Abstract
Hantavirus cardiopulmonary syndrome (HCPS) is an emerging infectious disease first reported in Chile in 1995. Andes hantavirus (ANDV) is responsible for the more than 500 cases of HCPS reported in Chile. Previous work showed that ANDV is genetically differentiated in Chile across contrasting landscapes. To determine whether the reservoir rodent (Oligoryzomys longicaudatus) populations are also geographically segregated, we conducted range-wide spatial genetic analyses of O. longicaudatus in Chile using the mitochondrial DNA cytochrome b gene. Given that landscape structure influences the incidence of hantavirus infections, we also tested 772 O. longicaudatus specimens for antibodies to ANDV captured during the period 2000 − 2006. Population genetic analyses of O. longicaudatus are largely congruent with those reported for ANDV, with the host primarily differentiated according to three defined ecoregions, Mediterranean, Valdivian rain forest and North Patagonian rain forest. Significant differences in the relative prevalence of anti-ANDV antibodies in rodent samples also were found across the three ecoregions. We relate these results to the number of reported human HCPS cases in Chile, and discuss the importance of landscape differences in light of ANDV transmission to humans and among rodent populations.
Keywords: Andes virus, Chile, Ecoregion, Landscape structure, HCPS, Oligoryzomys longicaudatus, Seropositivity, Spatial Genetics
1. Introduction
Hantavirus cardiopulmonary syndrome (HCPS) is an emerging infectious disease mainly known in North America from the outbreak of Sin Nombre virus (SNV) in the Four Corners region of the United States in 1993 (Nichol et al., 1993; Hjelle et al., 1994). The disease produces a variable number of human cases every year, with a case-fatality ratio between 30−50%. Rodents (Muridae and Cricetidae) are considered the reservoirs of hantavirus; however, recent discoveries have also documented a number of species of shrews (Soricomorpha) from at least three continents as hosts of viruses of unknown pathogenicity (Arai et al., 2007; Klempa et al., 2007; Song et al., 2009). Hantaviruses are transmitted to humans through rodent excreta and secretions (Botten et al., 2002; Padula et al., 2004). In Chile, the related Andes virus (ANDV) is responsible for all cases of HCPS, and the long-tailed pygmy rice rat (Oligoryzomys longicaudatus) appears to be the main reservoir (Toro et al., 1998; Medina et al., 2009). ANDV epidemiology is complicated slightly because person-to-person transmission has been well documented in Argentina and Chile although this is unique among hantaviruses (Martinez et al., 2005; Ferres et al., 2007). Since the first documented outbreak in Chile in 1995, serological surveys of hantavirus have confirmed the presence of ANDV from 30° 56'S to 53° 37'S (Toro et al., 1998; Torres-Pérez et al., 2004; Belmar-Lucero et al., 2009). This wide latitudinal range spans contrasting geographic features and landscapes ranging from a Mediterranean heterogeneous vegetation mosaic (Mediterranean ecoregion) to mixed evergreen-deciduous Temperate Forests (Valdivian and North Patagonian rain forest ecoregions) (Armesto et al., 2007; Veblen, 2007). Across these diverse ecotypes, strong differences in population structure and density have been documented for several species of small mammals (Murua et al., 1986; Simonetti and Aguero, 1990; Cofre et al., 2007). In a previous study, we reported that ANDV in Chile is segregated into distinct lineages that correspond to the limits of ecoregions (Medina et al., 2009). Populations of O. longicaudatus, however, seem to show a relatively homogeneous genetic structure (Palma et al., 2005), although ecogeographic subdivision has not been fully assessed. The observation that there exist areas wherein genetic lineages are subdivided according to different ecotypes may be the result of complex geographical, historical, and/or ecological processes with differential selective regimes or random genetic drift acting on populations (Wright, 1931; Hartl and Clark, 2007).
Environmental features influence the geographic distribution of diseases by affecting ecological properties of host and vectors (Linard et al., 2007). Landscape structure influences the incidence of hantavirus infections (Langlois et al., 2001; Glass et al., 2007; Heyman et al., 2009) and when combined with other environmental factors, may prove to be a strong determinant in the transmission of the virus to humans (Linard et al., 2007; Zhang et al., 2009). Variable host population structure has implications for viral population demographics and transmission (Adler et al., 2008). Given differences in ecology of O. longicaudatus populations across their latitudinal range in Chile, transmission rates of ANDV may also differ, resulting in differences in potential rates of infection of rodent hosts. Consequently, humans may be differentially exposed to viral infection.
In this study, we used molecular data from the rodent (O. longicaudatus) to explore genetic discontinuities across Chile, and discuss the importance of host population structure in the transmission of virus among rodents and to humans. We also compare the seroprevalence of ANDV in O. longicaudatus across the latitudinal gradient, and provide a quantitative assessment of the distribution of incident and fatal cases of HCPS during the period 2000 − 2006 in Chile. We predict differences in O. longicaudatus population structure across the ecogeographic regions in southcentral Chile (congruent with ANDV genetic structure), and that prevalence of ANDV-seropositive O. longicaudatus differs across those ecoregions. Our study highlights the value of combining information from host population structure, epidemiology, viral phylogeography, and geography to gain insights into the transmission and persistence of infectious diseases.
2. Materials and methods
2.1. Mitochondrial DNA Sequences and Spatial Genetic Analyses
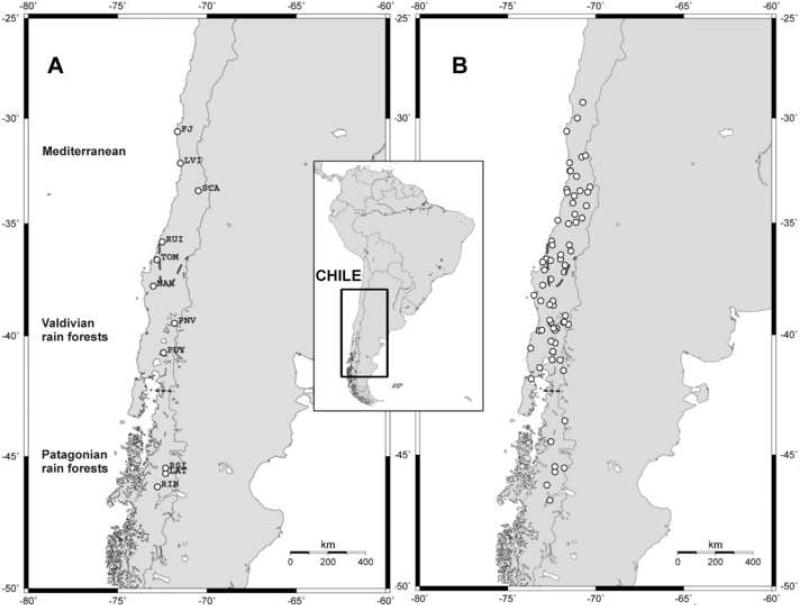
A total of 108 O. longicaudatus from 11 localities in Chile ranging from 30°S to 46°S (Fig. 1A) were used for mitochondrial DNA cytochrome b (cyt-b) amplification and sequencing following procedures explained previously (Palma et al., 2005). Sequences were edited using BioEdit Sequence Alignment Editor (Hall, 1999), and aligned using Clustal W implemented in BioEdit. Sequences are deposited in GenBank (Accession numbers GQ282502-GQ282603, AF346566, AF346568, AY275692, AY275693, AY275698, AY275699).
Figure 1.
Map of the sampled localities of O. longicaudatus used for (A) molecular analyses, and (B) seropositivity to ANDV in Chile. Locality names are explained in Table 1. Dashed lines represent approximate limits of ecoregions. Maps were generated using Online Map Creator (www.aquarius.ifm-geomar.de).
A spatial analysis of molecular variance was performed in SAMOVA v.1.0 (Dupanloup et al., 2002). This method uses a simulated annealing approach to identify groups of populations (K), which are geographically homogeneous and maximally differentiated by maximizing Fct (the proportion of the total genetic variance due to differences among groups of populations). Fct values were calculated by running the program sequentially (100 random initial conditions), and forcing the data into k groups (where k = 2 to 9). Population subdivision was estimated using the fixation index (Fst) between all population pairs generated in Arlequin 3.1 (Excoffier et al., 2005). For detecting dissimilar patterns among O. longicaudatus groups (ecoregions), population pairwise Fst values were used to perform a non-metric multidimensional scaling (MDS) analysis using Systat v.12 (SYSTAT Software, Inc); the analysis provides a visual representation of the pattern of genetic distances (Kruskal and Wish, 1978).
We used the Median Joining method (Bandelt et al., 1999) to perform a phylogenetic network analysis with O. longicaudatus haplotypes as implemented in Network 4.2.0.1 software (http://www.fluxus-engineering.com/sharenet.htm) to assess intraspecific relationships.
2.2. Serology, human hantavirus cases and data analysis
We sampled 76 sites throughout the distribution of O. longicaudatus in Chile from 28°S to 47°S between 2000 and 2006 (Fig. 1B). A total of 772 O. longicaudatus serum samples were subjected to screening for antibodies against the ANDV N protein using a strip immunoblot assay (SIA) as previously described (Yee et al., 2003). The relative anti-ANDV antibody prevalence O. longicaudatus was standardized using the ratio between the number of seropositive O. longicaudatus at each sampled locality and the trapping success (number of captures by the number of total trap-nights) of O. longicaudatus captured at that locality (Torres-Pérez et al., 2004).
Between 1995 and 2006, a total of 492 HCPS cases were reported in Chile (with three additional retrospective cases in 1975, 1993, and 1994; http//:epi.minsal.cl). All HCPS cases were located between 30° 56'S and 46° 46'S. Of these, 391 cases occurred between 2000 and 2006. The number of HCPS cases in each group was expressed as the ratio of the number of cases and the rural population (× 1,000) (information available in the Instituto Nacional de Estadisticas de Chile, http://www.ine.cl). The rural population (people inhabiting, working in, or visiting rural areas) is reported to be at highest risk of contracting HCPS (Castillo et al., 2002; Riquelme et al., 2003), due to closer contact with the habitat of the rodent host (O. longicaudatus). We acknowledge that an index based on rural population may be slightly underestimated due to a few HCPS cases that are not related to people inhabiting rural areas. Finally, we obtained data reflecting the number of fatal HCPS cases during the same period, which are reported standardized by the number of HCPS and the rural population. Seropositivity analyses and HCPS incidence and fatal cases are reported by grouping data according to the three ecoregions Mediterranean, Valdivian rain forests and North Patagonian rain forest. These clusters follow the results reported by Medina et al. (2009) and spatial genetic analyses derived from SAMOVA (see below).
3. Results
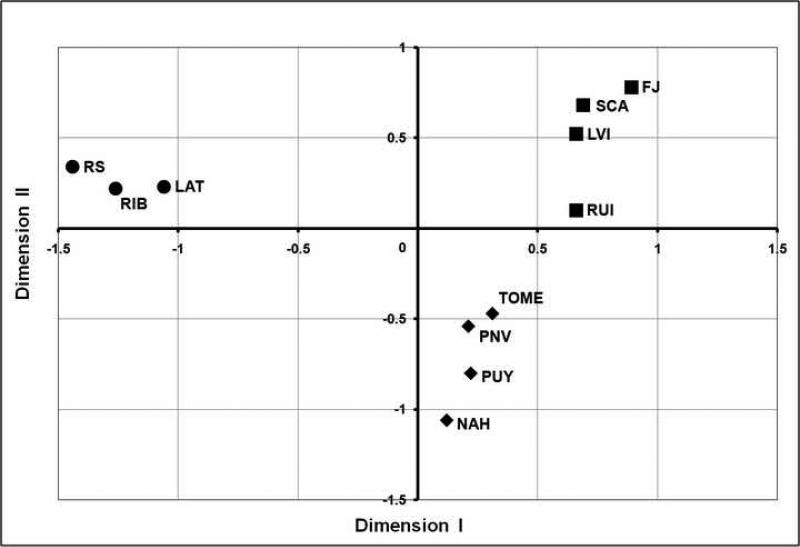
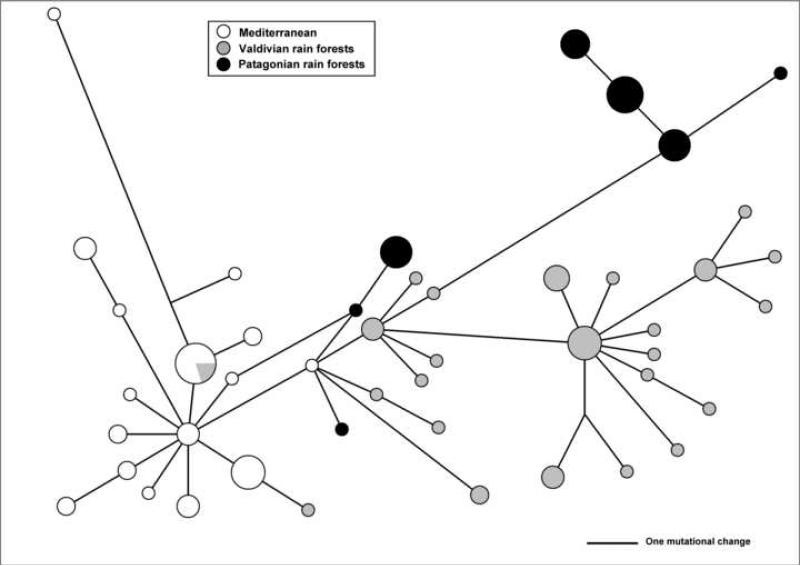
Using mitochondrial cyt-b sequences, spatial analysis of molecular variance was performed to assess substructure within the O. longicaudatus populations in Chile. FCT values ranged from 0.4703 to 0.4832, with the group structure maximized at k = 3. Collection sites from Fray Jorge to Los Ruiles (ca. 30° − 36° S) formed the first group, Tome to Puyehue (ca. 37° − 41° S) clustered into a second group, and three localities (ca. 45° − 46° S) clustered into a third group. These groups correspond to the major ecogeographic regions Mediterranean, Valdivian rain forests and North Patagonian rain forests respectively, that differ in climate, geology, and vegetation composition (Armesto et al., 2007; Veblen, 2007). Fst-values among O. longicaudatus populations in Chile (Table 1) ranged between 0.002 (Tome vs Nahuelbuta and P.N.Villarrica) to 0.670 (Fray Jorge vs Lago Atravesado), with the highest values within an ecogeographic region found between Mediterranean populations (range 0.063 − 0.234). Non-hierarchical analysis (MDS) using the fixation index (Fst) derived from cyt-b sequences (Fig. 2) showed a strong dispersal of O. longicaudatus populations into multidimensional space (Stress = 0.03), segregated in three groups that correspond to ecoregions. The unrooted phylogeographic network (Fig. 3) was concordant with SAMOVA and MDS analyses showing haplotypes primarily associated with ecogeographic distribution (Mediterranean, Valdivian and Patagonian), although some groups shared haplotypes across different ecoregions.
Table 1.
Fixation index (Fst) among O. longicaudatus sampling localities using mitochondrial DNA cytochrome b. (N = sample size). Numbers in bold represent significant differences (P ≤ 0.05) after 10,000 permutations.
| Ecoregion | Map number | Locality | FJ | LVI | SCA | RUI | TOM | NAH | PNV | PUY | RSI | RIB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mediterranean | 1 | FJ (N=10) | --- | |||||||||
| 2 | LVI (N=10) | 0.123 | --- | |||||||||
| 3 | SCA (N=11) | 0.234 | 0.157 | --- | ||||||||
| 4 | RUI (N=10) | 0.214 | 0.154 | 0.063 | --- | |||||||
| Valdivian | 5 | TOM (N=10) | 0.357 | 0.283 | 0.306 | 0.119 | --- | |||||
| 6 | NAH (N=7) | 0.540 | 0.426 | 0.493 | 0.334 | −0.002 | --- | |||||
| 7 | PNV (N=12) | 0.507 | 0.399 | 0.467 | 0.306 | −0.002 | −0.057 | --- | ||||
| 8 | PUY (N=10) | 0.448 | 0.359 | 0.405 | 0.266 | 0.021 | 0.020 | 0.016 | --- | |||
| Patagonian | 9 | RSI (N=9) | 0.623 | 0.524 | 0.544 | 0.530 | 0.440 | 0.522 | 0.525 | 0.481 | --- | |
| 10 | RIB (N=10) | 0.646 | 0.545 | 0.567 | 0.554 | 0.466 | 0.547 | 0.549 | 0.505 | −0.081 | --- | |
| 11 | LAT (N=9) | 0.670 | 0.571 | 0.593 | 0.588 | 0.516 | 0.592 | 0.590 | 0.546 | −0.049 | −0.081 |
Localities: Fray Jorge (FJ), Los Vilos (LVI), San Carlos de Apoquindo (SCA), Los Ruiles (RUI), Tome (TOM), Parque Nacional Nahuelbuta (NAH), Parque Nacional Villarrica (PNV), Puyehue (PUY), Reserva Nacional Río Simpson (RSI), Río Ibáñez (RIB), Lago Atravesado (LAT).
Figure 2.
Bidimensional ordination of pairwise Fst values for O. longicaudatus populations using a non-metric multidimensional scaling method (Stress = 0.03). Squares represent Mediterranean localities: Fray Jorge (FJ), Los Vilos (LVI), San Carlos de Apoquindo (SCA), Los Ruiles (RUI); Diamonds are Valdivian rain forests localities: Tome (TOM), Parque Nacional Nahuelbuta (NAH), Parque Nacional Villarrica (PNV), Puyehue (PUY); Circles are North Patagonian rain forests localities: Reserva Nacional Río Simpson (RSI), Río Ibáñez (RIB), Lago Atravesado (LAT).
Figure 3.
Unrooted network of O. longicaudatus using cytochrome b sequences with haplotypes depicted according to ecoregions. Size of circles represents the number of individuals per haplotypes.
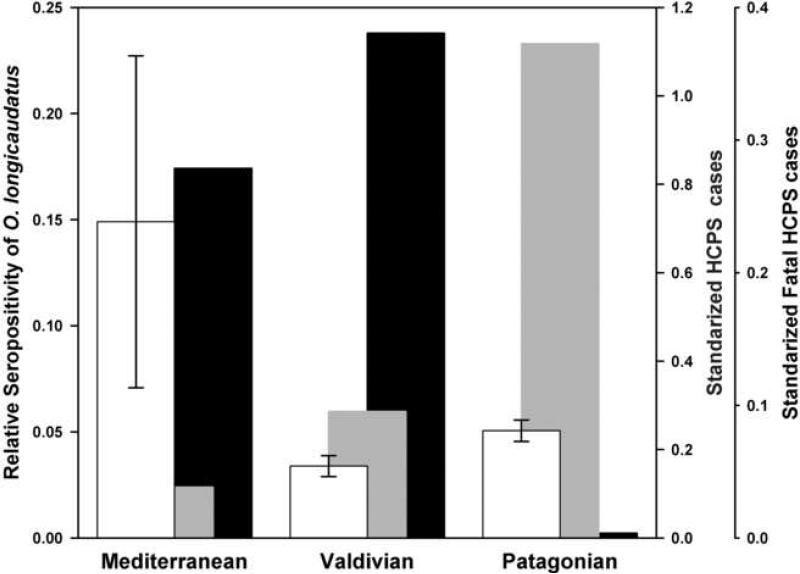
A total of 772 O. longicaudatus were captured during the years 2000 − 2006. Of these, 38 were positive to anti-ANDV antibodies, representing an overall seroprevalence of 4.9%. For the Mediterranean region, 171 O. longicaudatus were captured with 19 (11.1%) seropositive for ANDV. For the Valdivian and North Patagonian rain forests regions, 512 and 89 O. longicaudatus were collected with 14 (2.73%) and five (5.62%) individuals seropositive for ANDV, respectively. Using these values, the relative seropositivity was significantly higher in the Mediterranean (0.1490 ± 0.0782) compared to Valdivian (0.0339 ±0.00496) and Patagonian regions (0.0506 ± 0.00509)(Fig. 4). The anti-ANDV antibody prevalence was significant higher in the Patagonian than Valdivian region. During the same period, 98, 273, and 20 human HCPS cases were reported for the Mediterranean, Valdivian and Patagonian regions, respectively. Patagonia shows the highest number of HCPS cases relative to rural population compared to those from Mediterranean and Valdivian regions. The highest absolute number of fatal HCPS cases in the same period occurred in the Valdivian region (109 cases), followed by the Mediterranean (32 cases) and Patagonian (4 cases). However, standardizing fatal HCPS cases by the number of HCPS infections / rural population, the difference between Valdivian and Mediterranean regions decrease (0.279 and 0.380, respectively), with Patagonia remaining significantly lower (0.0036).
Figure 4.
O. longicaudatus relative seropositivity (white bars), and standardized incidence (grey bars) and fatal (black bars) HCPS cases in three Chilean ecoregions during the period 2000− 2006.
4. Discussion
The distribution of O. longicaudatus in Chile spans nearly 4,000 km of latitude and encompasses contrasting landscapes ranging from semi-desert thorn-scrub in the north to southern Magellanic rain forest. Population dynamics and rodent-to-rodent interactions necessary to transmit ANDV also vary considerably across this latitudinal span. The Mediterranean region is characterized by a highly heterogeneous vegetation mosaic (Armesto et al., 2007), with less suitable habitat for O. longicaudatus than the Temperate rain forest region (Murua, 1996). Differences in landscape structure and climate result in distinctive ecological parameters and differences in population genetic structure (Pilot et al., 2006). Close correspondence between population genetic structure and habitat subdivision (corresponding to distinct ecoregions) was reported in coyotes (Canis latrans) in California (Sacks et al., 2008). These differences were attributable mainly to the effects of random genetic drift in ecotypes with greater population subdivision. Population genetic structure (based on mtDNA cyt-b) of O. longicaudatus differs across the latitudinal span of Chile. Genetic divergence within Mediterranean populations was higher than those within either Valdivian or Patagonian rain forests populations, reflecting higher genetic substructure in northern populations. This rodent shows a tight association with mesic habitats (Cortes et al., 1988; Kelt et al., 1994), so greater spatial heterogeneity in the Mediterranean region due to a more xeric environment is likely a major cause of interregional differences. However, geographic factors (such as natural barriers or spatial distance) and behavioral differences, as well as historical processes (such as population expansion, bottlenecks) may also contribute to difference in genetic substructure as they also influence gene flow among populations (Hartl and Clark, 2007). We have not evaluated all of these processes, but greater population subdivision in the Mediterranean region may allow both deterministic and stochastic genetic processes to be prominent drivers in differentiating O. longicaudatus populations within ecoregions (as in the coyote example). No evident geographic barriers or behavioral features are responsible for producing genetic discontinuity in O. longicaudatus at the boundaries of these ecoregions, although isolation-by-distance and postglacial colonization have been suggested as important factors determining the current spatial distribution of this species in Chile (Palma et al., 2005).
Given that ecological dynamics and genetic structure of O. longicaudatus differs across the ecoregions in Chile, we speculated that transmission rates of ANDV may also vary by region, resulting in differences in O. longicaudatus seroprevalences. Mean relative seropositivity, the standardized incident, and number of fatal HCPS cases differed across ecoregions in Chile. An unexpected result of our study is that the highest relative seroprevalence of O. longicaudatus is in the Mediterranean region. Hantavirus seroprevalence in rodent populations depends on many factors affecting ecological dynamics of the virus and the host. Variables like landscape structure (Glass et al., 2007), density-based and/or frequency-based dependence (Madhav et al., 2007; Adler et al., 2008; Lehmer et al., 2008), dilution effect (Keesing et al., 2006; Tersago et al., 2008; Suzan et al., 2009), viral recrudescence (Kuenzi et al., 2005), climatic variability (Hjelle and Glass, 2000), habitat loss and fragmentation (Suzan et al., 2008), may all contribute to complex interactions that make predicting relative seropositivity to ANDV difficult. Our study spans seven years of rodent sampling, therefore factors related to changes through time (viral recrudescence, climatic variability, changes in abundances through time) although not assessed, are implicitly accounted. Transmission rates depend on contact rates among individuals and the proportion of susceptible individuals (McCallum et al., 2001; Clay et al., 2009). These parameters scale with host population density which differs across ecoregions in Chile. We postulate that spatial features such as landscape structure and habitat fragmentation are major components in differences in serological rates of this rodent in Chile, but alternative models should be explored. Suzan et al. (2009) showed that high species richness in a rodent community influences both abundance of reservoir hosts and infection prevalence and leads to decreased encounter rates between infected and susceptible hosts. Species richness of mammalian communities varies in a non-linear pattern across ecoregions in Chile (Cofre et al., 2007) so this factor should be evaluated in the future.
Differences in rodent seropositivity rates across ecoregions suggest that humans may be differentially exposed to infection with ANDV. People inhabiting the Mediterranean region are therefore potentially at higher risk of contracting HCPS. However, more HCPS cases (per rural population) have been reported in the southern regions (http//:epi.minsal.cl), and our study corroborates that finding. We can parsimoniously hypothesize that more HCPS cases in southern regions are related to a higher rural population. In the Temperate forest regions (Valdivian and Patagonia), despite a lower absolute number of people compared to the Mediterranean, there is a proportionally larger rural population. People associated with rural environments (farmers, residents and also visitors) are at higher risk of contracting HCPS in Chile (Castillo et al., 2002; Torres-Pérez et al., 2004), as is also evidenced in other countries within South America (Nichol et al., 1993; Williams et al., 1997; Bayard et al., 2004). O. longicaudatus is found closer to human settlements in rural and periurban areas, increasing the likelihood of exposure to the virus (Ortiz et al., 2004; Torres-Pérez et al., 2004). Also, the higher host abundance of ANDV in the southern regions of Chile is a major factor contributing to disease transmission risk, due to increased contact with humans.
Our study assesses factors affecting rodent host dynamics that directly or indirectly impact the distribution of ANDV and ultimately the incidence of HCPS. Ecology and geography may significantly influence spatial and temporal dynamics of the ANDV host reservoir. The spatial epidemiology of O. longicaudatus requires further investigation to determine more accurately transmission rates of ANDV within/among rodent populations, and ultimately to humans.
Acknowledgments
We thank Ministry of Health in Chile and local health services for providing information on sampling sites where HCPS cases occurred, and the Servicio Agrícola y Ganadero (SAG) and Corporacion Nacional Forestal (CONAF) for trapping permits. We also thank Hantavirus field crew, and the Centro de Investigaciones Médicas (PUC) for fieldwork or lab support. Financial support was provided by the Fogarty International Center Research Grant # D43 TW007131, the NIH-ICIDR Chilean Hantavirus Grant 1 U19 AI45452-01, FONDECYT 1070331 and FONDECYT-CASEB 1501-0001. A. Hope kindly reviewed the draft manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler FR, Pearce-Duvet JMC, Dearing MD. How host population dynamics translate into time-lagged prevalence: An investigation of Sin Nombre virus in deer mice. Bull. Math. Biol. 2008;70:236–252. doi: 10.1007/s11538-007-9251-8. [DOI] [PubMed] [Google Scholar]
- Arai S, Song JW, Sumibcay L, Bennett SN, Nerurkar VR, Parmenter C, Cook JA, Yates TL, Yanagihara R. Hantavirus in northern short-tailed shrew, United States. Emerg. Infect. Dis. 2007;13:1420–1423. doi: 10.3201/eid1309.070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesto JJ, Arroyo MTK, Hinojosa LF, Veblen TT, Young KR, Orme AR. The Mediterranean Environment of Central Chile. In: Veblen TT, Orme AR, Young KR, editors. The Physical Geography of South America. Oxford University Press; Oxford: 2007. pp. 184–199. [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bayard V, Kitsutani PT, Barria EO, Ruedas LA, Tinnin DS, Munoz C, de Mosca IB, Guerrero G, Kant R, Garcia A, Caceres L, Gracia FG, Quiroz E, Castillo ZC, Armien B, Libel M, Mills JN, Khan AS, Nichol ST, Rollin PE, Ksiazek TG, Peters CJ. Outbreak of hantavirus pulmonary syndrome, Los Santos, Panama, 1999−2000. Emerg. Infect. Dis. 2004;10:1635–1642. doi: 10.3201/eid1009.040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmar-Lucero S, Godoy P, Ferres M, Vial P, Palma RE. Range expansion of Oligoryzomys longicaudatus (Rodentia, Sigmodontinae) in Patagonian Chile, and first record of Hantavirus in the region. Rev. Chil. Hist. Nat. 2009 (in press) [Google Scholar]
- Botten J, Mirowsky K, Ye CY, Gottlieb K, Saavedra M, Ponce L, Hjelle B. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J. Virol. 2002;76:7587–7594. doi: 10.1128/JVI.76.15.7587-7594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo C, Sanhueza L, Tager M, Munoz S, Ossa G, Vial P. Seroprevalence of antibodies against hantavirus in 10 communities of the IX Region of Chile where hantavirus cardiopulmonary syndrome cases were reported. Rev. Med. Chile. 2002;130:251–258. [PubMed] [Google Scholar]
- Clay CA, Lehmer EM, Previtali A, St. Jeor S, Dearing MD. Contact heterogeneity in deer mice: implications for Sin Nombre virus transmission. Proc. R. Soc. B. 2009;276:1305–1312. doi: 10.1098/rspb.2008.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cofre HL, Samaniego H, Marquet PA. Patterns of small mammal species richness in Mediterranean and Temperate Chile. In: Kelt D, Lessa EP, Salazar-Bravo J, Patton JL, editors. The Quintessential Naturalist: Honoring the Life and Legacy of Oliver P. Pearson. University of California Publications in Zoology; California: 2007. pp. 275–302. [Google Scholar]
- Cortes A, Zuleta C, Rosenmann M. Comparative water economy of sympatric rodents in a Chilean semi-arid habitat. Comp. Biochem. Physiol. A. 1988;91:711–714. doi: 10.1016/0300-9629(88)90954-1. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1081:39–56. [PMC free article] [PubMed] [Google Scholar]
- Ferres M, Vial P, Marco C, Yanez L, Godoy P, Castillo C, Hjelle B, Delgado I, Lee SJ, Mertz GJ. Prospective evaluation of household contacts of persons with hantavirus cardiopulmonary syndrome in Chile. J. Infect. Dis. 2007;195:1563–1571. doi: 10.1086/516786. [DOI] [PubMed] [Google Scholar]
- Glass GE, Shields T, Cai B, Yates TL, Parmenter R. Persistently highest risk areas for hantavirus pulmonary syndrome: Potential sites for refugia. Ecol. Appl. 2007;17:129–139. doi: 10.1890/1051-0761(2007)017[0129:phrafh]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates, Inc.; Sunderland, MA: 2007. [Google Scholar]
- Heyman P, Mele RV, Smajlovic L, Dobly A, Cochez C, Vandenvelde C. Association between habitat and prevalence of hantavirus infections in bank voles (Myodes glareolus) and wood mice (Apodemus sylvaticus). Vector-Borne Zoonotic Dis. 2009;9:141–146. doi: 10.1089/vbz.2007.0155. [DOI] [PubMed] [Google Scholar]
- Hjelle B, Glass GE. Outbreak of hantavirus infection in the four corners region of the United States in the wake of the 1997−1998 El Nino-southern oscillation. J. Infect. Dis. 2000;181:1569–1573. doi: 10.1086/315467. [DOI] [PubMed] [Google Scholar]
- Hjelle B, Jenison S, Torrez-Martinez N, Yamada T, Nolte K, Zumwalt R, MacInnes K, Myers G. A novel hantavirus associated with an outbreak of fatal respiratory disease in the southwestern United States: evolutionary relationships to known hantaviruses. J.Virol. 1994;68:592–596. doi: 10.1128/jvi.68.2.592-596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecol. Lett. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- Kelt DA, Meserve PL, Lang BK. Quantitative habitat associations of small mammals in a Temperate Rain Forests in southern Chile - empirical patterns and the importance of ecological scale. J. Mammal. 1994;75:890–904. [Google Scholar]
- Klempa B, Fichet-Calvet E, Lecompte E, Auste B, Aniskin V, Meisel H, Barriere P, Koivogui L, ter Meulen J, Kruger DH. Novel hantavirus sequences in Shrew, Guinea. Emerg. Infect. Dis. 2007;13:520–522. doi: 10.3201/eid1303.061198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal JB, Wish M. Multidimensional Scaling. Sage Publications, Inc; California: 1978. [Google Scholar]
- Kuenzi AJ, Douglass RJ, Bond CW, Calisher CH, Mills JN. Long-term dynamics of Sin Nombre viral RNA and antibody in deer mice in Montana. J. Wildl. Dis. 2005;41:473–481. doi: 10.7589/0090-3558-41.3.473. [DOI] [PubMed] [Google Scholar]
- Langlois JP, Fahrig L, Merriam G, Artsob H. Landscape structure influences continental distribution of hantavirus in deer mice. Landscape Ecol. 2001;16:255–266. [Google Scholar]
- Lehmer EM, Clay CA, Pearce-Duvet J, Jeor SS, Dearing MD. Differential regulation of pathogens: the role of habitat disturbance in predicting prevalence of Sin Nombre virus. Oecologia. 2008;155:429–439. doi: 10.1007/s00442-007-0922-9. [DOI] [PubMed] [Google Scholar]
- Linard C, Tersago K, Leirs H, Lambin EF. Environmental conditions and Puumala virus transmission in Belgium. Int. J. Health Geogr. 2007;6:55. doi: 10.1186/1476-072X-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhav NK, Wagoner KD, Douglass RJ, Mills JN. Delayed density-dependent prevalence of Sin Nombre virus antibody in Montana deer mice (Peromyscus maniculatus) and implications for human disease risk. Vector-Borne Zoonotic Dis. 2007;7:353–364. doi: 10.1089/vbz.2006.0605. [DOI] [PubMed] [Google Scholar]
- Martinez VP, Bellomo C, San Juan J, Pinna D, Forlenza R, Elder M, Padula PJ. Person-to-person transmission of Andes virus. Emerg. Infect. Dis. 2005;11:1848–1853. doi: 10.3201/eid1112.050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H, Barlow N, Hone J. How should pathogen transmission be modelled? Trends Ecol. Evol. 2001;16:295–300. doi: 10.1016/s0169-5347(01)02144-9. [DOI] [PubMed] [Google Scholar]
- Medina RA, Torres-Perez F, Galeno H, Navarrete M, Vial PA, Palma RE, Ferres M, Cook JA, Hjelle B. Ecology, genetic diversity, and phylogeographic structure of Andes virus in humans and rodents in Chile. J. Virol. 2009;83:2446–2459. doi: 10.1128/JVI.01057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murua R. Comunidades de mamíferos del bosque templado de Chile. In: Armesto JJ, Villagrán C, Arroyo MTK, editors. Ecología de los bosques nativos de Chile. Editorial Universitaria; Santiago: 1996. pp. 113–133. [Google Scholar]
- Murua R, Gonzales LA, Meserve PL. Population ecology of Oryzomys longicaudatus Philippii (Rodentia, Cricetidae) in southern Chile. J. Anim. Ecol. 1986;55:281–293. [Google Scholar]
- Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- Ortiz JC, Venegas W, Sandoval JA, Chandia P, Torres-Pérez F. Hantavirus in rodents of the VIII Region of Chile. Rev. Chil. Hist. Nat. 2004;77:251–256. [Google Scholar]
- Padula P, Figueroa R, Navarrete M, Pizarro E, Cadiz R, Bellomo C, Jofre C, Zaror L, Rodriguez E, Murua R. Transmission study of Andes hantavirus infection in wild sigmodontine rodents. J. Virol. 2004;78:11972–11979. doi: 10.1128/JVI.78.21.11972-11979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma RE, Rivera-Milla E, Salazar-Bravo J, Torres-Pérez F, Pardinas UFJ, Marquet PA, Spotorno AE, Meynard AP, Yates TL. Phylogeography of Oligoryzomys longicaudatus (Rodentia : Sigmodontinae) in temperate South America. J. Mammal. 2005;86:191–200. [Google Scholar]
- Pilot M, Jedrzejewski W, Branicki W, Sidorovich VE, Jedrzejewska B, Stachura K, Funk SM. Ecological factors influence population genetic structure of European grey wolves. Mol. Ecol. 2006;15:4533–4553. doi: 10.1111/j.1365-294X.2006.03110.x. [DOI] [PubMed] [Google Scholar]
- Riquelme R, Riquelme M, Torres A, Rioseco ML, Vergara JA, Scholz L, Carriel A. Hantavirus pulmonary syndrome, southern Chile. Emerg. Infect. Dis. 2003;9:1438–1443. doi: 10.3201/eid0911.020798. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Bannasch DL, Chomel BB, Ernest HB. Coyotes demonstrate how habitat specialization by individuals of a generalist species can diversify populations in a heterogeneous ecoregion. Mol. Biol. Evol. 2008;25:1384–1394. doi: 10.1093/molbev/msn082. [DOI] [PubMed] [Google Scholar]
- Simonetti JA, Aguero T. Assessing Independence of Animal Movements - Spatiotemporal Variability in Oryzomys longicaudatus. Mammalia. 1990;54:316–320. [Google Scholar]
- Song JW, Kang HJ, Gu SH, Moon SS, Bennett SN, Song KJ, Baek LJ, Kim HC, O'Guinn ML, Chong ST, Klein TA, Yanagihara R. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura). J. Virol. 2009;83:6184–6191. doi: 10.1128/JVI.00371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzan G, Marce E, Giermakowski JT, Armien B, Pascale J, Mills J, Ceballos G, Gomez A, Aguirre AA, Salazar-Bravo J, Armien A, Parmenter R, Yates T. The effect of habitat fragmentation and species diversity loss on hantavirus prevalence in panama. Ann. N. Y. Acad. Sci. 2008;1149:80–83. doi: 10.1196/annals.1428.063. [DOI] [PubMed] [Google Scholar]
- Suzan G, Marce E, Giermakowski JT, Mills JN, Ceballos G, Ostfeld RS, Armien B, Pascale JM, Yates TL. Experimental Evidence for Reduced Rodent Diversity Causing Increased Hantavirus Prevalence. PLoS ONE. 2009;4:e5461. doi: 10.1371/journal.pone.0005461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersago K, Schreurs A, Linard C, Verhagen R, Van Dongen S, Leirs H. Population, Environmental, and Community Effects on Local Bank Vole (Myodes glareolus) Puumala Virus Infection in an Area with Low Human Incidence. Vector-Borne Zoonotic Dis. 2008;8:235–244. doi: 10.1089/vbz.2007.0160. [DOI] [PubMed] [Google Scholar]
- Toro J, Vega JD, Khan AS, Mills JN, Padula P, Terry W, Yadon Z, Valderrama R, Ellis BA, Pavletic C, Cerda R, Zaki S, Wun-Ju S, Meyer R, Tapia M, Mansilla C, Baro M, Vergara JA, Concha M, Calderon G, Enria D, Peters CJ, Ksiazek TG. An outbreak of hantavirus pulmonary syndrome, Chile, 1997. Emerg. Infect. Dis. 1998;4:687–694. doi: 10.3201/eid0404.980425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Pérez F, Navarrete-Droguett J, Aldunate R, Yates TL, Mertz GJ, Vial PA, Ferres M, Marquet PA, Palma RE. Peridomestic small mammals associated with confirmed cases of human hantavirus disease on southcentral Chile. Am. J. Trop. Med. Hyg. 2004;70:305–309. [PubMed] [Google Scholar]
- Veblen TT. Temperate Forests of the Southern Andean Region. In: Veblen TT, Orme AR, Young KR, editors. The Physical Geography of South America. Oxford University Press; Oxford: 2007. pp. 217–231. [Google Scholar]
- Williams RJ, Bryan RT, Mills JN, Palma RE, Vera I, De Velasquez F, Baez E, Schmidt WE, Figueroa RE, Peters CJ, Zaki SR, Khan AS, Ksiazek TG. An outbreak of hantavirus pulmonary syndrome in western Paraguay. Am. J. Trop. Med. Hyg. 1997;57:274–282. doi: 10.4269/ajtmh.1997.57.274. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian Populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee J, Wortman IA, Nofchissey RA, Goade D, Bennett SG, Webb JP, Irwin W, Hjelle B. Rapid and simple method for screening wild rodents for antibodies to Sin Nombre hantavirus. J. Wildl. Dis. 2003;39:271–277. doi: 10.7589/0090-3558-39.2.271. [DOI] [PubMed] [Google Scholar]
- Zhang YZ, Dong X, Li X, Ma C, Xiong HP, Yan GJ, Gao N, Jiang DM, Li MH, Li LP, Zou Y, Plyusnin A. Seoul virus and hantavirus disease, Shenyang, People's Republic of China. Emerg. Infect. Dis. 2009;15:200–206. doi: 10.3201/eid1502.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]