to site I is somewhat narrower than the pathway to site II (Fig. 2B), in accord with the overall conclusion that the main pathway passes through site II but is linked to cation binding site I via a narrower connection17. In addition, the relatively abrupt change in radius of all the tunnels (~20 Å from site II; Fig. 2B) in the vicinity of residue T806, which is at the outermost end of TM6 within the floor of the extracellular-facing vestibule (Fig. 1A), corroborates our earlier electrophysiological results13. In those findings, reaction of mutant G805C with variously charged MTS reagents yielded correspondingly varied electrostatic effects on Na+ current through palytoxin-bound Na+,K+ pump-channels. In contrast, those same reagents all, regardless of charge, diminished Na+ current upon reaction with a cysteine introduced at the adjacent position, in mutant T806C, presumably because the adducts all sterically impeded Na+ ion flow13. This suggested that the wide extracellular pathway suddenly narrows at position T806.
Despite this reasonable correspondence between scanning results and model based on the SERCA BeF3− structure, the tunnel analysis implies that although a 1.9-Å diameter Na+ ion could reach site II (though not site I) the at least two times wider MTS reagents should not. Moreover, as the cytoplasmic-side pathway in the SERCA E2-BeF3− structure is closed by a 15-20 Å barrier1, we attempted tunnel analysis using a homology model of Na+,K+-ATPase based on the E1·2Ca2+ structure (PDB code 1SU4; ref. 20) in which the cytoplasmic pathway appears open. However, whether starting from site I or site II most of the tunnels calculated for this model exited the transmembrane domain at the level of the lipid bilayer, with several emerging between TM1 and TM2. Only one tunnel led to the cytoplasm, and it started in site II, passed between TM1, TM2, and TM4, and contacted the deepest MTSET+-responsive positions, L339 (TM4), T145 (TM2), and G100 (TM1). Unfortunately, the significance of that tunnel remains unclear because two comparable tunnels, following almost the same route and also contacting L339, T145, and G100, were identified in a model based on a different E1·2Ca2+ structure (PDB code 1T5S), with bound AMPPCP, in which the cytoplasmic pathway appears closed21.
We conclude that the homology model of Na+,K+-ATPase built on the SERCA E2-BeF3− structure provides a reliable representation of the extracellular access pathway to the ion binding sites, but that a structural model of the Na+,K+ pump's cytoplasmic access pathway for Na+ and K+ ions remains elusive. The incomplete nature of the atomic scaffold notwithstanding, the outline of the ion pathway through palytoxin-bound Na+,K+ pump channels obtained by combining electrophysiological data with homology modeling and cavity search analysis affords a structural framework for further understanding ion translocation by the Na,K-ATPase and by related P-type pumps, such as the Ca- and H,K-ATPases.
Acknowledgements
Supported by NIH HL36783 (to D.C.G) and a fellowship from the Vicente Trust (to P.A.); N.R. is presently a Jane Coffin Fund Fellow.
References
- 1.Olesen C, Picard M, Winther AML, Gyrup C, Morth JP, Oxvig C, Møller JV, Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 2.Toyoshima C, Norimatsu Y, Iwasawa S, Tsuda T, Ogawa H. How processing of aspartylphosphate is coupled to luminal gating of the ion pathway in the calcium pump. Proc Natl Acad Sci USA. 2007;104:19831–19836. doi: 10.1073/pnas.0709978104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Post RL, Sen AK, Rosenthal AS. A phosphorylated intermediate in adenosine triphosphate-dependent sodium and potassium transport across kidney membranes. J Biol Chem. 1965;240:1437–1445. [PubMed] [Google Scholar]
- 4.Albers RW. Biochemical aspects of active transport. Ann Rev Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 5.Scheiner-Bobis G, Meyer zu Heringdorf D, Christ M, Habermann E. Palytoxin induces K+ efflux from yeast cells expressing the mammalian sodium pump. Mol Pharmacol. 1994;45:1132–1136. [PubMed] [Google Scholar]
- 6.Artigas P, Gadsby DC. Na+/K+-pump ligands modulate gating of palytoxin-induced ion channels. Proc. Natl. Acad. Sci. USA. 2003;100:501–505. doi: 10.1073/pnas.0135849100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artigas P, Gadsby DC. Large diameter of palytoxin-induced Na/K pump channels and modulation of palytoxin interaction by Na/K pump ligands. J Gen Physiol. 2004;123:357–376. doi: 10.1085/jgp.200308964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- 9.Guennoun S, Horisberger JD. Structure of the 5th transmembrane segment of the Na,K-ATPase α subunit: a cysteine-scanning mutagenesis study. FEBS Lett. 2000;482:144–148. doi: 10.1016/s0014-5793(00)02050-0. [DOI] [PubMed] [Google Scholar]
- 10.Guennoun S, Horisberger JD. Cysteine-scanning mutagenesis study of the sixth transmembrane segment of the Na,K-ATPase α subunit. FEBS Lett. 2002;513:277–281. doi: 10.1016/s0014-5793(02)02323-2. [DOI] [PubMed] [Google Scholar]
- 11.Horisberger JD, Kharoubi-Hess S, Guennoun S, Michielin O. The fourth transmembrane segment of the Na,K-ATPase α subunit: a systematic mutagenesis study. J. Biol. Chem. 2004;279:29542–29550. doi: 10.1074/jbc.M400585200. [DOI] [PubMed] [Google Scholar]
- 12.Artigas P, Gadsby DC. Ouabain affinity determining residues lie close to the Na/K pump ion pathway. Proc Natl Acad Sci USA. 2006;103:12613–12618. doi: 10.1073/pnas.0602720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes N, Gadsby DC. Ion permeation through the Na+,K+-ATPase. Nature. 2006;443:470–474. doi: 10.1038/nature05129. [DOI] [PubMed] [Google Scholar]
- 14.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 15.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 16.Harmel N, Apell HJ. Palytoxin-induced effects on partial reactions of the Na,K-ATPase. J Gen Physiol. 2006;128:103–118. doi: 10.1085/jgp.200609505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi A, Reyes N, Artigas P, Gadsby DC. The ion pathway through the opened Na+,K+-ATPase pump. Nature. 2008;456:413–416. doi: 10.1038/nature07350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadsby DC, Takeuchi A, Artigas P, Reyes N. Peering into an ATPase ion pump with single-channel recordings. Philos Trans R Soc Lond B Biol Sci. 2009;364:229–238. doi: 10.1098/rstb.2008.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petřek M, Košinová P, Koča J, Otyepka M. MOLE: A voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 2007;15:1357–1363. doi: 10.1016/j.str.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 21.Sørensen TLM, Møller JV, Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]