Abstract
Regeneration of mineralized tissues affected by chronic diseases comprises a major scientific and clinical challenge. Periodontitis, one such prevalent disease, involves destruction of the tooth-supporting tissues, alveolar bone, periodontal-ligament and cementum, often leading to tooth loss. In 1997, it became clear that, in addition to their function in enamel formation, the hydrophobic ectodermal enamel matrix proteins (EMPs) play a role in the regeneration of these periodontal tissues. The epithelial EMPs are a heterogeneous mixture of polypeptides encoded by several genes. It was not clear, however, which of these many EMPs induces the regeneration and what mechanisms are involved. Here we show that a single recombinant human amelogenin protein (rHAM+), induced in vivo regeneration of all tooth-supporting tissues after creation of experimental periodontitis in a dog model. To further understand the regeneration process, amelogenin expression was detected in normal and regenerating cells of the alveolar bone (osteocytes, osteoblasts and osteoclasts), periodontal ligament, cementum and in bone marrow stromal cells. Amelogenin expression was highest in areas of high bone turnover and activity. Further studies showed that during the first 2 weeks after application, rHAM+ induced, directly or indirectly, significant recruitment of mesenchymal progenitor cells, which later differentiated to form the regenerated periodontal tissues. The ability of a single protein to bring about regeneration of all periodontal tissues, in the correct spatio-temporal order, through recruitment of mesenchymal progenitor cells, could pave the way for development of new therapeutic devices for treatment of periodontal, bone and ligament diseases based on rHAM+.
Keywords: amelogenin, periodontal disease, bone regeneration, cementum regeneration, PDL regeneration, CD105, STRO-1
Introduction
Periodontal diseases are prevalent diseases and are substantial public health burden worldwide. They involve the chronic and progressive destruction of the tooth supporting tissues, mainly the alveolar bone and periodontal ligament (PDL). These diseases often lead to tooth loss. A major discovery was the finding that periodontal tissue regeneration could be achieved by application of acidic extract of enamel matrix proteins (EMPs) [1, 2], and later – by the FDA approved medical device termed Emdogain. EmdogainR is an acidic extract of extracellular enamel matrix, and includes a heterogeneous mixture of mainly hydrophobic polypeptides encoded by several genes [1, 3]. It was not clear, however, which of the EMPs induce the regeneration, and what molecular mechanisms are involved [3].
Enamel, is an ectodermal tissue covering vertebrate teeth. Mature enamel is composed of highly organized tightly packed hydroxy-apatite crystallites. The major enamel proteins produced during enamel development by the ameloblast cells (a specialized cell layer of the enamel organ) are the amelogenins. They comprise 90% of the developing extracellular EMPs [4] and play a major role in the biomineralization and structural organization of enamel [5, 6]. Amelogenins are hydrophobic molecules that self-assemble in vitro and in vivo into nanos-pheric structures which regulate the oriented and elongated growth, shape and size of the enamel mineral crystal [7, 8]. During enamel development and mineralization, the abundant secreted amelogenins in the extracellular enamel are sequentially and discretely degraded by specific proteases, the metalloprotease, enamelysin (MMP-20) and the serine protease, EMSP-1 (KLK-4) [9]. Other enamel extracellular matrix proteins include: ameloblastin, enamelin (ENAM), tuftelin, dentin phosphoprotein (DPP) and amelotin; these proteins have been implicated in various steps of enamel formation [10–12].
The EMPs undergo post-translational modifications and post-secretory processing [13–16]. These factors, as well as alternative mRNA splicing, give rise to the heterogeneous mixture of polypeptides in the enamel matrix. The amelogenins are eventually, together with other EMPs, replaced by mineral ions, calcium and phosphorus, the enamel finally becoming hard, fully mineralized (96%) and mature [17].
The amelogenin gene contains 7 exons, which undergo alternative mRNA splicing. The most abundant isoform of the native protein secreted into the enamel matrix lacks the internal region encoded by exon 4. In rodents, a rare isoform including two additional exons termed exons 8 and 9 and lacking exon 7 was reported [18, 19]. Mutations in the X-chromosomal copy of the amelogenin gene [20] have been associated with the hereditary disease, amelogenesis imperfecta (AI), which illustrates the importance of amelogenin in developing enamel. To date, 15 mutations in AMELX, leading to different phenotypes of AI, have been identified. Mutations in other genes encoding for EMPs, such as ENAM, enamelysin (MMP-20) and kallikrein 4 (KLK-4), were also identified in AI patients [21–23]. Abnormal enamel formation and mineralization have also been demonstrated by knockout (KO) of amelogenin expression. The teeth of the amelogenin KO mice expressed a hypoplastic enamel phenotype with reduced enamel thickness [24].
For decades, amelogenin was thought to be exclusively an enamel (epithelial origin) protein. However, in more recent years, amelogenin has also been detected in dentin matrix [2, 25] and odontoblasts [26], during cementogenesis in remnants of Hertwig’s root sheath and in PDL cells [27, 28]. Very recently, we have described amelogenin expression in long bone cells; osteocytes, osteoblasts and osteoclasts, in cartilage chondrocytes and differentially in growth plate cells. We have identified amelogenin expression in long bone marrow stromal cells, some of which are multi-potent stem cells [29]. Amelogenin expression was also identified in non-mineralizing tissues cells such as brain cells and haematopoetic cells [30]. The relatively large number of amelogenin alternatively spliced mRNA translated polypeptides and the fact that amelogenin is expressed in different tissues (calcifying and soft tissues) and of different embryonic origin, possibly reflect different functions of amelogenin.
More recently, amelogenin, its splice products and degradation products, have been suggested to be associated also with cell signalling. Veis et al. showed that specific low molecular mass amelogenin splice products, designated as [A+4] and [A-4] (which are composed of exons 2, 3, 4, 5, 6d, 7 and exons 2, 3, 5, 6d, 7, respectively. [A-4] is also known as LRAP (leucine rich amelogenin peptide), has the ability to interact with immature cells, both in culture and in in vivo implants. In vitro, in rat embryonic muscle fibroblasts culture, [A-4] up-regulated the transcription factor Runx2, that is associated with osteoblast differentiation, whereas [A+4] more prominently up-regulated Sox9, which is associated with chondrogenesis [31]. In vivo, in rat muscle, [A-4] containing implants became profusely mineralized within a 4-week period, whereas [A+4] implants were mineralized more focally. Both types of implants were infiltrated by cells, became vascularized and formed islands of extracellular matrix [31]. The matrix that developed after 4 weeks contained proteins characteristic of mineralized tissues. When [A+4] and [A-4] were implanted into the mucosa of mouse cheeks, they induced recruitment of cells that expressed osteochondrogenic markers [32]. Thus, the cell signalling activities of these amelogenin peptides are related to the formation of mineralized tissues. Furthermore, [A-4] and [A+4] were shown to have different signalling effects on ameloblast and odontoblast differentiation in a developing tooth culture model [33, 34], and when implanted in the pulp [35]. In embryonic stem cell cultures, [A-4] was also shown to induce significant increase in bone sialoprotein and osterix gene expression and further mineral matrix formation [36]. Viswanathan et al. demonstrated that the full length recombinant murine amelogenin regulates bone-sialoprotein expression in a cementoblast cell line in a dose-dependent manner [37]. Tyrosine-rich amelogenin peptide, a specific proteolytic cleavage product of amelogenin, was shown to regulate osteocalcin and osteopontin expression in the same cell line [38].
A progressive deterioration of cementum (a mineralized tissue covering the tooth root surface) was later observed in the amelogenin KO mice. The defects in cementum were characterized by the increased presence of osteoclasts. These defects were also associated with an increased expression of receptor activator of nuclear factor-κB ligand (RANKL) near the cementum, suggesting that amelogenin may play a key role in osteoclastogenesis through the RANKL/RANK mediated pathway [39, 40].
The purpose of the present study was to find out whether the recombinant human amelogenin protein (rHAM+), which comprises 90% of the extracellular EMPs, could alone bring about the regeneration of the tooth supporting tissues (periodontium) after induction of experimental periodontitis in the dog.
Here we identify the amelogenin protein as an active component of the EMP which is responsible for the regeneration of the tooth-supporting tissues. We show that in vivo application of the rHAM+ alone [41], caused significant and progressive regeneration of all three tooth supporting tissues; alveolar bone, PDL and cementum, after induction of experimental periodontitis in the dog. Further immunohistochemistry studies using markers for stem cells, combined with the above findings, suggested that amelogenin induces, directly or indirectly, recruitment of stem cells during the regeneration of the tooth supporting tissues.
Materials and methods
Production of the rHAM+
The rHAM+[41], produced in our laboratory, was used in the present regeneration studies. rHAM+ corresponds to the most abundant human amelogenin mRNA transcript in the developing enamel [20], an isoform that lacks the 14 amino acids encoded by exon 4. In brief, human cDNA coding for a 175 amino acid amelogenin protein was subcloned into the FASTBAC™ HTb plasmid (Invitrogen, CA, USA). This system adds a hexahistidine tag to the amino terminus of the expressed protein enabling efficient purification of the recombinant protein. The recombinant protein, rHAM+, was expressed in eukaryotic Spodoptera frugiperda (Sf9) insect cells, and was characterized by SDS-PAGE Western blot, ESI-TOF mass spectrometry restriction mapping and MS/MS sequencing.
In vivo induction of periodontal tissues regeneration using the rHAM+, in the dog model
All in vivo dog experiments were carried out in the animal experimentation facilities of the Charite-Universitatsmedizin, Berlin, according to the regulations of animal experimentation at this institution.
Six female Beagle dogs aged 13–16 months and weighing 9–10 kg were used in these experiments. The dogs underwent two surgeries: In the first surgery under anaesthesia, experimental periodontitis was induced in all four quadrants of the dog mouth. A bony defect was created in the furcation area (between the tooth roots) creating III degree furcation. Orthodontic wire ligatures were then tied around the defect area in order to induce chronic periodontitis state. The second third and fourth pre-molar teeth were operated in both quadrants of the lower jaws. In the two quadrants of the upper jaws only the second and the third pre-molar teeth were operated. Immediately after the surgery the dogs were given analgesic therapy for 8 days. No antibiotics were used, only tooth cleaning with 0.12% chlorhexidine fluid once a day was performed. Six weeks after the first surgery the ligatures were removed under sedation, together with ultrasonic cleaning. A week later, a second surgery was performed under anaesthesia. The surgery included periodontal flap operation, scaling and root planning, removal of granulation tissue, demineralizing the root surface with 24% ethylenediaminetetraacetic acid (EDTA) gel, and application of the purified rHAM+ on the root surface, into the furcation defect. A notch was created on the surface of the root marking the height of the bone – termed ‘the marking notch’ just prior to protein application. The rHAM+ protein was dissolved in propylene glycol alginate carrier (PGA, Biora), and was applied to one quadrant of three dogs (100 μg rHAM+ per tooth, dissolved in 0.1 ml sterile aqueous solution of PGA). PGA carrier alone (control) was applied to one quadrant in each of six dogs. Enriched, but not purified, extracted fractions of bovine EMPs were applied to other quadrants. This manuscript describes the results obtained after treatment with the purified rHAM+ (in PGA carrier), compared to the control (PGA carrier only).
The dogs were killed under anaesthesia, 2, 4 and 8 weeks after the reconstructive surgery. The jaws were dissected into blocks containing the pre-molar teeth and the surrounding alveolar bone, and were immediately placed in 4% paraformaldehyde (PFA). Two pre-molars in each quadrant were prepared for demineralization for histology immunology and in situ hybridization studies, and one was left mineralized in 100% ethanol for future micro-computerized tomography (micro-CT) studies.
Micro-CT analysis
Micro-CT (MicroCT40, Scanco Medical, Bassersdorf, Switzerland) was used to study the regeneration of the mineralized tooth supporting tissues of the experimental (application of rHAM+ dissolved in PGA carrier) and control (application of PGA carrier only) teeth. Samples were placed in a cylindrical sample holder filled with 100% ethanol to preserve the tooth during the measurement. The scanning parameters were: 30-μm resolution, energy of 50 kVp and a 160 μA current. Three-dimensional reconstruction was performed (with Scanco software) with the following filtration parameters: σ−0.8, support −1 and threshold −220. Measurements of the regenerated mineralized tissues were performed by contouring the area of furcation between the tooth roots, above the marking notch, in the selected central coronal slice. The same contouring was performed on 30 coronal slices from each side of the central slice. Three-dimensional evaluation of the volume limited between these 60 slices was performed with the above software, with filtration parameters of: σ− 0.8, support − 1 and threshold − 200. The total volume measured is the volume of the defect at the time of the reconstructive surgery and the bone volume is a measure of the regenerated calcified tissue (bone and cementum) above the marking notch.
Histological analysis
One tooth from each quadrant (experimental and control) was demineralized using acid formaldehyde solution and paraffin embedded. Several slides from each tooth were stained with haematoxylin and eosin.
For morphometric analysis, the 10 coronal most central slides of each experimental and control demineralized tooth were Azan stained. These slides were then subjected to morphometric analysis using Scion Image software (Scion Corp., USA). As described above, the total area measured in each slide is the area above the marking notch and between the tooth roots, and the regenerated area – the regenerated tissues above the marking notch. For each tooth, the values used for the statistical analysis is the average area measured from these 10 central slides.
Statistical analysis
For statistical analysis we combined the percent of regeneration (regenerated volume per total volume) observed by the micro-CT scan measurements and by the histological measurements for each time-point, after demonstrating positive correlation between the results obtained by these two methods (not shown).
Two Wilcoxon non-parametric tests were performed: The first (not shown) compared the ratio of regenerated mineralized tissue volume to total defect volume, between the experimental group (average of 2, 4 and 8 weeks after treatment with rHAM+, n = 6), and the control group (average of 2, 4 and 8 weeks after treatment with the PGA carrier only, n= 10). The second Wilcoxon non-parametric test compared the results obtained 4 and 8 weeks after treatment with rHAM+ (n = 4), to the control group, 4 and 8 weeks after application of only PGA carrier (n = 7). For detailed explanation – see ‘Results’ and ‘Discussion’.
Preparation of rat tissues
All rat experiments were approved by Hadassah Medical School animal care ethical committee, Hadassah – Hebrew University, Jerusalem. Mandibles were dissected from 5- and 10-week-old Sabra male rats weighting 100 and 350 g, respectively. For immunohistochemistry the tissues were fixed in 4% PFA for 1 hr at room temperature and for in situ hybridization – for 24 hrs. The tissues were then decalcified at 4°C for two months in 0.5 M EDTA, pH 7.4, replaced every 2 days, then washed in PBS and dehydrated. The tissues were embedded in paraffin and sagitally sectioned (5 μm). Pictures were taken from molars and the surrounding periodontal tissues.
Preparation of human mandibular sections
Paraffin embedded sections of human fetal mandibles were prepared by K.H. The Institutional Review Board of the Department of Obstetrics and Gynecology, University of Helsinki, Finland, had approved the study [42].
Indirect immunohistochemistry
Slides were deparaffinized, hydrated, rinsed in PBS and endogenous peroxidase activity was blocked by 3% H2O2 (diluted in methanol) for 10 min. Slides were blocked in non-immune goat serum for 15 min. (Histostain-SP kit, Zymed Laboratories, Inc., San Francisco, CA, USA) followed by overnight incubation in the first antibody (diluted in PBS) at 4°C in a humidified chamber. The primary antibodies used were: (1) LF-108 – a polyclonal rabbit amelogenin antibody, raised against a synthetic peptide corresponding to 10 amino acids at the C-terminus of human amelogenin (cross reacts with dog and rat) conjugated to horseshoe crab haemocyanin (LPH), and affinity purified using protein G [29]. The antibody was diluted to 1/1000 in PBS for rat tissues, 1/2000 for dog tissues. (2) C-term polyclonal rabbit antibody, raised against bovine amelogenin C-terminus, diluted in PBS to 1/500. (3) 270-polyclonal rabbit antibody, raised against amelogenin N-terminus (MPLPPHPG), diluted in PBS to 1/500. (4) 110BQ – a monoclonal mouse antibody raised against human amelogenin [43], diluted to 1/1800 in PBS for rat tissues, 1/2000 for human tissues. (5) CD105 endoglin (H-300) – a polyclonal rabbit antibody, raised against a recombinant protein corresponding to amino acids 27–326 of human endoglin (Santa Cruz Biotechnology, Inc., CA, USA), diluted in PBS to 1/200. (6) STRO-1 (MAB1038) – Monoclonal mouse antibody, raised against human STRO-1 (R&D Systems, Inc., Minneapolis, MN, USA), diluted in PBS to 1 μg/ml. After rinsing, slides were treated according to the Histostain-SP kit protocol, Picture plus kit or Super Picture kit (Zymed Laboratories, Inc.). Negative controls included: (i) the corresponding pre-immune mouse serum. (ii) non-immune rabbit serum and (iii) PBS replacing the first antibody. All slides were examined by Axioskop (Zeiss, Gttingen, Germany) and pictures were taken using Coolpix 990 digital camera (Nikon, Tokyo, Japan), and ProgRes C10 (Jenoptik, Jena, Germany).
Expression of CD105 and STRO-1 in the regenerating tissues: granulation tissue, PDL, bone and bone marrow, 2 weeks after treatment with rHAM+ compared to the control
Five slides from the experimental tooth 2 weeks after treatment with rHAM+, and five slides from each of two control teeth 2 weeks after treatment with PGA carrier only, were reacted with CD105 antibody. The same procedure was also carried out for STRO-1 antibody. Five serial consecutive, non-overlapping pictures were taken from each slide for each tissue studied; granulation tissue, PDL, bone and bone marrow (magnification x480). The number of positively stained cells in each picture was counted manually using Image Pro-Plus software (Media Cybernetics, Inc., Bethesda, MD, USA). The average number (mean ± S.D.) of positively stained cells in each of the tissues was calculated from all pictures of the same tissue.
In situ hybridization
Production of anti-sense and sense (control) RNA-probes: male Sabra rat incisor enamel organ was carefully dissected using microsurgery tools under a stereo-dissecting microscope [29]. The cells were lysed and total cDNA was produced, using Cells-to-cDNA™ II (Ambion, Inc., Austin, USA). The following primers were designed according to the rat amelogenin sequence, gi 9506380, to PCR amplify the cDNA obtained from rat enamel organ: forward – exon 2 (25–47): 5′-TGGATCTTGTTTGCCTGCCTCCT-3′ and reverse – end of exon 6 (622–645): 5′-ATCCACTTCTTCCCGCTTGGTCTT-3′. Sequencing of the PCR product revealed the sequence of LRAP. The PCR product was cloned into pGEM-T Easy Vector (Promega, Madison, USA). Antisense and sense RNA probes were generated using a DIG RNA labelling kit (SP6/T7) (Roche Diagnostics, Mannheim, Germany).
Keeping RNase-free conditions, slides were deparaffinized, hydrated and treated according to the InnoGenex universal ISH kit protocol (InnoGenex, San-Ramon, CA, USA). The slides were incubated at 80°C for 5 min. then cooled to the hybridization temperature; 54°C. Hybridization was carried out for 12 hrs.
Results
Micro-CT analyses reveal regeneration of dog alveolar bone by rHAM+ after induction of experimental periodontitis
Application of the rHAM+ alone [41], brought about significant and progressive regeneration of all three periodontal tissues; alveolar bone, PDL and cementum (Figs 1 and 2). Figure 1A presents a 3D reconstruction image of micro-CT scan of normal dog tooth without any treatment, and a micro-CT cross section of the tooth. Significant regeneration of the alveolar bone was observed in the 3D reconstruction image of micro-CT scan of experimental tooth 8 weeks after treatment with rHAM+, while no such regeneration was detected in the control (PGA carrier) treated tooth (Fig. 1B). In Fig. 1C, cross sections of the experimental and control teeth 2, 4 and 8 weeks after treatment were compared. The yellow dotted line connects all the marking notches, which indicate the levels of the alveolar bone just after the second surgery, when rHAM+ (in PGA carrier) and PGA carrier alone were applied to the experimental and control teeth, respectively. In the experimental teeth, 2 weeks after application of rHAM+, the beginning of regeneration of the mineralizing alveolar bone can be observed. Four and 8 weeks after treatment, there is a significant (P-value < 0.002) (Fig. 1D) and progressive (Fig. 1C) regeneration of the mineralized alveolar bone. No significant regeneration was observed in the control, PGA treated, teeth.
Figure 1.
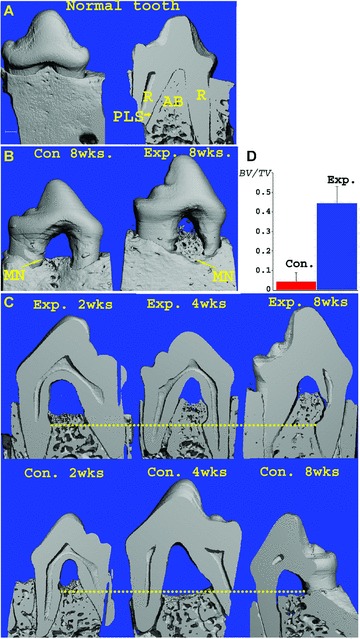
(A) Three-dimensional reconstruction of micro-CT scans of normal dog tooth, without any treatment and a micro-CT cross section of the tooth (R – root, PLS – periodontal ligament space, AB – alveolar bone) (bar = 1 mm). (B) Three-dimensional reconstruction of micro-CT scans, 8 weeks after application of rHAM+ in PGA carrier (Exp.) or PGA carrier only (Con.). The yellow arrows points to the marking notches, indicating the bone height at the time of application of rHAM+ or PGA, respectively. (C) Coronal micro-CT cross-sections of the 3D image of the experimental and control teeth 2, 4 and 8 weeks after treatment. The yellow line connects the marking notches. In the experimental tissues the regeneration started at 2 weeks, after 4 and 8 weeks there is significant and progressive regeneration. In the control almost no regeneration is seen. (D) Statistical analysis of regenerated bone volume (BV) divided by total volume of the furcation (TV) (above the marking notch and between the tooth roots) 4 and 8 weeks (pooled results) after treatment with rHAM+ in PGA carrier (Exp.) or PGA carrier alone (Con.), P-value < 0.002.
Figure 2.
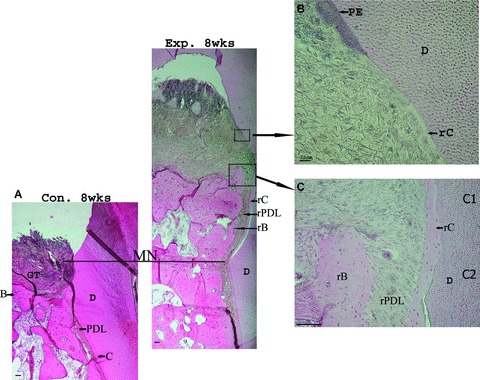
(A) Histological pictures of the dog experimental tooth and dog control tooth 8 weeks after treatment with rHAM+ in PGA carrier (Exp.) or PGA carrier alone (Con.) (PDL = periodontal ligament, D = dentin, GT = granulation tissue, MN = marking notch, r = regenerated, B = bone, C = cementum, bar = 100 μm). (B) Higher magnification (bar = 20 μm) of the regenerating front showing the penetrating oral epithelium (PE) from above and the ascending regenerating cementum from below. Organized PDL and bone are not apparent yet. (C) Enlargement of the regenerated area (bar = 100 μm) showing newly regenerated cementum, PDL and bone (lower part – C2). In the younger regenerating area (upper part – C1), only organized regenerating cementum and PDL are apparent.
Histological analysis revealed regeneration of all three periodontal tissues; cementum, PDL and alveolar bone in the experimental tooth
Histological studies (Fig. 2) revealed regeneration of all three periodontal tissues; cementum, PDL and alveolar bone in the experimental tooth, while no regeneration occurred in the control tooth, and only granulation tissue was seen at the level of the marking notch. Magnifications of the micro-CT images revealed that there was no separation between dentin and regenerating cementum, indicating that the gap observed in the histological images is an artefact of preparation. Higher magnification of the regenerated tissues showed that the regeneration process occurred in the correct spatio-temporal order, restoring the morphology of the tooth attachment apparatus. In the youngest regenerating tissues (Fig. 2B), only regenerating cementum, covering the dentin surface, was observed. A region slightly beneath the youngest regenerating tissues (Fig. 2C, upper part – C1) contained regenerated cementum and PDL, but no apparent regenerating bone could yet be observed. In older regenerating periodontal tissues (Fig. 2C, lower part – C2), all three regenerating tissues; alveolar bone, PDL and cementum, were observed. Electron microscopy analysis 8 weeks after application of rHAM+, showed regenerating cementum and PDL. The PDL was anchored as Sharpey’s fibres in the newly formed cementum (not shown).
Surprisingly, endogenous expression of amelogenin protein was detected in normal (untreated) rat, dog and human periodontal tissues; cementum, PDL and alveolar bone and also in bone marrow (Fig. 3.1–3.4). Amelogenin protein was expressed in different types of rat connective tissue cells and in specific bone marrow cells; in rat alveolar bone osteocytes, in osteoblasts lining the alveolar bone trabecules, in cells surrounding blood vessels (Fig. 3.1A), in osteoclasts (Fig. 3.1C) and in few distinct bone marrow cells (Fig. 3.1E). As was previously reported, amelogenin expression was also detected in PDL cells and in clusters of cementoblasts along the root (Fig. 3.1G) [27, 28]. No staining was detected in the corresponding controls (Fig. 3.1B, D, F and H). Different staining methods were used, yielding red (Fig. 3.1C and E) to brown (Fig. 3.1A and G) reactions. In normal dog periodontal tissues, staining was detected in PDL cells, in cementoblasts, in osteocytes and in osteoblasts (Fig. 3.2A and C), while no staining was detected in the corresponding control (Fig. 3.2B and D). In human embryonic mandibular tissue (Fig. 3.3), amelogenin expression in the epithelial pre-ameloblast cells (Fig. 3.3B) was much higher than that in the alveolar bone cells (Fig. 3.3A). No staining was detected in the corresponding controls (Fig. 3.3D and C), respectively.
Figure 3.
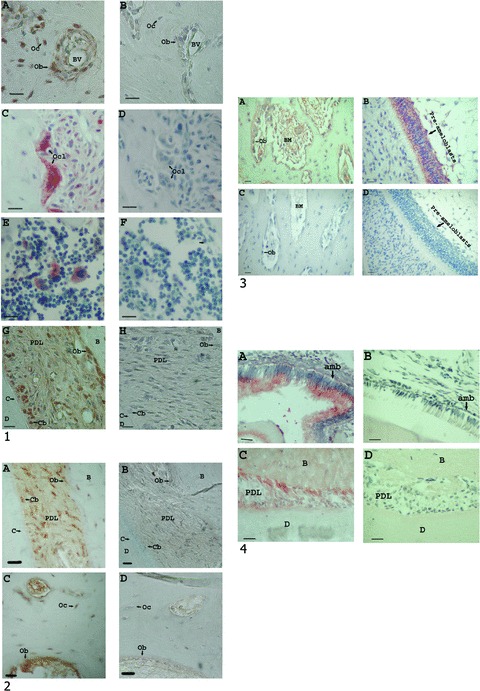
(1) – (A–H) Examples of immunohistochemical analyses of rat normal periodontal tissues using different amelogenin antibodies (bar = 20 μm). All amelogenin antibodies identified the same cell types. (A) Amelogenin expression in alveolar bone osteoblasts (Ob), osteocytes (Oc), and cells surrounding the blood vessels (BV). Using the amelogenin polyclonal rabbit antibody (270), raised against amelogenin N-terminus, diluted in PBS to 1/500. (B) Control – PBS replacing the first antibody. (C) Amelogenin expression in alveolar bone ostoeclasts (Ocl), using a polyclonal rabbit amelogenin antibody (LF108), raised against the 10 amino acids at the C-terminus of human amelogenin, diluted in PBS to 1/1000. (D) Control – non-immune rabbit serum replacing the first antibody, diluted in PBS to 1/500. (E) Amelogenin expression in specific alveolar bone marrow cells using a monoclonal mouse antibody raised against human amelogenin (110BQ), diluted in PBS to 1/1800. (F) Control – pre-immune mouse serum replacing the first antibody, diluted in PBS to 1/1800. (G) Amelogenin expression in the periodontal ligament (PDL), mainly in osteoblasts and cementoblasts (dentin = D, cementoblast = Cb, C = cementum, B = bone), using a polyclonal rabbit amelogenin antibody, raised against bovine amelogenin C-terminus, diluted in PBS to 1/500. (H) Control – PBS replacing the first antibody. (2) – Examples of immunohistochemical analyses of dog normal periodontal tissues, using the polyclonal rabbit amelogenin antibody (LF108), diluted in PBS to 1/2000, and the corresponding controls, non-immune rabbit serum replacing the first antibody, diluted in PBS to 1/500 (bar = 20 μm). (A) Amelogenin expression in the PDL. (B) The PDL control. (C) Amelogenin expression in the alveolar bone cells. (D) The alveolar bone control. (3) – Immunohistochemical analysis of human embryonic mandibular tissues, using a monoclonal mouse antibody raised against human amelogenin (110BQ), diluted in PBS to 1/2000 (bar = 20 μm). A comparison between amelogenin expression, in the same section, in the alveolar bone (A) and enamel organ (pre-ameloblasts) (B), revealed much higher expression of amelogenin in the enamel organ. Parts (C) and (D) are controls, PBS replacing the first antibody. No staining was observed within the control section (Ob = osteoblast, BM = bone marrow). (4) – Amelogenin antibody reaction with WT mouse and amelogenin KO mouse mandibular sections. Using amelogenin polyclonal rabbit antibody (270), raised against amelogenin N-terminus, diluted in PBS to 1/1000 (bar = 20 μm). (A) Amelogenin expression in WT mouse enamel organ (amb = ameloblasts). (B) Amelogenin expression in KO mouse enamel organ. (C) Amelogenin expression in WT mouse periodontium (B = bone, D = dentin). (D) Amelogenin expression in KO mouse periodontium.
The amelogenin antibodies generally identified the same cell types. To further determine the specificity of the amelogenin antibodies which reacted with the periodontal tissues, mouse mandibular sections, including the enamel organ and periodontal tissues, from wild-type (WT) and amelogenin KO mouse [24] were used (Fig. 3.4). No reaction was detected with the KO mouse enamel organ (Fig. 3.4B) and periodontal tissues (Fig. 3.4D), from the same section, but very strong reaction was detected in the corresponding WT enamel organ (Fig. 3.4A), and to a lesser extent with the periodontal tissues (Fig. 3.4C) from the same section, respectively.
Amelogenin mRNA (Fig. 4A-F) and protein (Fig. 4G–L) expression in rat periodontal tissues was studied. Amelogenin mRNA expression was highest in the alveolar bone adjacent to the PDL, as compared to bone cells distal to the PDL, in the same section (Fig. 4A and E, respectively). The same expression pattern was observed for the amelogenin protein (Fig. 4G and K, respectively). Higher amelogenin mRNA expression in the periosteal cells was observed, compared to amelogenin expression in bone cells distal to the periosteum (Fig. 4C and E, respectively). Higher amelogenin protein expression was also observed in the periosteum, as compared to bone cells distal to the periosteum (Fig. 4I and K, respectively). No staining was detected in the mRNA and protein corresponding controls, respectively.
Figure 4.
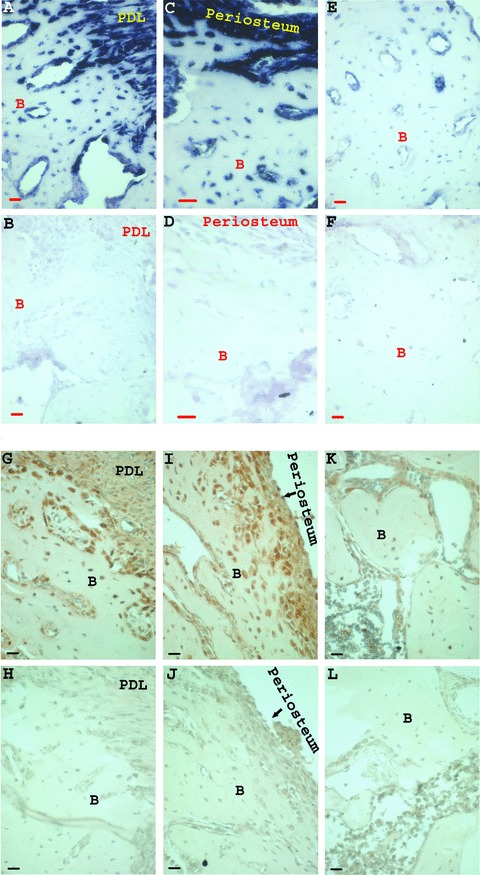
(A–F) In situ hybridization of rat periodontal tissues using rat amelogenin probe. The anti-sense (experimental) and sense (control) probes were diluted to 1/1000. In the same slide, a much higher amelogenin mRNA expression was detected in bone cells adjacent to the PDL (A) and in periosteal cells (C) compared to bone cells distal to the PDL and periosteum (E). Parts (B), (D) and (F) are the corresponding sense probe (controls). (G-L) Immunohistochemistry analysis using the amelogenin polyclonal rabbit antibody (270), raised against amelogenin N-terminus, diluted in PBS to 1/500. Similar to the in situ hybridization results, a much higher amelogenin protein expression was detected in bone cells adjacent to the PDL (G) and in periosteal cells (I) as compared with bone cells distal to the PDL and periosteum (K) (in the same slide). Parts (H), (J) and (L) are the corresponding controls, PBS replacing the first antibody (bar = 20 μm, B = bone).
In the regenerating dog bone, amelogenin was highly expressed in bone endosteal cells, osteoblasts and cells surrounding blood vessels (Fig. 5A), and to a much lesser extent in bone cells distal to the regenerating area (Fig. 5B). No staining was detected in the control (Fig. 5C).
Figure 5.
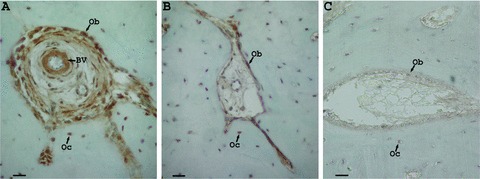
(A–C) Immunohistochemistry analysis of regenerating dog periodontal tissues using a polyclonal rabbit amelogenin antibody LF108, diluted in PBS to 1/2000. Comparison between amelogenin expression 8 weeks after treatment with rHAM+, in bone cells in the regenerating area (A) and distal to the regenerating area (under the marking notch) (B), in the same section. Part (C) is the controlnon-immune rabbit serum replacing the first antibody, diluted in PBS to 1/500 (bar = 20 μm, Ob = osteoblast, Oc = osteocyte, BV = blood vessel).
The number of CD105 and STRO-1 positive cells, both known markers of bone marrow stromal stem cells [44–55], in the granulation tissue, PDL, alveolar bone and bone marrow of the experimental tooth and surrounding bone 2 weeks after treatment with rHAM+, was compared to the number of CD105 and STRO-1 positive cells in the control tooth and surrounding bone treated with PGA carrier (Fig. 6.1 and 6.2). Significantly more CD105 (Fig. 6.1) and STRO-1 (Fig. 6.2) positive cells were detected in the granulation tissue, in the PDL, as well as in the bone and bone marrow cells of the experimental tooth treated with rHAM+ dissolved in PGA, compared to control tooth, treated only with PGA. Four and 8 weeks after treatment, the differences between the amount of cells expressing CD105 and STRO-1 in the experimental tooth and surrounding bone compared to the control tooth and surrounding bone were less prominent (not shown).
Figure 6.
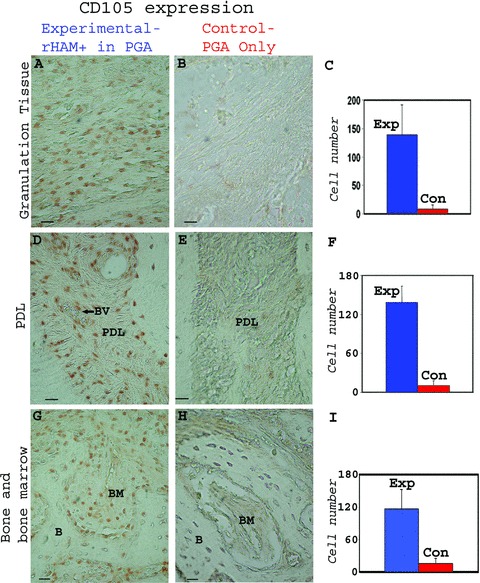
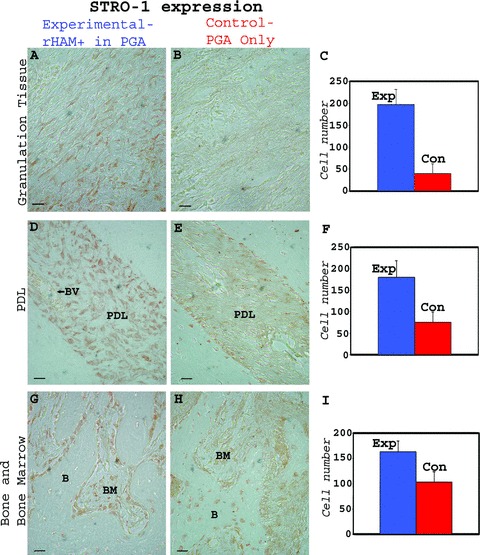
Immunohistochemical analysis of dog periodontal tissues comparing the number of cells expressing CD105 (endoglin) in teeth 2 weeks after treatment with rHAM+ dissolved in PGA, with the control, 2 weeks after treatment with PGA carrier only. The polyclonal CD105 antibody was diluted in PBS to 1/200. CD105 expression in the (A) granulation tissue, (D) PDL, (G) alveolar bone (B) and bone marrow (BM) of the experimental tooth. CD105 expression in the (B) granulation tissue, (E) PDL, (H) alveolar bone (B) and bone marrow (BM) of the control tooth Statistical analysis comparing the average number of cells expressing CD105 in the (C) granulation tissue, (F) PDL, (I) bone and bone marrow cells of the experiment tooth with the control tooth, 2 weeks after the treatment. Many more cells expressing CD105 were observed in the experimental tooth; in the granulation tissue, periodontal ligament, and in bone and bone marrow cells as compared to control. (2) – Immunohistochemical analysis of dog periodontal tissues comparing the number of cells expressing STRO-1 in teeth 2 weeks after treatment with rHAM+ dissolved in PGA, with the control, 2 weeks after treatment with PGA carrier only. The monoclonal STRO-1 antibody was diluted in PBS to 1 μg/ml. STRO-1 expression in the (A) granulation tissue, (D) PDL (BV = blood vessel), (G) alveolar bone (B) and bone marrow (BM) of the experimental tooth. STRO-1 expression in the (B) granulation tissue, (E) PDL, (H) alveolar bone (B) and bone marrow (BM) of the control tooth Statistical analysis comparing the average number of cells expressing STRO-1 in the (C) granulation tissue, (F) PDL, (I) bone and bone marrow cells of the experiment tooth with the control tooth, 2 weeks after the treatment. Many more cells expressing STRO-1 were observed in the experimental tooth in granulation tissue and periodontal ligament as compared to control. In the alveolar bone and bone marrow of the control teeth, relatively large numbers of cells were expressing STRO-1. Hence the difference between the experimental and control group is smaller.
Discussion
Periodontal diseases cause chronic and progressive destruction of the tooth supporting tissues, which attaches the tooth to the alveolar bone of the jaw. The results of the present study show, that the rHAM+ alone [41], which does not contain any other enamel matrix component, caused significant and progressive regeneration of all three periodontal tissues, after induction of experimental periodontitis in the dog model. Hence, most probably, the amelogenins are the active component of EMPs, as well as EmdogainR, in the regeneration process of the periodontium. This does not exclude the possibility that other components of the EMPs also contribute to the regeneration process.
Our histological observations suggest that the regeneration process mimics the normal pattern of periodontal development, in which the cementum is first produced, in the youngest regenerated tissue, followed by the PDL and alveolar bone. Electron micrographs of the regenerated cementum and PDL showed that the structure of the PDL Sharpey’s fibres, penetrated into the regenerated cementum, as in normal functional periodontium.
Surprisingly, amelogenin expression was detected in normal tissues of the periodontium; cementum, PDL and alveolar bone. Hence, application of the recombinant amelogenin protein represents overexpression rather than ectopic expression. The expression of amelogenin in the epithelial ameloblast cells was much higher as compared to the cementum, PDL and alveolar bone cells. The large difference in amelogenin expression could explain why the presence of amelogenin protein in bone cells was overlooked in the past. This difference in expression level most probably also represent different role(s) for amelogenin in these tissues; in the forming enamel, amelogenin mainly serves as an extracellular structural protein that self-assembles into nanospheric structures which regulate the oriented and elongated growth, shape and size of the enamel mineral crystal [8], and hence, is highly expressed. The relatively small quantities of amelogenin protein expression in the other tissues, may suggest that amelogenin might have other functions in the process of normal development and regeneration of the periodontal tissues.
While studying amelogenin mRNA and protein expression in normal rat and dog periodontal tissues, we noticed that amelogenin expression is highest in bone cells adjacent to the PDL and in the periosteal cells, as compared to bone cells distal to the PDL and the periosteum. The bone adjacent to the PDL absorbs all the mastication forces and bone turnover and remodelling is much higher in this area, as compared to bone remote to the PDL region. In addition, the periosteum covers the actively growing bone of the jaw. It is the main site of bone growth and remodelling, and it contains mainly progenitor cells. These findings suggest that in the normal uninjured animal, amelogenin expression is increased at sites of high bone activity and remodelling. A similar phenomenon was seen in the regenerated alveolar dog bone; amelogenin was highly expressed in endosteal cells, osteoblasts, and cells surrounding blood vessels in the regenerated area, and to a much lesser extent in bone cells distal to the regenerated area.
Interestingly, we detected amelogenin expression both in osteoclasts and in osteoblasts lining the alveolar bone trabecules. Sequencing of a macrophage cell line, which shares a common progenitor with osteoclasts, revealed the expression of the amelogenin mRNA isoform – M180 in macrophages [30], suggesting the endogenous expression of amelogenin in osteoclasts. Bone resorption during osteoclastogenesis requires the expression of RANKL and RANK. RANKL is produced by osteoblasts and bone marrow stromal cells and interacts by cell-to-cell contact with its receptor RANK on the osteoclast precursors, promoting osteoclastogenesis [39]. A progressive deterioration of tooth cementum was observed in the amelogenin KO mice. The defect in cementum was characterized by increased presence of osteoclasts, and was also associated with an increased expression of RANKL in the region of the cementum [39, 40, 56]. Our findings of amelogenin expression in osteoclasts, osteoblasts and bone marrow stromal cells [29], and the increase in osteoclast activity in the amelogenin KO mice, suggest that amelogenin might function in the fine balance between bone formation by the osteoblasts and bone resorption by the osteoclasts, possibly through the RANKL/RANK mediated pathway [39, 40]. During alveolar bone regeneration after induced inflammation, amelogenin might act by increasing bone formation over the resorption process.
The regeneration process requires recruitment and differentiation of a variety of cells. Several studies indicated that amelogenin might be involved in cell signalling. Indeed a cell receptor for LRAP [A–4], which was previously suggested to have signalling activity (see ‘Introduction’), was identified as LAMP-1 [57]. In addition, the full length amelogenin lacking exon 4 (M180), which corresponds to rHAM+[41], that we used in the present regeneration study, and the short isoform LRAP, were found to bind to LAMP-1, as well as LAMP-2 and CD63 [58]. The exact binding regions of these three proteins for specific amelogenin (M180 and LRAP) sequences were reported [58]. These are ubiquitously expressed lysosomal integral membrane proteins which are also localized to the plasma membrane. Shapiro et al. found that exogenously added amelogenin moves rapidly into established LAMP-1 positive vesicles that subsequently localize to the perinuclear region of the cell [59]. These results showed that M180, and hence the corresponding human recombinant amelogenin, might act directly or indirectly as signalling molecules.
Formation, remodelling and regeneration of tissues generally require recruitment of different types of multipotent cells. CD105 (also termed endoglin) is a cell membrane glycoprotein (transforming growth factor-β receptor) and is considered as a marker for bone marrow mesenchymal stem cells (MSCs) [44–49]. Aslan et al.[49] immunoisolated CD105 positive MSCs and demonstrated that they can differentiate into osteogenic, chondrogenic and adipogenic lineages. CD105 positive cells are also detected within the vascular system, and CD105 is also one of the markers for neovascularization [60, 61]. Several studies have also used STRO-1 as a marker for bone marrow stromal stem cells [50–55]; Dennis et al. showed that STRO-1 positive cells have the ability to differentiate into myofi-broblasts with a vascular smooth muscle phenotype, adipocytes, osteoblasts and chondrocytes [55]. Previously, using immunohistochemistry and confocal microscopy, we reported that some of the long bone marrow CD105 positive cells also express amelogenin [29]. Sequencing of mRNA from dog and rat long bone marrow revealed the expression of full length amelogenin lacking exon 4 and of LRAP (equivalent to mouse M180 and M59, respectively) [29].
In the present study, we examined the number of cells expressing the MSC markers, CD105 and STRO-1, in the granulation tissue, PDL, bone and bone marrow, of the experimental and control teeth, 2 weeks after treatment with rHAM+ or with PGA carrier respectively. We found that 2 weeks after treatment, the number of CD105 and STRO-1 positive cells in the tissues treated with rHAM+ was much higher than in the tissues treated with PGA carrier only. Four and 8 weeks after treatment, the differences between the amount of cells expressing CD105 and STRO-1 in the experimental compared to the control teeth were less obvious. These findings suggest that rHAM+ induced, directly or indirectly recruitment of MSC/progenitor cells to the injured site. During the first 2 weeks after treatment, substantial recruitment and proliferation of these cells was induced by rHAM+, which, in turn, brought about the significant regeneration of the periodontal tissues, as seen 4 and 8 weeks after treatment. Lacerda-Pinheiro et al.[33] implanted ectopically collagen beads coated with amel-ogenin splice products ([A+4] and [A–4]) into the oral mucosa of mice and showed that amelogenin splice products have the ability to recruit, among inflammatory cells, mesenchymal progenitors that eventually differentiate into osteochondrogenic cells. Goldberg et al. showed that LRAP [A-4] has the ability to recruit cells from the dental pulp with the potential to proliferate and differentiate into osteoblast-like cells [62], further strengthening our findings. The origin of the MSC/progenitor cells recruited during periodontal tissues regeneration could be the adjacent, native periodontal tissues or bone marrow cells. King and Hughes [63] demonstrated, in the periodontal regeneration model, migration of cells from the adjacent unwounded PDL into the wounded area. Kawaguchi et al. [64] showed that bone marrow stromal cells have the potential to regenerate the periodontium.
Very recently, we showed that application of amelogenin (rHAM+) beads together with DiI, on E13.5 and E14.5 mouse embryonic mandibular mesenchyme and on mouse embryonic tooth germ, revealed recruitment of mesenchymal cells [65], suggesting, again, that amelogenin is involved, directly or indirectly in cellular signalling.
CD105 is also a marker for neovascularization [47, 60, 61], and STRO-1 positive cells have the potential to differentiate into myofibroblasts with a vascular smooth muscle phenotype [50, 55]. The overexpression of CD105 and STRO-1 in the regenerating tissues, suggests that amelogenin promotes the regeneration not only by local MSC/progenitor cells recruitment, but also perhaps by inducing, directly or indirectly, angiogenesis for metabolic support of the developing tissues. These findings correlate with our observations of the rich vascularization of the regenerating tissues, and amelogenin expression in cells normally surrounding blood vessels. The cells surrounding regenerated blood vessels might be pericytes, which are involved in regulating angiogenesis and can differentiate, along several lineages including osteogenic and chondrogenic phenotypes [66]. The finding that amelogenin might promote angiogenesis in the regenerating tissues explains Collett et al.[66] and Yuan et al.[67]in vivo and in vitro findings demonstrating chemotaxis of endothelial cells and induction of blood vessel formation by EMPs. As discussed above, ectopic application of LRAP to rat muscle induced mesenchymal cells in-growth into the implants followed by vascularization [68].
Our current data indicate that amelogenin stimulates a cascade of events enhancing remodelling and regeneration of periodontal tissues. The observed regeneration of all periodontal tissues using the externally applied amelogenin, probably reflects enhancement of its normal endogenous activity in local remodelling of tissues. Amelogenin induces, directly or indirectly, MSC/progenitor cell recruitment at the site of the injury. Application of amelogenin induces local neovascularization to support the developing tissues, and possibly might change the fine balance between osteoclast and osteoblast activity, favouring regeneration over the natural process of degeneration after induction of chronic periodontitis. This degeneration process is characterized by connective tissue and epithelial proliferation into the injured area, an event thought to prevent proper regeneration of the periodontal tissues [1].
The results presented in this manuscript, together with our previous results describing amelogenin expression in long bone cells, long bone stromal cells, and its differential expression in the epiphyseal growth plate cells, articular cartilage cells and cells of the periosteal bone [29], suggest that bone phenotypes should be investigated in families affected by X-linked AI and in the amelogenin KO mouse. The existence of bone phenoptypes in AI and in amelogenin KO mice, have not been thoroughly investigated, since the distribution of amelogenin in cells of alveolar and long bones, were only recently published [29, 30, 69]. Furthermore, the significant progressive induction of peri-odontal regeneration by the recombinant amelogenin protein, and amelogenin expression in long bone cells, bone marrow and cartilage cells, may possibly offer, in the future, treatment to some of the widespread periodontal, bone, ligament and joint diseases, which are of major clinical, scientific and financial challenges.
Acknowledgments
This study was partially supported by DFG grant no. Be 1142/4–1 (J.P.B. and D.D.), by BSF grant no. 2003–239 (D.D. and M.F.Y.) and by the IRP, NIH, DHHS (M.F. Y. and L.W.F.).
References
- 1.Hammarstrom L. Enamel matrix, cementum development and regeneration. J Clin Periodontol. 1997;24:658–68. doi: 10.1111/j.1600-051x.1997.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 2.Hammarstrom L, Heijl L, Gestrelius S. Periodontal regeneration in a buccal dehiscence model in monkeys after application of enamel matrix proteins. J Clin Periodontol. 1997;24:669–77. doi: 10.1111/j.1600-051x.1997.tb00248.x. [DOI] [PubMed] [Google Scholar]
- 3.Saito K, Konishi I, Nishiguchi M, Hoshino T, Fujiwara T. Amelogenin binds to both heparan sulfate and bone morphogenetic protein 2 and pharmacologically suppresses the effect of noggin. Bone. 2008;43:371–6. doi: 10.1016/j.bone.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 4.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;25:9760–8. [PubMed] [Google Scholar]
- 5.Robinson C, Brookes SJ, Shore RC, Kirkham J. The developing enamel matrix: nature and function. Eur J Oral Sci. 1998;106:282–91. doi: 10.1111/j.1600-0722.1998.tb02188.x. [DOI] [PubMed] [Google Scholar]
- 6.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126:270–99. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 7.Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. Self-assembly of a recombinant amel-ogenin protein generates supramolecular structures. J Struct Biol. 1994;112:103–9. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 8.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–4. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 9.Simmer JP, Hu JC. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43:441–9. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- 10.Zeichner-David M. Is there more to enamel matrix proteins than biomineralization? Matrix Biol. 2001;20:307–16. doi: 10.1016/s0945-053x(01)00155-x. [DOI] [PubMed] [Google Scholar]
- 11.Paine ML, Luo W, Wang HJ, Bringas P, Jr, Ngan AY, Miklus VG, Zhu DH, MacDougall M, White SN, Snead ML. Dentin sialoprotein and dentin phosphoprotein overexpression during amelogenesis. J Biol Chem. 2005;280:31991–8. doi: 10.1074/jbc.M502991200. [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki K, Bajenova E, Somogyi-Ganss E, Miller M, Nguyen V, Nourkeyhani H, Gao Y, Wendel M, Ganss B. Amelotin- a novel secreted, ameloblast-specific protein. J Dent Res. 2005;84:1127–32. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- 13.Fincham AG, Moradian-Oldak J. Recent advances in amelogenin biochemistry. Connect Tissue Res. 1995;32:119–24. doi: 10.3109/03008209509013713. [DOI] [PubMed] [Google Scholar]
- 14.Fincham AG, Moradian-Oldak J. Amelogenin post-translational modifications: carboxy-terminal processing and the phosphorylation of bovine and porcine “TRAP” and “LRAP” amelogenins. Biochem Biophys Res Commun. 1993;197:248–55. doi: 10.1006/bbrc.1993.2468. [DOI] [PubMed] [Google Scholar]
- 15.Fincham AG, Moradian-Oldak J, Sarte PE. Mass-spectrographic analysis of a porcine amelogenin identifies a single phosphorylated locus. Calcif Tissue Int. 1994;55:398–400. doi: 10.1007/BF00299322. [DOI] [PubMed] [Google Scholar]
- 16.Salih E, Huang JC, Strawich E, Gouverneur M, Glimcher MJ. Enamel specific protein kinases and state of phos-phorylation of purified amelogenins. Connect Tissue Res. 1998;38:225–35. doi: 10.3109/03008209809017041. [DOI] [PubMed] [Google Scholar]
- 17.Deutsch D, Catalano-Sherman J, Dafni L, David S, Palmon A. Enamel matrix proteins and ameloblast biology. Connect Tissue Res. 1995;32:97–107. doi: 10.3109/03008209509013710. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Mathews C, Gao C, DenBesten PK. Identification of two additional exons at the 3’ end of the amelogenin gene. Arch Oral Biol. 1998;43:497–504. doi: 10.1016/s0003-9969(98)00013-2. [DOI] [PubMed] [Google Scholar]
- 19.Baba O, Takahashi N, Terashima T, Li W, DenBesten PK, Takano Y. Expression of alternatively spliced RNA transcripts of amelogenin gene exons 8 and 9 and its end products in the rat incisor. J Histochem Cytochem. 2002;50:1229–36. doi: 10.1177/002215540205000910. [DOI] [PubMed] [Google Scholar]
- 20.Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet. 1992;50:303–16. [PMC free article] [PubMed] [Google Scholar]
- 21.Stephanopoulos G, Garefalaki ME, Lyroudia K. Genes and related proteins involved in amelogenesis imperfecta. J Dent Res. 2005;84:1117–26. doi: 10.1177/154405910508401206. [DOI] [PubMed] [Google Scholar]
- 22.Wright JT. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet A. 2006;140:2547–55. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu JC, Simmer JP. Developmental biology and genetics of dental malformations. Orthod Craniofac Res. 2007;10:45–52. doi: 10.1111/j.1601-6343.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 24.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–5. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 25.Nebgen DR, Inoue H, Sabsay B, Wei K, Ho CS, Veis A. Identification of the chondrogenic-inducing activity from bovine dentin (bCIA) as a low-molecular-mass amelogenin polypeptide. J Dent Res. 1999;78:1484–94. doi: 10.1177/00220345990780090201. [DOI] [PubMed] [Google Scholar]
- 26.Papagerakis P, MacDougall M, Hotton D, Bailleul-Forestier I, Oboeuf M, Berdal A. Expression of amelogenin in odontoblasts. Bone. 2003;32:228–40. doi: 10.1016/s8756-3282(02)00978-x. [DOI] [PubMed] [Google Scholar]
- 27.Fong CD, Hammarstrom L. Expression of amelin and amelogenin in epithelial root sheath remnants of fully formed rat molars. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2000;90:218–23. doi: 10.1067/moe.2000.107052. [DOI] [PubMed] [Google Scholar]
- 28.Janones DS, Massa LF, Arana-Chavez VE. Immunocytochemical examination of the presence of amelogenin during the root development of rat molars. Arch Oral Biol. 2005;50:527–32. doi: 10.1016/j.archoralbio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Haze A, Taylor AL, Blumenfeld A, Rosenfeld E, Leiser Y, Dafni L, Shay B, Gruenbaum-Cohen Y, Fermon E, Haegewald S, Bernimoulin JP, Deutsch D. Amelogenin expression in long bone and cartilage cells and in bone marrow progenitor cells. Anat Rec. 2007;290:455–60. doi: 10.1002/ar.20520. [DOI] [PubMed] [Google Scholar]
- 30.Deutsch D, Haze-Filderman A, Blumenfeld A, Dafni L, Leiser Y, Shay B, Gruenbaum-Cohen Y, Rosenfeld E, Fermon E, Zimmermann B, Haegewald S, Bernimoulin JP, Taylor AL. Amelogenin, a major structural protein in mineralizing enamel, is also expressed in soft tissues: brain and cells of the hematopoietic system. Eur J Oral Sci. 2006;114:183–9. doi: 10.1111/j.1600-0722.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- 31.Veis A, Tompkins K, Alvares K, Wei K, Wang L, Wang XS, Brownell AG, Jengh SM, Healy KE. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem. 2000;275:41263–72. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- 32.Tompkins K, Alvares K, George A, Veis A. Two related low molecular mass polypeptide isoforms of amelogenin have distinct activities in mouse tooth germ differentiation in vitro. J Bone Miner Res. 2005;20:341–9. doi: 10.1359/JBMR.041107. [DOI] [PubMed] [Google Scholar]
- 33.Lacerda-Pinheiro S, Septier D, Tompkins K, Veis A, Goldberg M, Chardin H. Amelogenin gene splice products A+4 and A-4 implanted in soft tissue determine the reorientation of CD45-positive cells to an osteo-chondrogenic lineage. J Biomed Mater Res A. 2006;79:1015–22. doi: 10.1002/jbm.a.30912. [DOI] [PubMed] [Google Scholar]
- 34.Le TQ, Zhang Y, Li W, Denbesten PK. The effect of LRAP on enamel organ epithelial cell differentiation. J Dent Res. 2007;86:1095–9. doi: 10.1177/154405910708601114. [DOI] [PubMed] [Google Scholar]
- 35.Lacerda-Pinheiro S, Jegat N, Septier D, Priam F, Bonnefoix M, Bitard J, Kellermann O, Tompkins K, Veis A, Goldberg M, Poliard A. Early in vivo and in vitro effects of amelogenin gene splice products on pulp cells. Eur J Oral Sci. 2006;114:232–8. doi: 10.1111/j.1600-0722.2006.00320.x. [DOI] [PubMed] [Google Scholar]
- 36.Warotayanont R, Zhu D, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem Biophys Res Commun. 2008;367:1–6. doi: 10.1016/j.bbrc.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viswanathan HL, Berry JE, Foster BL, Gibson CW, Li Y, Kulkarni AB, Snead ML. Somerman MJ. Amelogenin: a potential regulator of cementum-associated genes. J Periodontol. 2003;74:1423–31. doi: 10.1902/jop.2003.74.10.1423. [DOI] [PubMed] [Google Scholar]
- 38.Swanson EC, Fong HK, Foster BL, Paine ML, Gibson CW, Snead ML, Somerman MJ. Amelogenins regulate expression of genes associated with cementoblasts in vitro. Eur J Oral Sci. 2006;114:239–43. doi: 10.1111/j.1600-0722.2006.00321.x. [DOI] [PubMed] [Google Scholar]
- 39.Hatakeyama J, Sreenath T, Hatakeyama Y. Thyagarajan T, Shum L, Gibson CW, Wright JT, Kulkarni AB. The receptor activator of nuclear factor-kappa B ligand-mediated osteoclastogenic pathway is elevated in amelogenin-null mice. J Biol Chem. 2003;278:35743–8. doi: 10.1074/jbc.M306284200. [DOI] [PubMed] [Google Scholar]
- 40.Hatakeyama J, Philp D, Hatakeyama Y, Haruyama N, Shum L, Aragon MA, Yuan Z, Gibson CW, Sreenath T, Kleinman HK, Kulkarni AB. Amelogenin-mediated regulation of osteoclastogenesis, and periodontal cell proliferation and migration. J Dent Res. 2006;85:144–9. doi: 10.1177/154405910608500206. [DOI] [PubMed] [Google Scholar]
- 41.Taylor AL, Haze-Filderman A, Blumenfeld A, Shay B, Dafni L, Rosenfeld E, Leiser Y, Fermon E, Gruenbaum-Cohen Y, Deutsch D. High yield of biologically active recombi-nant human amelogenin using the bac-ulovirus expression system. Protein Expr Purif. 2006;45:43–53. doi: 10.1016/j.pep.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Heikinheimo K, Jee KJ, Niini T, Aalto Y, Happonen RP, Leivo I, Knuutila S. Gene expression profiling of ameloblastoma and human tooth germ by means of a cDNA microarray. J Dent Res. 2002;81:525–30. doi: 10.1177/154405910208100805. [DOI] [PubMed] [Google Scholar]
- 43.Catalano-Sherman J, Laskov R, Palmon A, David S, Deutsch D. Production of a monoclonal antibody against human amel-ogenin. Calcif Tissue Int. 1994;54:76–80. doi: 10.1007/BF00316294. [DOI] [PubMed] [Google Scholar]
- 44.Barry FP, Boynton RE, Haynesworth S, Murphy JM, Zaia J. The monoclonal antibody SH-2, raised against human mes-enchymal stem cells, recognizes an epitope on endoglin (CD105) Biochem Biophys Res Commun. 1999;265:134–9. doi: 10.1006/bbrc.1999.1620. [DOI] [PubMed] [Google Scholar]
- 45.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 46.Tuli R, Tuli S, Nandi S, Wang ML, Alexander PG, Haleem-Smith H, Hozack WJ, Manner PA, Danielson KG, Tuan RS. Characterization of multipotential mesenchymal progenitor cells derived from human trabecular bone. Stem Cells. 2003;21:681–93. doi: 10.1634/stemcells.21-6-681. [DOI] [PubMed] [Google Scholar]
- 47.Oswald J, Boxberger S, Jorgensen B, Feldmann S, Ehninger G, Bornhauser M, Werner C. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells. 2004;22:377–84. doi: 10.1634/stemcells.22-3-377. [DOI] [PubMed] [Google Scholar]
- 48.Alsalameh S, Amin R, Gemba T, Lotz M. Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum. 2004;50:1522–32. doi: 10.1002/art.20269. [DOI] [PubMed] [Google Scholar]
- 49.Asian H, Zilberman Y, Kandel L, Liebergall M, Oskouian RJ, Gazit D, Gazit Z. Osteogenic differentiation of noncul-tured immunoisolated bone marrow-derived CD105+ cells. Stem Cells. 2006;24:1728–37. doi: 10.1634/stemcells.2005-0546. [DOI] [PubMed] [Google Scholar]
- 50.Simmons PJ, Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood. 1991;78:55–62. [PubMed] [Google Scholar]
- 51.Gronthos S, Simmons PJ. The growth factor requirements of STRO-1-positive human bone marrow stromal precursors under serum-deprived conditions in vitro. Blood. 1995;85:929–40. [PubMed] [Google Scholar]
- 52.Gronthos S, Zannettino AC, Hay SJ, Shi S, Graves SE, Kortesidis A, Simmons PJ. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci. 2003;116:1827–35. doi: 10.1242/jcs.00369. [DOI] [PubMed] [Google Scholar]
- 53.Chen SC, Marino V, Gronthos S, Bartold PM. Location of putative stem cells in human periodontal ligament. J Periodontal Res. 2006;41:547–53. doi: 10.1111/j.1600-0765.2006.00904.x. [DOI] [PubMed] [Google Scholar]
- 54.Nagatomo K, Komaki M, Sekiya I, Sakaguchi Y, Noguchi K, Oda S, Muneta T, Ishikawa I. Stem cell properties of human periodontal ligament cells. J Periodont Res. 2006;41:303–10. doi: 10.1111/j.1600-0765.2006.00870.x. [DOI] [PubMed] [Google Scholar]
- 55.Dennis JE, Carbillet JP, Caplan Al, Charbord P. The STRO-1 + marrow cell population is multipotential. Cells Tissues Organs. 2002;170:73–82. doi: 10.1159/000046182. [DOI] [PubMed] [Google Scholar]
- 56.Nishiguchi M, Yuasa K, Saito K, Fukumoto E, Yamada A, Hasegawa T. Yoshizaki K, Kamasaki Y, Nonaka K. Fujiwara T, Fukumoto S. Amelogenin is a negative regulator of osteoclastogenesis via downregulation of RANKL, M-CSF and fibronectin expression in osteoblasts. Arch Oral Biol. 2007;52:237–43. doi: 10.1016/j.archoralbio.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 57.Tompkins K, George A, Veis A. Characterization of a mouse amelogenin [A-4]/M59 cell surface receptor. Bone. 2006;38:172–80. doi: 10.1016/j.bone.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 58.Zou Y, Wang H, Shapiro JL, Okamoto CT. Brookes SJ, Lyngstadaas SP, Snead ML. Paine ML. Determination of protein regions responsible for interactions of amelogenin with CD63 and LAMP1. Biochem J. 2007;408:347–54. doi: 10.1042/BJ20070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shapiro JL, Wen X, Okamoto CT, Wang HJ, Lyngstadaas SP, Goldberg M, Snead ML, Paine ML. Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cell Mol Life Sci. 2007;64:244–56. doi: 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duff SE, Li C, Garland JM, Kumar S. CD105 is important for angiogenesis: evidence and potential applications. FASEB J. 2003;17:984–92. doi: 10.1096/fj.02-0634rev. [DOI] [PubMed] [Google Scholar]
- 61.Fonsatti E, Sigalotti L, Arslan P, Altomonte M, Maio M. Emerging role of endoglin (CD105) as a marker of angio-genesis with clinical potential in human malignancies. Curr Cancer Drug Targets. 2003;3:427–32. doi: 10.2174/1568009033481741. [DOI] [PubMed] [Google Scholar]
- 62.Goldberg M, Lacerda-Pinheiro S, Jegat N, Six N, Septier D, Priam F, Bonnefoix M, Tompkins K, Chardin H, Denbesten P, Veis A, Poliard A. The impact of bioactive molecules to stimulate tooth repair and regeneration as part of restorative dentistry. Dent Clin North Am. 2006;50:277–98. doi: 10.1016/j.cden.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 63.King GN, Hughes FJ. Bone morphogenetic protein-2 stimulates cell recruitment and cementogenesis during early wound healing. J Clin Periodontol. 2001;28:465–75. doi: 10.1034/j.1600-051x.2001.028005465.x. [DOI] [PubMed] [Google Scholar]
- 64.Kawaguchi H, Hirachi A, Hasegawa N, Iwata T, Hamaguchi H, Shiba H, Takata T, Kato Y, Kurihara H. Enhancement of peri-odontal tissue regeneration by transplantation of bone marrow mesenchymal stem cells. J Periodontol. 2004;75:1281–7. doi: 10.1902/jop.2004.75.9.1281. [DOI] [PubMed] [Google Scholar]
- 65.Gruenbaum-Cohen Y, Tucker AS, Haze A, Shilo D, Taylor AL, Shay B, Sharpe PT, Mitsiadis TA, Ornoy A, Blumenfeld A, Deutsch D. Amelogenin in cranio-facial development: the tooth as a model to study the role of amelogenin during embryogenesis. J Exp Zoolog B Mol Dev Evol. 2008;312B:445–457. doi: 10.1002/jez.b.21255. [DOI] [PubMed] [Google Scholar]
- 66.Collett GD, Canfield AE. Angiogenesis and pericytes in the initiation of ectopic calcification. Circ Res. 2005;96:930–8. doi: 10.1161/01.RES.0000163634.51301.0d. [DOI] [PubMed] [Google Scholar]
- 67.Yuan K, Chen CL, Lin MT. Enamel matrix derivative exhibits angiogenic effect in vitro and in a murine model. J Clin Periodontol. 2003;30:732–8. doi: 10.1034/j.1600-051x.2003.00413.x. [DOI] [PubMed] [Google Scholar]
- 68.Veis A. Amelogenin gene splice products: potential signaling molecules. Cell Mol Life Sci. 2003;60:38–55. doi: 10.1007/s000180300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Yuan ZA, Aragon MA, Kulkarni AB, Gibson CW. Comparison of body weight and gene expression in amelogenin null and wild-type mice. Eur J Oral Sci. 2006;114:190–3. doi: 10.1111/j.1600-0722.2006.00286.x. [DOI] [PubMed] [Google Scholar]


