Abstract
Protein splicing is a posttranslational modification in which an intein domain excises itself out of a host protein. Here, we investigate how the steps in the splicing process are coordinated so as to maximize the production of the final splice products and minimize the generation of undesired cleavage products. Our approach has been to prepare a branched intermediate (and analogs thereof) of the Mxe GyrA intein using protein semi-synthesis. Kinetic analysis of these molecules indicates that the high fidelity of this protein splicing reaction results from the penultimate step in the process (intein-succinimide formation) being rate-limiting. NMR experiments indicate that formation of the branched intermediate affects the local structure around the amide bond cleaved during succinimide formation. We propose that this structural change reflects a re-organization of the catalytic apparatus to accelerate succinimide formation at the C-terminal splice junction.
Introduction
Protein splicing is a post-translational process in which an intervening sequence, termed an intein, is removed from a host protein, the extein [1, 2]. Over 375 members of this protein domain family have been identified in unicellular organisms from all three phylogenetic domains (for a complete listing see; www.neb.com/neb/inteins.html) [3]. In addition, multicellular organisms contain autoprocessing domains orthologous to inteins at the structural and/or mechanistic levels [4-6].
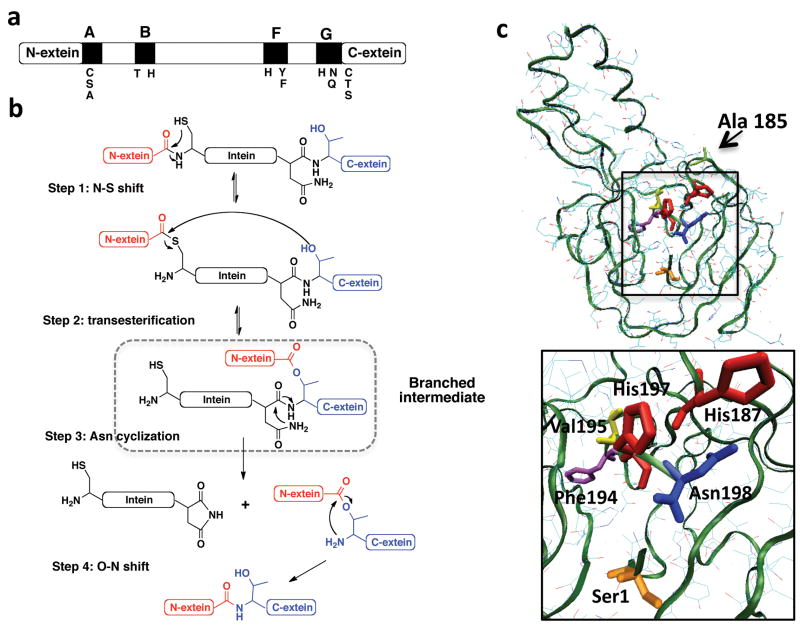
Members of the intein family share conserved sequence motifs that contain residues critical to the splicing reaction (Fig. 1a). A wealth of biochemical data indicates that protein splicing is a multi-step process (reviewed in refs 1, 2, 7). The first step in the standard protein splicing mechanism involves an N→S (or N→O) acyl shift in which the N-extein unit is transferred to the side-chain SH or OH group of a Cys/Ser residue (Fig. 1b). In the next step, the entire N-extein unit is transferred to a second conserved Cys/Ser/Thr residue at the intein-C-extein boundary (+1 position) in a transesterification step. The resulting branched intermediate is then resolved through a cyclization reaction involving a conserved asparagine residue at the C-terminus of the intein [8]. The intein is thus excised as a C-terminal succinimide derivative. In the final step, an amide bond is formed between the two exteins following an S→N (or O→N) acyl shift. The final step is known to be a spontaneous chemical reaction [9] and presumably does not require the structured intein.
Figure 1.
The mechanism of protein splicing. (a) Schematic illustrating conserved regions within the intein family. Conserved sequences (A, B, F and G) are indicated by filled boxes. Residues involved in the splicing reaction are shown below the bar. (b) Schematic of the mechanism of protein splicing. (c) Upper panel; ribbon representation of the Mxe GyrA intein structure (pdb code 1AM2) showing the location of Ala185 ligation site used for EPL. Lower panel; close up of the Mxe GyrA active site. His187 and 197 are shown in red, Asn198 in blue, Ser 1 in orange, Phe194 in purple and Val195 in yellow.
Although we have a reasonable description of the chemical steps in protein splicing, the mechanistic details of autocatalysis are incomplete. The high-resolution structures of several protein splicing precursors (i.e. intein embedded in exteins) have been solved by x-ray crystallography [10-13] and NMR methods [14, 15] and reveal a conserved β-sheet intein fold which positions the key catalytic residues proximal to the N- and C-terminal splice junctions (Fig. 1c). However, all intein structures reported to date have, by necessity, used inteins inactivated through mutation – the kinetics of protein splicing is rapid relative to the time required for high-resolution structural analyses. Thus, we currently have no high-resolution structural information on an active protein-splicing precursor, and by extension of any splicing intermediate. This caveat aside, structural analyses provide some surprising insights into how inteins might accelerate certain of the steps. For instance, the scissile peptide bond at the amino-terminal splice junction (-1 amide) has been found in a variety of conformations ranging from normal trans [13] to twisted-trans [10] to a cis-configuration [16]. This has led to the hypothesis that the scissile bond is activated by distortion away from the normal trans-peptide plane. Consistent with this distortion idea, we demonstrated using NMR methods that the -1 amide has an abnormally low one bond dipolar coupling (1JNC = 12.3 ± 0.3 Hz) in a fully active protein splicing precursor containing the Mycobacterium xenopi DNA gyrase A intein (Mxe GyrA) [17]. Intriguingly, the scissile peptide bond at the C-terminal splice junction (+1 amide) was also found to be distorted in the crystal structures of a mutant Sce VMA intein [10] and a mutant Synechocystis sp (Ssp) DnaB intein [11]. However, it remains to be established whether, by analogy to the -1 scissile amide, peptide bond distortion at the C-terminal splice junction is required for the cleavage reaction.
The overall fidelity of protein splicing hinges on succinimide formation occurring after branched intermediate formation (Fig. 1b). Premature cleavage of the C-extein would be a competing reaction were that not the case. Although mutant inteins have been generated with C-extein cleavage activity [18-20], this represents only a very minor side-reaction in the context of wild-type inteins embedded in native extein flanking sequences [21-23]. It is currently unclear how the steps in protein splicing are coordinated so as to ensure that succinimide formation occurs only in the presence of the branched intermediate. The simplest explanation would be if succinimide formation were the rate-limiting step in the process – thus, there would be a build up of branched intermediate. While there have been a number of kinetic studies performed on inteins [24-26], the rate of succinimide formation in the context of a branched intermediate has not been reported. Thus, it remains to be seen if the high efficiency of splicing in a native context is explained by the differential kinetics of the steps. A second possibility, which is not mutually exclusive from the first, is that formation of the branched intermediate induces a local conformational change in the C-terminal splice site that accelerates succinimide formation. In other words, branched intermediate formation and succinimide formation are coupled. This triggering model is attractive, but in the absence of any structural information on a branched intermediate, it is currently unclear whether any reorganization of the intein active site occurs upon branched intermediate formation.
Here we employ semi-synthetic versions of the Mxe GyrA intein to show that succinimide formation is indeed the rate-determining step in the splicing reaction of this intein, and that conserved histidine residues in box F and G are essential for this step. Selective incorporation of NMR active-isotopes into the +1 amide bond allowed us to directly show that branched intermediate formation affects the local structure around the +1 amide bond. We propose that this structural change reflects a re-organization of the catalytic apparatus, perhaps including distortion of the +1 amide itself, to favor succinimide formation at the C-terminal splice junction.
Results
As in our previous studies, we chose to work on the Mxe GyrA intein due to its accessibility to semi-synthesis and suitability for NMR studies [17].
Semisynthesis of an active branched intermediate
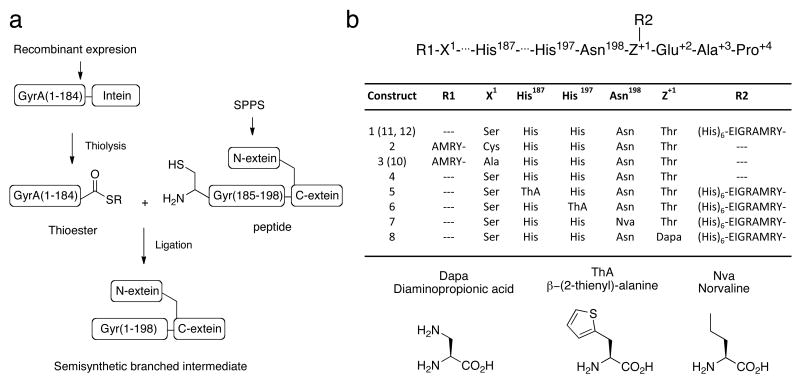
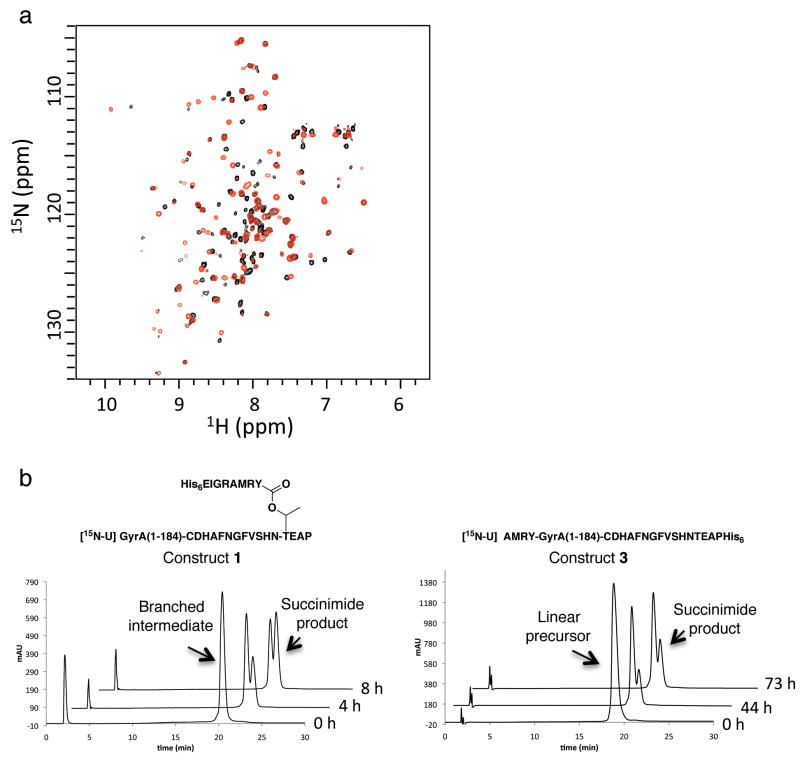
The goal of this study was to perform biochemical and biophysical studies on an active branched intermediate. Since this is an evanescent species in the splicing process, it cannot be easily isolated in pure form from protein splicing reactions. Consequently, we undertook to prepare this molecule using expressed protein ligation (EPL) [27]. We envisioned the synthetic route outlined in Fig. 2a in which a premade branched synthetic peptide, corresponding to the C-terminus of the intein and including both native N- and C-extein residues, would be ligated to a recombinant polypeptide corresponding to the remainder of the intein (Fig. 2a). Importantly, this intein fragment would contain a Cys1 to Ser mutation to prevent the reverse reaction (branched intermediate back to linear precursor) in the final product, which would otherwise complicate our analysis. In preliminary studies, we identified Ala185 as a suitable ligation site; the A185C mutant of Mxe GyrA had comparable splicing activity to the wild-type (Supplementary Fig. 1). Construct 1 was generated by ligation of GyrA(1-184)-α-thioester and a branched peptide corresponding to the remainder of the intein (residues 185-198) linked to four native C-extein residues (TEAP) as well as four native N-extein residues (AMRY) attached through the side-chain of the threonine in the C-extein sequence (Fig. 2b). The former building block was prepared by thiolysis of the corresponding recombinant intein fusion, whereas the latter was chemically synthesized using an orthogonal protection strategy (Supplementary Fig. 2 & 3). In addition, the recombinant fragment was uniformly labeled with 15N to assess the folded state of the final product. The ligation reaction was performed under denaturing conditions in order to prevent the product from undergoing in situ succinimide formation. Analysis by HPLC and mass spectrometry indicated the reaction was complete after five days, after which the product was purified (Supplementary Fig. 5 & 6). Folding was achieved by a stepwise dialysis protocol at 4 °C (see Methods). Note, succinimide formation was found to be very slow at this temperature (Table 1), allowing the branched intermediate to be studied by NMR. The 1H{15N} HSQC spectrum of refolded 1 was found to be well dispersed and indicative of a defined tertiary fold (Fig. 3a). This semi-synthesis and refolding protocol was used to prepare a series of branched intermediate and linear precursor analogs containing unnatural amino acid substitutions and heavy-isotope labels (Fig. 2b, Supplementary Fig. 5 & 6).
Figure 2.
Semi-synthesis of Mxe GyrA intein constructs 1-12. (a) Schematic of the semi-synthetic route. (b) Summary of constructs used in the study, where R1 is the sequence N-terminal to the intein, X is the first intein residue, Z is the first C-extein residue and R2 is the sequence at the branch site. The chemical structures of the unnatural amino acids incorporated into the constructs are shown at bottom.
Table 1.
First order rate constants for asparagine cyclization reaction and O to N acyl shift reaction. Data represent mean values ±s.d.
| Construct | pH | T (°C) | Kobs (s-1) | T½ |
|---|---|---|---|---|
| 1 | 7.5 | 25 | 2.1 (±0.4) × 10-5 | ∼8 h |
| 1 | 4.5 | 25 | inactive | --- |
| 1 | 7.5 | 4 | 3.8 (±0.5) × 10-6 | ∼47 h |
| 2 | 7.5 | 25 | 1.9 (±0.3) × 10-5 * | ∼8 h |
| 3 | 7.5 | 25 | 1.7 (±0.2) × 10-6 | ∼110 h |
| 4 | 7.5 | 25 | 2.1 (±0.1) × 10-6 | ∼100 h |
| 5 | 7.5 | 25 | inactive | --- |
| 5 | 7.5 | 25 | inactive | --- |
| 7 | 7.5 | 25 | Ester hydrolysis | ∼128 h |
| 8 | 7.5 | 25 | 1.9 (±0.4) × 10-5 | ∼8.5 h |
| 9 | 6.5 | 25 | 2.1 (±0.3) × 10-3 | ∼3 min |
| 9+intein | 6.5 | 25 | 1.9 (±0.4) × 10-3 | ∼3 min |
Overall apparent first order rate constant
Figure 3.
Functional characterization of branched and linear Mxe GyrA intein constructs. (a) Solution 1H{15N} HSQC spectra of constructs 1 (red) and 3 (black) recorded on a 900 MHz spectrometer at 4 °C. (b) Kinetics of succinimide formation for branched construct 1 (left) and linear construct 3 (right) at 25 °C and pH 7.5. Shown are a series of RP-HPLC chromatograms of the reaction mixtures at different time-points. Identity of starting materials and product were determined by ms.
Branched intermediate resolution is the rate-limiting step
An HPLC-based assay was developed to measure the efficiency and kinetics of succinimide formation from the various refolded semi-synthetic constructs. The expected splice products, intein-succinimide and amide-linked exteins, were efficiently generated when branched intermediate construct 1 was incubated at 25 °C and pH 7.5 (Fig. 3b, Supplementary Fig. 7). Succinimide formation from branched intermediate 1 obeyed first-order kinetics with a calculated rate constant of 2.1 × 10-5 s-1 (t1/2 ∼ 8h) (Table 1, Supplementary Fig. 8). The rate of this step is substantially slower than that of the initial N→S acyl shift for this intein (step 1 in Fig. 1b), which we have previously shown proceeds with an apparent first-order rate constant of at least 1 × 10-3 s-1 (t1/2 ∼11 min) [17]. The overall rate of the Mxe GyrA splicing reaction was measured using linear precursor construct 2 (Fig. 2b). Splice product accumulation could be fit well to a 1st-order reaction with a rate constant of 1.9 × 10-5 s-1 at 25 °C. This value is in the range measured for other inteins [24, 25]. Assuming a sequential kinetic reaction of the type U→BI→SP (where U is the linear precursor intein, BI is the branched intermediate and SP the spliced product), we can easily deduce that the rate constant k1 for the first reaction (U→BI) that includes the N→S acyl shift and the transesterification steps, must be much larger than the rate constant for the second reaction k2 (BI→SP) (see Methods). Therefore, succinimide formation is the rate-limiting step in the overall Mxe GyrA splicing reaction.
Linear precursor construct 3, which cannot form the branched intermediate due to a C1A mutation, also gave rise to the intein-succinimide product (in this case with concomitant cleavage of the C-extein) when incubated at 25 °C and pH 7.5 (Fig. 3b, Supplemental Fig. 9). However, the rate of succinimide formation was over an order of magnitude slower than for construct 1 (Table 1). There are two ways to interpret this result in terms of mechanism; either succinimide formation is accelerated by the formation of the branched intermediate, or it is inhibited when the N-extein is still linked to the intein N-terminus. To discriminate between these possibilities we generated a construct, 4, lacking an N-extein and asked whether it would support C-terminal extein cleavage via succinimide formation. We observed no significant difference in the rate of succinimide formation between constructs 3 and 4 (Table 1). From this we conclude that the branch intermediate structure directly stimulates the rate of intein-succinimide formation.
Previous studies have implicated the conserved blocks F and G histidine residues (His187 and His197 in the Mxe GyrA intein, Fig. 1a) in the asparagine cyclization reaction [11, 18, 28]. Synthetic access to the branched intermediate allowed us to directly test whether these histidines are required for succinimide formation in the context of the Mxe GyrA intein. Two constructs were generated, 5 and 6, in which His187 and His197 were individually replaced with the histidine isostere, β-(2-thienyl)-alanine (Fig. 2b), which due to the extremely low basicity of the thiophene sidechain, is unable to donate a hydrogen bond or participate in proton transfer chemistry at physiological pH [29]. Neither of these branched intermediate analogs possessed any splicing activity, even though both were folded (Supplementary Fig. 10). Thus, the imidazole side-chains of the histidines are required for asparagine cyclization. We also explored the effect of replacing the asparagine residue (Asn198) with the isosteric amino acid, norvaline (construct 7). Surprisingly, we observed hydrolysis of the ester linkage at the branch site with liberation of the N-extein (Supplementary Fig. 11). This hydrolysis reaction was slow at pH 7.5 (t1/2 ∼ 128 hrs), but, importantly, it was not observed when construct 7 was denatured. Therefore, it appears to be catalyzed by the folded intein structure.
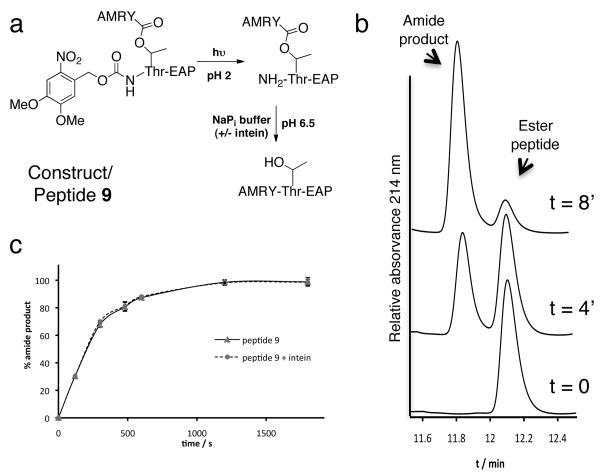
We also asked whether succinimide formation is sensitive to changes at the branch site. Thus, the first residue of the C-extein, Thr+1, was replaced by diaminopropionic acid (Dapa), thereby giving branched intermediate analog, 8, containing a β-isopeptide linkage. This modification had no effect on the rate of intein-succinimide formation (Table 1). This is consistent with the idea that the penultimate and final steps in the splicing reaction are independent. Lastly, we designed an experiment to determine if the intein plays any role in the final step of splicing, the S(O)→N acyl shift (Fig. 1b). This is known to be a rapid chemical rearrangement and it has always been assumed that the intein plays no role in this step. To test this, we prepared O-acyl peptide, 9, containing an N-extein sequence linked through an ester to the Thr side-chain of a C-extein sequence (Fig. 4a). The α-amino group of the Thr was protected with a photocleavable 6-nitroveratryloxycarbonyl (Nvoc) group. To measure the kinetics of O→N shift, we first irradiated an acidic solution of peptide 9 and then diluted this deprotected peptide into splicing buffer at pH 6.5 to trigger the rearrangement. The slower kinetics of O→N acyl shift at pH 6.5 (compared to pH 7.5) allowed us to follow the conversion of the O-acyl peptide into amide-linked product using HPLC (Fig. 4b). We observed no difference in the rate of acyl shift in the presence or absence of the intein-succinimide (previously isolated from a protein splicing reaction) in the splicing buffer (Table 1 and Fig. 4c). Thus, we conclude that the final step of the splicing reaction is not accelerated by the intein.
Figure 4.
Kinetics of the O-N acyl migration reaction. (a) Scheme showing the photoprotected O-acyl peptide (9) and its reaction. (b) Following irradiation peptide 9 was incubated in splicing buffer at pH 6.5. The conversion of unprotected ester peptide into amide product was followed by RP-HPLC. Shown are a series of RP-HPLC chromatograms of the reaction mixtures at different time-points in the absence of intein (c) Plot of the fraction amide product formed at different timepoints in the presence or absence of added intein.
Branched intermediate formation causes a structural change
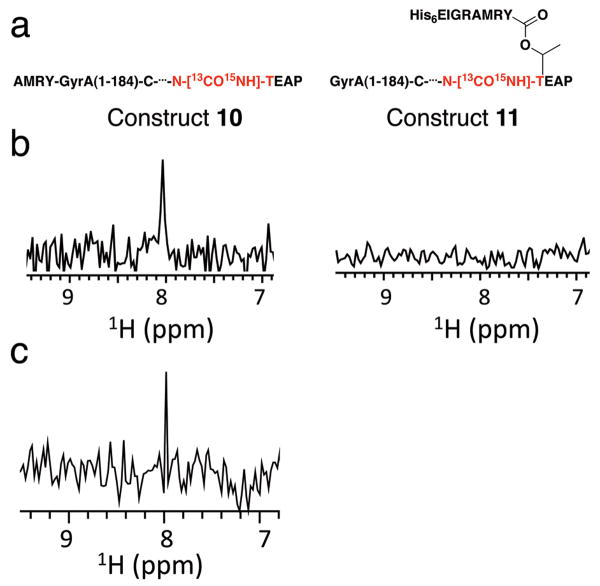
Our kinetic studies indicate that the presence of the branched intermediate stimulates succinimide formation. We wondered whether this could be explained by a structural change, induced by the branched intermediate. To explore this, we used NMR spectroscopy to compare the local structure around the scissile +1 peptide bond in the context of the linear precursor and the branched intermediate. Linear and branched constructs 10 and 11 were generated in which 13C and 15N isotopes were specifically incorporated into the scissile +1 peptide bond. In addition, 15N was uniformly incorporated into the recombinant component of the constructs (Fig. 5a). This segmental labeling scheme allowed the global fold of the inteins to be assessed using 1H{15N} HSQC experiments and provided a unique spectral handle on the scissile amide through the use of HNCO-type experiments. As expected, the HNCO spectrum of linear precursor 10, acquired at pH 7.5 and 4 °C, contained a single signal (Fig. 5b). In contrast, no signal was observed for branched construct 11 under the same conditions (Fig. 5b). We were surprised at the lack of signal(s) in the HNCO spectrum of 11 and wondered whether the NMR sample had undergone succinimide formation prior to or during the experiment – cleavage of the dual-labeled amide would account for the lack of signal. However, HPLC and mass spectrometry analysis indicated that minimal splicing had occurred during the NMR experiment. Proof that a dual labeled amide was in fact present came when the HNCO experiment was repeated on a sample of 11 that had been denatured. In this case, a clear HNCO signal was obtained (Fig. 5c).
Figure 5.
Solution NMR analysis of isotopically labeled constructs. (a) Schematic representation of linear (10) and branched (11) constructs with the scissile +1 peptide bond site-specifically labeled with 13C and 15N. The recombinant component of the constructs was also labeled with 15N. (b) 1D HNCO spectra of construct 10 (left) and construct 11 (right) at pH 7.5 and 4 °C. The signal corresponding to the scissile +1 peptide bond could only be observed for the linear construct 10. Spectra were recorded on a 900 MHz spectrometer. (c) 1D HNCO spectrum of construct 11 under denaturing conditions (10% ACN/D2O + 0.1 %TFA).
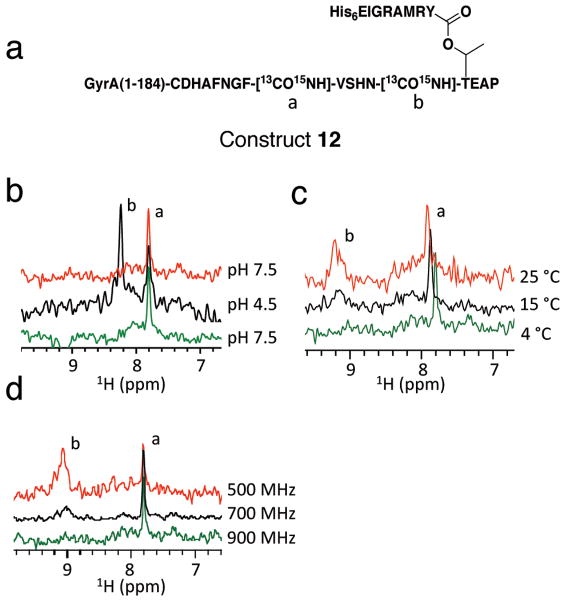
Based on the above, we hypothesized that the lack of signals in the HNCO spectrum of 11 at pH 7.5 reflected some kind of exchange process, either chemical and/or conformational, around the labeled amide. Comparison of the 1H{15N} HSQC spectra of 10 and 11 suggested that this was probably a localized phenomenon because only a subset of signals were perturbed (Supplementary Fig. 12). To explore this idea further, we generated an additional labeled construct, 12, in which two peptide bonds were labeled with 13C and 15N - the scissile +1 amide as well, as the nearby amide connecting Phe194 and Val195 (Fig. 6a). In this case, no 15N labels were incorporated into the remainder of the intein so as to simplify subsequent analyses. A single signal was observed in the HNCO spectrum of 12 at pH 7.5 with a 1H chemical shift of 7.89 ppm (Fig. 6b). By contrast, we observed two signals (1H δ = 8.29 and 7.89 ppm) when the HNCO experiment was performed on the same sample acidified to pH 4.5 (Fig. 6b), a pH at which the intein is inactive (Table 1). Use of a 1H{15N} HSQC-TOCSY experiment allowed us to assign the downfield signal to the scissile +1 amide proton and the upfield signal to the Phe194-Val195 amide proton. Upon raising the pH back to 7.5, we again obtained a single signal in the HNCO spectrum with a 1H δ = 7.89 ppm (Fig. 6b). Thus, the signal from the scissile +1 amide is highly sensitive to the pH, whereas that from the Phe194-Val195 amide is not. The 1JNC′ coupling constants for the scissile and Phe194-Val195 amides at pH 4.5 were found to be 15.4 ± 0.5 and 15.5 ± 0.2 Hz, respectively, which is indicative of a normal trans-planar conformation (Supplementary Fig. 12) [17, 30].
Figure 6.
Effect of changing pH, temperature and magnetic field on the solution structure of the +1 amide bond in the branched intermediate. (a) Schematic representation of site-specifically labeled construct 12. The protein contains 13C and 15N isotopes at both the scissile +1 amide bond (labeled b) and the amide connecting Phe194 and Val195 (labeled a). (b) 1D HNCO spectra of construct 12 at different pH recorded on a 900 MHz spectrometer. In the series, the temperature was kept constant at 4 °C, but the pH switched between pH 7.5 (green), pH 4.5 (black) and then back to pH 7.5 (red). (c) 1D-1H{15N} HSQC spectra of 12 at different temperatures recorded on a 900 MHz spectrometer. In the series, the pH was kept constant at pH 7.5, but the temperature increased from 4 °C (green), to 15 °C (black) and then to 25 °C (red). Peak assignments were based on a 1H{15N} HSQC-TOCSY experiment. (d) 1H{15N} HSQC spectra of 12 at different field strengths. In the series, the temperature and the pH were kept constant at 4 °C and 7.5 respectively, but the spectrum was acquired on a 500 MHz (red), 700 MHz (black) or 900 MHz (green) spectrometer.
We next performed a series of 1H{15N} HSQC experiments on construct 12 at different temperatures, all at pH 7.5. A second signal (1H δ = 9.22 ppm) appeared as the temperature increased, although this was broad compared to the other signal whose line width was largely unchanged over this temperature range (Fig. 6c). This new signal corresponds to the scissile +1 amide. This result is consistent with the idea that there is localized conformational and/or chemical exchange around the +1 amide that is in the intermediate exchange regime on the NMR time-scale [31]. In further support of this interpretation, this same second signal was also observed when the 1H{15N} HSQC spectrum of 12 was acquired on a lower field spectrometer (500 MHz and 700 MHz versus 900 MHz) (Fig. 6d). Attempts to directly measure relaxation time (R2) data at the lower field strengths using a series of R2 CPMG experiments were unsuccessful due to the low peak intensity and rapid transverse relaxation, which further decays over the duration of the experiment (several hours) due to concurrent splicing of the sample.
Discussion
Inteins offer some intrinsic advantages for studying multi-step enzymatic reactions. Firstly, they are small in size, making them amenable to detailed structure-function analysis, including the use of protein semi-synthesis approaches as demonstrated here. Secondly, intein-mediated protein splicing is an autocatalytic process (there is no turnover), which greatly simplifies kinetic dissection of the individual chemical steps in the process. In this study, we have been concerned with how the steps in the splicing process are coordinated so as to maximize the production of the final splice products and minimize the generation of undesired extein cleavage products.
Several kinetic studies have been performed on protein splicing reactions [24-26], however, ours is the first to measure the kinetics of succinimide formation directly from a branched intermediate, which was assembled using EPL (Fig. 2a). The branched intermediate used in our studies contained native N- and C-extein sequences appended to the intein. This is important since several studies indicate that the nature of the extein amino acids at the splice junctions can affect the efficiency and kinetics of protein splicing reactions [21-23]. We find that for the Mxe GyrA intein, the rate of succinimide formation is, within error, the same as the overall splicing rate (Table 1). Thus, succinimide formation represents the slow step for the Mxe GyrA intein-mediated splicing reaction. Additional kinetic studies with construct 3 and 4, allowed us to deduce that the presence of the branched intermediate directly stimulates intein-succinimide formation (Table 1). Thus, our kinetic studies show that the fidelity of the Mxe GyrA intein rests on two key attributes of the mechanism; (i) succinimide formation is the slow step in the process and thus occurs almost exclusively in the context of a branched intermediate, (ii) the presence of the branched intermediate greatly accelerates succinimide formation which ensures that slower C-extein cleavage side-reaction (which bypasses the branch intermediate) cannot compete significantly.
Why is intein-succinimide formation faster in the context of the branched intermediate? It seems reasonable to postulate that transfer of the N-extein sequence to the +1 Thr side-chain would lead to some repositioning of catalytic residues in the intein. Our NMR studies using selectively labeled splicing precursors and intermediates provide the first evidence for this. The 1H{15N} HSQC fingerprint spectra of branched intermediate 1 and linear precursor 3 are not identical, indicating some differences in the structure (Fig. 3a). Further, use of a unique isotopic labeling scheme allowed us to show that the key +1 scissile amide experiences a very different environment upon branched intermediate formation. Specifically, we observe severe line broadening of the amide signal upon shifting the branched intermediate to a pH at which it is active (Fig. 5b). By contrast, no line broadening occurs in the linear splicing precursor 3 or when the pH of the branched intermediate sample is lowered to 4.5 (Fig. 6b), a pH where the intein is still folded, but where no splicing occurs. Additional experiments at higher temperatures and different spectrometer field strengths indicate that the line broadening results from an intermediate exchange phenomenon on the NMR timescale. We stress that this exchange is not felt by the nearby Phe194-Val195 amide (Fig. 6). This argues for a highly localized process. In principle, the line broadening of the +1 amide could reflect chemical and/or conformational exchange. The former might involve protonation of the amide nitrogen, which would lower the π character of the amide, whereas the latter could relate to the dynamics of the local catalytic apparatus and/or the conformation of the amide bond itself. Unfortunately, our efforts thus far to tease this apart have been frustrated by the active nature of the branched intermediate – the signal intensity from the +1 amide rapidly decays during the course of the NMR relaxation analysis. However, the key point remains that this exchange, whatever its nature, is only observed for the +1 amide and only in the context of the branched intermediate under conditions where it is active.
It is most intriguing that exchange around this amide is observable only after the branched intermediate is formed. There is accumulating evidence that enzymes fluctuate between active and inactive (or less active) conformations irrespective of the position on a given reaction coordinate [32, 33]. Thus, it may be more accurate to think of enzyme catalysis in the context of an ensemble of interconverting enzyme conformers leading to a large number of parallel reaction paths, some of which may be more favorable than others [34]. Given that succinimide formation is more favorable after the generation of the branched intermediate and that exchange around the scissile +1 amide is only observed in this context, it seems likely that the scissile amide is sampling one or more high energy states at this point of the reaction coordinate.
We have previously shown that the -1 amide in the Mxe GyrA intein is highly distorted in solution and that the conserved block B histidine is required for this distortion [17]. We believe that this distortion helps drive the first step in the splicing process since strained amides are more susceptible to nucleophilic attack [35-37]. Interestingly, the scissile +1 amide bond has been found in a distorted trans conformation in one crystal structure of a mutant Sce VMA intein [10] and in a mutant of Ssp DnaB intein [11]. In these cases, it was suggested that the distortion might promote succinimide formation and, in the former, that the conserved block G histidine might play a role in protonation of the +1 amide nitrogen. Interestingly, we find that replacement of either of the conserved histidines (His187, His197) with the isosteric residue, β-(2-thienyl)-alanine, abolishes succinimide formation from the branched intermediate. Analysis of the Mxe GyrA crystal structure reveals that the imidazole sidechain of His197 is within 3 Å of the C-terminal Asn carboxyl group (the construct used in the crystal structure does not contain C-extein residues) and so could interact with the +1 amide [16]. The side-chain of His187 appears poised to interact with the side-chain amide of the Asn and may play a role in deprotonation of this group by analogy to other systems [10, 11]. It seems plausible then that the +1 amide is interconverting between multiple strained (i.e. more reactive) conformers, perhaps as a result of an H-bonding interaction involving the imidazole ring from the histidines. The pH-dependence of the exchange phenomena as well as the fact that 1H chemical shift of the +1 amide changes from 8.29 ppm at pH 4.5 to 9.22 ppm at pH 7.5 are both consistent with this idea. Based on this, we propose that formation of the branched intermediate alters the dynamics of the intein active site such that the +1 amide fluctuates between multiple states, some of which may involve distortion of the bond itself. By analogy with the -1 amide at the N-terminal splice junction, such distorted conformers would be more susceptible to nucleophilic attack by the Asn side-chain.
Methods
Peptide synthesis
All peptides were synthesized on Rink-amide ChemMatrix resin using Fmoc/tBu protection chemistry. Synthesis of branched peptides relied on the incorporation of a sidechain unprotected Thr residue into the linear chain. The branched chain (i.e. the N-extein sequence) was then elongated off the β-hydroxyl group of this residue using optimized protocols described in SI Methods (Supplementary Fig. 3 & 4).
Cloning and protein expression
Standard PCR-based cloning procedures were used to generate constructs in which a fragment of the Mxe GyrA intein (residues 1-184, either with or without N-extein residues) was fused to the N-terminus of full length GyrA intein (SI Methods for details). Fusion proteins were expressed in E. coli (BL21(DE3)) cells either grown in M9 minimal media supplemented with [15N]NH4Cl for NMR labeling experiments or LB (Luria Bertani) for kinetic studies. Protein expression was induced by the addition of IPTG for 3 hours. Inclusion bodies were washed, extracted and the fusion proteins purified by Ni-NTA affinity chromatography under denaturing conditions. Pure fusion protein fractions were pooled and folded by stepwise dialysis at 4 °C.
Expressed protein ligation
Thiolysis of the purified intein fusion protein was initiated with 200 mM MESNA followed by incubation for 2 days at 25 °C. Depending on the desired construct, this purified α-thioester intein fragment was mixed with one of several different lyophilized peptides in ligation buffer and incubated for 5 days at 4 °C. The ligation product was purified by Ni-NTA affinity chromatography, followed by semi-preparative C4 RP-HPLC. Products were characterized by analytical C4 RP-HPLC (Supplementary Fig. 5), and ESI-MS spectrometry (Supplementary Fig. 6). All ligated proteins were >90 % pure based on RP-HPLC.
Splicing activity assay and kinetic analysis
Purified branched intermediate constructs were folded at 4 °C by stepwise dialysis from denaturing buffer into the splicing assay buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl and 1 mM TCEP). The samples were then incubated at 25 °C or 4 °C and aliquots of the cleavage reaction were taken at several time points and analyzed by RP-HPLC on a C4 analytical column (Supplementary Fig. 7). The reaction progress was monitored by integrating the branched intermediate and the succinimide product peaks. This data was then fit to a first-order rate equation [BI]=[BI]oe-k2t (Supplementary Fig. 8) which described the reaction: Branched intermediate (BI)Π Spliced product (SP).
The overall splicing reaction was modelled as 3-distict states. The linear precursor intein (2) was folded as above, and the sample was then incubated at 25 °C. The integrated peaks from the HPLC-based splicing assay were fit to the sequential 3-state model: Unspliced (U) →Branched intermediate (BI) → Spliced product (SP) Data from four independent experiments were normalized and fit collectively to the following solutions for the differential equations that describe the splicing reaction:
[U]=[U]oe-k1t, [BI]=(k1[U]o/(k2-k1))(e-k1t-e-k2t), [SP]=[U]o(1-(k2e-k1t)/(k2-k1)+(k1e-k2t)/(k2-k1))
where [U]o is the concentration of initial unspliced reactants, [BI] is the concentration of the branched intermediate, [SP] is the concentration of the spliced product, k1 is the rate of U→BI, k2 is the rate of BI→SP, and t is the time elapsed from the start of the reaction. Using this 3-state model, k1 and [U]o were determined and we observed that k1≫k2.
The O→N acyl shift reaction of peptide 9 was initiated by irradiation of the sample (9:1 acetonitrile/H2O) with a He-Cd laser (325 nm, 4.74 W/cm2, Kimmon Electric Co., Englewood, CO) 3 times for 15 seconds. The peptide was then diluted in splicing buffer (100 mM NaPi pH 6.5, 100 mM NaCl and 1mM TCEP) with or without the intein succinimide product (1 eq. relative to the peptide). The reaction was monitored by analytical C18 RP-HPLC and the data fit to a first-order rate equation (Fig. 4b). All kinetic measurements were carried out at least in duplicate.
NMR spectroscopy
Protein samples for NMR spectroscopy were folded at 4 °C by stepwise dialysis from denaturing buffer into the splicing assay buffer. The protein was concentrated to ∼200 μM for constructs 11 and 12 and ∼70 μM for construct 10. 1H{15N} HSQC and HNCO spectra were collected on a Bruker Avance-500, 700, 800 or 900 spectrometer with a cryo-probe. 2D HNCO spectra were collected on a a Bruker Avance 900 spectrometer (Supplementary Fig. 13) The HSQC-TOCSY experiment was performed on a Bruker Avance-500 spectrometer.
Supplementary Material
Acknowledgments
We thank Steve W. Lockless for helpful discussions. This work is supported by National Institutes of Health Grants GM086868 and GM55843 (to T.W.M.) and by the Generalitat de Catalunya (Spain) for the postdoctoral fellowships Beatriu de Pinos (S.F).
Footnotes
Author Contributions: Experiments were designed by S.F. and T.W.M. Semi-synthetic constructs and kinetic studies were performed by S.F. NMR experiments were carried out by S.F. and M.G. Preliminary studies with A185C mutant were performed by B.G. Data were analyzed by S.F., M.G., D.C., T.W.M. The manuscript was prepared by S.F and T.W.M.
Additional details are presented in SI methods.
References
- 1.Noren CJ, Wang J, Perler FB. Dissecting the chemistry of protein splicing and its applications. Angew Chem Int Ed. 2000;39:450–466. [PubMed] [Google Scholar]
- 2.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 3.Perler FB. InBase: The intein database. Nucleic Acids Res. 2002;30:383–3384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter JA, et al. Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 5.Xu Q, Buckley D, Guan C, Guo H. Structural insights into the mechanism of intramolecular proteolysis. Cell. 1999;98:651–661. doi: 10.1016/s0092-8674(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 6.Rosenblum JS, Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc Natl Acad Sci USA. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu MQ, et al. Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation. EMBO J. 1994;13:5517–5522. doi: 10.1002/j.1460-2075.1994.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu MQ, Southworth MW, Mersha FB, Hornstra LJ, Perler FB. In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 9.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 10.Poland BW, Xu MQ, Quiocho FA. Structural insight into protein splicing mechanism of PI-SceI*. J Biol Chem. 2000;275:16408–16413. doi: 10.1074/jbc.275.22.16408. [DOI] [PubMed] [Google Scholar]
- 11.Ding Y, et al. Crystal structure of a Mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing. J Biol Chem. 2003;278:39133–39142. doi: 10.1074/jbc.M306197200. [DOI] [PubMed] [Google Scholar]
- 12.Sun P, et al. Crystal structures of an intein from the split dnaE gene of Synechocystis sp PCC6803 reveal catalytic model without the penultimate histidine and the mechanism of Zn ion inhibition of protein splicing. J Mol Biol. 2005;353:1093–1105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Mizutani R, et al. Protein-splicing reaction via a thiazolidine intermediate: crystal structure of the VMA-1 derived endonuclease bearing the N and C-terminal propeptides. J Mol Biol. 2002;316:919–929. doi: 10.1006/jmbi.2001.5357. [DOI] [PubMed] [Google Scholar]
- 14.Johnson M, et al. NMR structure of a KlbA intein predursor from Methanococcus jannaschii. Protein Science. 2007;16:1316–1328. doi: 10.1110/ps.072816707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oeemig JS, Aranko AS, Djupsjöbacka J, Heinämäki K, Iwaï H. Solution structure of DnaE intein from Nostoc punctiforme: Structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS letter. 2009;583:1451–1456. doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 16.Klabunde T, Sharma S, Telenti A, Jacobs WR, Sacchettini J. Crystal structure of GyrA intein from Mycobacterium xenopi reveal structural basis of protein splicing. Nat Struct Biol. 1998;5:31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 17.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Semisynthesis of segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at N-extein-intein junction. Proc Natl Acad Sci USA. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu MQ, Perler FB. The mechanism of protein splicing and its modulation by mutation. EMBO J. 1996;15:5146–5153. [PMC free article] [PubMed] [Google Scholar]
- 19.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi Gyrase A intein. Biotechniques. 1999;27:110–120. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 20.Wood DW, Wu W, Belfort G, Derbyshire V, Belfort MA. A genetic system yields self-cleaving inteins for bioseparations. Nat Biotechnol. 1999;17:889–892. doi: 10.1038/12879. [DOI] [PubMed] [Google Scholar]
- 21.Telenti A, et al. The Mycobacterium xenopi GyrA protein splicing element; characterization of a minimal intein. J Bacteriol. 1997;179:6378–6382. doi: 10.1128/jb.179.20.6378-6382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogami S, Satow Y, Ohya Y, Anraku Y. Probing novel elements for protein splicing in the yeast Vma1 protozyme: A study of replacement mutagenesis and intragenic suppression. Genetics. 1997;147:73–85. doi: 10.1093/genetics/147.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amitai G, Callahan BP, Stanger MJ, Belfort G, Belfort M. Modulation of intein activity by its neighboring extein substrates. Proc Natl Acad Sci USA. 2009;106:11005–11010. doi: 10.1073/pnas.0904366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin DD, Xu MQ, Evans TC. Characterization of a naturally occurring trans-splicing intein from Synechocystis sp. PCC6803. Biochemistry. 2001;40:1393–1402. doi: 10.1021/bi001786g. [DOI] [PubMed] [Google Scholar]
- 25.Mills KV, Dorval DM, Lewandowski KT. Kinetic Analysis of the Individual Steps of Protein Splicing for the Pyrococcus abyssi PolII Intein. J Biol Chem. 2005;280:2714–2720. doi: 10.1074/jbc.M412313200. [DOI] [PubMed] [Google Scholar]
- 26.Zettler J, Schuetz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Letters. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawasaki M, Nogami S, Satow Y, Ohya Y, Anraku Y. Identification of three core regions essential for protein splicing of the yeast Vma1 protozyme. A random mutagenesis study of the entire Vma1-derived endonuclease sequence. J Biol Chem. 1997;272:15668–15674. doi: 10.1074/jbc.272.25.15668. [DOI] [PubMed] [Google Scholar]
- 29.Streitwieser A, Scannon PJ. Acidity of hydrocarbons. I. Equilibrium ion pair acidities of thiophene, benzothiophene, thiazole, benzothiazole, and benzofuran toward cesium cyclohexylamine in cyclohexylamine. J Am Chem Soc. 2002;95:6273–6276. [Google Scholar]
- 30.Delaglio F, Torchia DA, Bax A. Mesurment of 15N-13C J coupling in staphylococcal nuclease. J Biomol NMR. 1991;1:439–446. doi: 10.1007/BF02192865. [DOI] [PubMed] [Google Scholar]
- 31.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR spectroscopy Principles and practice. Academic Press; 1996. [Google Scholar]
- 32.Loria JP, Berlow RB, Watt ED. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc Chem Res. 2008;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 33.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry. 2008;47:3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 34.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 35.Harrison RK, Stein RL. Mechanistic studies of peptidyl prolyl cys-trans isomerase: Evidence for catalysis by distortion. Biochemistry. 1990;29:1684–1689. doi: 10.1021/bi00459a003. [DOI] [PubMed] [Google Scholar]
- 36.Lopez X, Mujika JI, Blackburn GM, Karplus M. Alkaline hydrolysis of amide bond twist and nitrogen pyramidalization. J Phys Chem. 2003;107:2304–2315. [Google Scholar]
- 37.Johansson DGA, Macao B, Sandberg A, Härd T. Protein Autoproteolysis: Conformational strain linked to the rate of peptide cleavage by the pH dependence of the N→O acyl shift reaction. J Am Chem Soc. 2009;131:9475–9477. doi: 10.1021/ja9010817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.