Abstract
The identification of hot spots, i.e. binding regions that contribute substantially to the free energy of ligand binding, is a critical step for structure-based drug design. Here we present the application of two fragment-based methods to the detection of hot spots for DJ-1 and glucocerebrosidase (GCase), targets for the development of therapeutics for Parkinson’s and Gaucher’s diseases respectively. While the structures of these two proteins are known, binding information is lacking. In this study we employ both the multiple solvent crystal structures (MSCS) method and the FTMap algorithm to identify regions suitable for the development of pharmacological chaperones for DJ-1 and GCase. Comparison of data derived via MSCS and FTMap also shows that FTMap, a computational method for the identification of fragment binding hot spots, is an accurate and robust alternative to the performance of expensive and difficult MSCS experiments.
Keywords: fragment-based drug design, structure-based drug design, hot spot identification, DJ-1, glucocerebrosidase, Parkinson’s disease, Gaucher’s disease, pharmacological chaperones
Introduction
The increasing application of fragment-based methods in drug discovery is motivated by the desire to develop smaller, yet potent therapeutics, i.e. molecules with increased ligand efficiency [1]. To achieve a commensurate level of potency as compared to larger molecules, fragment-based strategies must exploit hot spots, i.e. sub-sites of binding regions that significantly contribute to the free energy of ligand binding [2]. However, in the absence of either biophysical data or extensive SAR studies, discernment of hot spots within a binding region is often not possible. As the number of drug targets identified increases for which structures have been determined but no other information is available, the ability to design therapeutics will be hindered greatly without the ability to identify hot spots based on structural data alone. Here we present the application of two fragment-based strategies that rely solely on structure for the identification of hot spots for DJ-1 and glucocerebrosidase (GCase), two targets for the development of treatments for neurological and neurodegenerative disorders.
Parkinson’s disease (PD, [3]) is a neurodegenerative disorder that results in impairment of motor skills and cognitive abilities [4], eventually leading to death in patients with advanced PD. Although most cases of PD are sporadic, multiple genes are implicated in the development of PD [5]. Among these is DJ-1, a recent target to emerge for the development of anti-PD agents [6]. DJ-1 consists of 189 amino acids and its function is currently unknown; however, several point mutations are associated with PD [7]. The best-characterized PD-associated mutation of DJ-1 is L166P, located on a C-terminal helix in the homodimer interface [8]. This mutant loses its ability to dimerize [9], resulting in structural instability and rapid degradation [10]. Hence, stabilization of DJ-1 may serve to prevent PD-associated degradation.
Gaucher disease (GD), the most prevalent of the lysosomal storage disorders, i.e. caused by deficient lysosomal glycolipid hydrolase activity [11], is caused by aberrations in acid-β-glucosidase (GCase), the enzyme that hydrolyzes glucosylceramide in the lysosome. GD affects one in 60,000 individuals in the general population and one in 800 in the Ashkenazi Jewish population [12]. Currently, GD patients are treated with enzyme replacement therapy (ERT, Cerezyme) or substrate reduction therapy (SRT, Zavesca). Missense mutations in GCase not localized to its active site exhibit defects in cellular trafficking [13], but do not abolish enzymatic activity of GCase [14]. A new small-molecule therapy currently in clinical trials for GD, pharmacological chaperone (PC) therapy [15], aims to rescue partially active GCase variants, such as the N370S mutant, for lysosomal trafficking and activity. To date, high affinity competitive inhibitors of GCase, which are expected to bind in the active site, have been tested as PCs for GD [16–21]. Although somewhat counterintuitive, enzyme inhibitors can increase steady-state cellular levels of active enzymes. One such pharmacological chaperone is isofagomine (IFG), a nanomolar inhibitor that binds to the active site of GCase [22] and increases cellular trafficking of the N370S-mutant GCase to the lysosome [16,22]. However, to eliminate competition with the substrate once in the lysosome, a small molecule that binds to an allosteric site remote from the active site in a mutant enzyme with the same effect would be preferable as a PC. The identification of several different solvent molecules bound to a single patch on the target protein can be used as the starting point for drug development, such as for a non active site-directed PC.
In this study we aim to identify non-catalytic binding regions that would serve as starting points for the development of pharmacological chaperones for DJ-1 and GCase. We employed two fragment-based methodologies that have been validated previously for the detection of hot spot regions on protein surfaces. First, the multiple solvent crystal structures (MSCS) method developed by Mattos and Ringe [23–25] was utilized for the detection of consensus solvent binding regions on protein surfaces. FTMap [26], a fragment-based method for the in silico detection of hot spots, was employed in conjunction with the MSCS experiments. Previous analyses using the MSCS method, where a crystalline protein is exposed to a series of organic solvents, have shown that consensus sites, i.e. regions of the proteins surface where multiple solvent molecules co-localize, are indicative of ligand binding regions [24]. Similar in concept to the MSCS approach, FTMap is used to determine consensus regions for the binding of fragment-sized molecules [26]. Studies using CSMap [27,28], the precursor to FTMap, showed that in addition to being able to replicate results of MSCS experiments [29], this approach is extremely accurate in the detection of hot spots [30,31] as compared to other biophysical approaches, such as calorimetry [2]. By using both MSCS and FTMap, in this study we detect multiple hot spots suitable for ligand design on the surface of DJ-1 and GCase. Comparison of data resulting from the MSCS experiments to hot spots derived from FTMap show that the computational method is able to accurately reproduce experimental findings, providing a faster and cheaper alternative to crystallographic experiments.
Methods
Multiple Solvent Crystal Structures of DJ-1 and GCase
Purification and Crystallization
Recombinant human DJ-1 and GCase were expressed and purified to homogeneity according to published protocols [8,22].
Crystallization of DJ-1 with solvents was accomplished using hanging drop vapor diffusion. The drops consisted of 2uL concentrated protein (30mg/mL) plus 2uL mother liquor. The mother liquor was comprised of 30% polyethylene glycol (PEG) 400 for cryoprotection, 100mM Tris-HCl pH 8.5, 200mM sodium citrate and, individually, 20%(w/w) Acetonitrile, 20% Ethanol, 10% dimethyl formamide (DMF), 20% dimethyl sulfoxide (DMSO), 10% Phenol, or 5% trifluoroethane (TFE). Crystals with dimensions of approximately 200um × 200um appeared overnight at 20°C.
Crystals of GCase were grown as described previously [22] using the hanging drop vapor diffusion technique. Crystals were crosslinked by placing a micro-bridge (Hampton Research) with 3μl of a 25% glutaraldehyde solution (Sigma) in the well below a crystal-containing drop and incubating for 30–45 min. For methanol soaking, crosslinked crystals were then transferred briefly to a solution containing 50% methanol, 5mM acetate pH 4.5 and 15% PEG 400, and flash cooled in liquid N2. For soaking with phenol, crystals were briefly soaked in a solution containing 23 mM phenol in 30% glycerol and 10 mM acetate buffer pH 4.5, and flash cooled in liquid N2.
Data Collection and Structure Determination
Diffraction data for DJ-1 were collected at the Stanford Synchrotron Radiadion Laboratory (SSRL) beamline 11-3. GCase diffraction data were collected at the GM/CA-CAT beamline at the Advanced Photon Source (Darien, IL). Data sets for both proteins were processed with the software program HKL2000 [32]. Solvent-bound DJ-1 structures were solved by molecular replacement with the program MolRep [33] using apo DJ-1 ([8], PDB entry 1SOA) as a search model, and subsequent refinment was performed using Refmac [34], both of the CCP4 package [35]. GCase structures were solved by rigid body refinement in Refmac5 using a model derived from PDB code 2NT0 [22] after removing all non-protein molecules. Model building for all DJ-1 and Gcase structures were carried out in Coot [36]. Ligand parameters for DJ-1 were obtained from the Dundee PRODRG2 server (http://davapc1.bioch.dundee.ac.uk/prodrg). Water molecules were added to the model of GCase after several rounds of fitting atomic models into electron density maps using Coot and including phosphates and carbohydrates. For the GCase structures each non-protein molecule was subsequently inspected and noted for additional Fo-Fc different electron density. Methanol or phenol molecules were then modeled, as appropriate. Data processing and refinement statistics for DJ-1 and GCase are given in Tables 1 and 2 respectively.
Table 1.
Data refinement and statistics for DJ-1 MSCS experiments
| Data collection | |||||
|---|---|---|---|---|---|
| Solvent | ACN | Ethanol | DMF | DMSO | TFE |
| Space group | P3121 | P3121 | P3121 | P3121 | P3121 |
| Wavelength (Å) | 1.03 | 1.03 | 1.03 | 1.03 | 1.03 |
| Resolution (Å) | 1.53 | 1.53 | 1.53 | 1.53 | 1.53 |
| Completeness (%) | 99.77 | 99.82 | 99.93 | 99.79 | 99.92 |
| Refinement | |||||
| Unique reflection | 34492 | 34940 | 34897 | 34651 | 35456 |
| R-factor (Rfree) (%)1 | 13.3 (15.6) | 11.8 (14.2) | 13.0 (15.7) | 15.7 (13.1) | 24.6 (25.7) |
| Average B-factor (Å2) | 17 | 16.8 | 13.7 | 15.6 | 10.5 |
R-factor = Σ||Fo| − |Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure-factor amplitudes. Rfree is monitored with 5% reflections excluded from refinement.
Table 2.
Data refinement and statistics for GCase MSCS experiments
| Bound Solvent | Methanol | Phenol |
|---|---|---|
| Data collection | ||
| Space group | P2(1) | P2(1) |
| Cell dimensions a, b, c (Å) | 108.4, 91.5, 152.5 | 113.2, 91.6, 153.2 |
| abg (°) | 90. 110.7, 90 | 90, 111.6, 90 |
| Resolution (Å)* | 20-2.40 (2.49-2.40) | 15-2.30 (2.38-2.30) |
| Rsym | 90(41.6) | 8.6(52.2) |
| I/sI | 16.1(2.6) | 24(2.8) |
| Completeness (%) | 93.2(79.8) | 89.3(95.7) |
| Redundancy | 3.0 | 3.5 |
| Refinement | ||
| Resolution (Å) | 20-2.40 | 15-2.30 |
| No. reflections | 78243 | 109635 |
| Rwork/Rfree* | 22.8/30.6 | 20.9/27.4 |
| No. atoms | ||
| Protein residues | 1988 | 1978 |
| N-acetyl-glucosamine (NAG) | 9 | 4 |
| Sulfate anion (SO42−) | 14 | 9 |
| Glycerol (GOL) | 4 | |
| Water | 432 | 662 |
| MeOH | 5 | |
| Phenol | 5 | |
| B-factors | ||
| Protein | 45.5 | 39.6 |
| NAG | 66.9 | 53.5 |
| SO43− | 82.1 | 74.5 |
| GOL | 52.6 | |
| Water | 39.6 | 43.3 |
| MeOH | 52.4 | |
| Phenol | 77.5 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.016 | 0.021 |
| Bond angles (°) | 1.865 | 2.016 |
Highest resolution shell is shown in parenthesis. 5% of reflections were selected for Rfree.
FTMap
FTMAP starts with sampling billions of fragment positions on a dense translational and rotational grid. The positions are scored using an energy function that includes attractive and repulsive van der Waals terms, electrostatic interaction energy based on Poisson-Boltzmann calculations, a cavity term to represent the effect of nonpolar enclosures, and a structure-based pairwise interaction potential. In spite of its relative complexity, the energy expression is written as sum of correlation functions with components defined on grids. This enables the use of the extremely efficient FFT correlation method for function evaluation [37]. The FTMAP algorithm consists of five steps as follows.
1. Rigid body docking of fragments
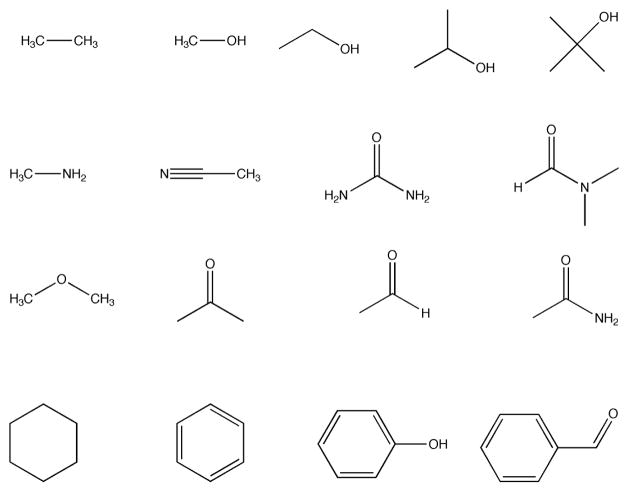
Prior to simulations all bound ligands and water molecules are removed from crystal structures. The 16 fragments shown in Fig 1 are used to probe the binding surface. Mapping requires only the atomic coordinates of the two molecules, i.e. no information on the binding site is used. For each fragment a special purpose rigid-body docking algorithm using the Fast Fourier Transform (FFT) correlation approach samples billions of conformations. The 2000 best poses for each fragment are retained for further processing.
Fig 1.
Fragment set used in FTMap simulations.
2. Minimization and re-scoring
The free energy of each of the 2000 complexes generated in Step 1 is minimized using the CHARMM potential with the Analytic Continuum Electrostatic (ACE, [38]) model representing the electrostatics and solvation terms as implemented in version 27 of CHARMM [39] using the parameter set from version 19 of the program. The ACE model includes a surface area dependent term to account for the solute-solvent van der Waals interactions. Minimizations are performed using an adopted basis Newton-Raphson method. During the minimization the protein atoms are held fixed while the atoms of fragments are free to move.
3. Clustering and ranking
Using a simple greedy algorithm, the minimized fragment conformations generated from Step 2 are grouped into clusters. The lowest energy conformer is selected and conformers within 3~ RMSD are joined in the first cluster. The members of this cluster are removed, and the next lowest energy conformer is selected to start the second cluster. This step is repeated until the entire set is exhausted. Clusters with less than 10 members are excluded from consideration thereby avoiding narrow energy minima with low entropy [40]. The retained clusters are ranked on the basis of their Boltzman averaged energies.
4. Determination of consensus sites
Similar to MSCS experiments, FTMAP utilizes a consensus clustering approach to detect hot spots, i.e. regions of the protein surface where clusters of different fragment types overlap. The six clusters with the lowest average free energies (see Step 3) are retained for each fragment type. Clusters of different fragments are clustered into consensus sites using the distance between the centers of mass of the cluster centers as the distance measure. FTMAP again employs a simple greedy algorithm to find the cluster with the maximum number of neighbors (defined as cluster centers within 4~ from each other), forming the first consensus site. Members of this site are then removed from consideration, and the procedure is repeated until all clusters are exhausted. Conformers are then redistributed among the consensus sites such that each conformer is closest to the center of its own consensus site, and finally the consensus sites are ranked based on the number of their clusters.
5. Characterization of the binding site
FTMAP first selects the largest consensus site (CS1), which is generally identifies the most important subsite (or hot spot) for ligand binding. CS1 forms the kernel of the binding site. The binding site is then expanded by adding any consensus site (irrespective of its size) within 7~ of any consensus site already in the binding site. This procedure continues until no further expansion is possible. The resulting set of consensus sites is used to describe the binding site.
Results and Discussion
Detection of hot spots for DJ-1
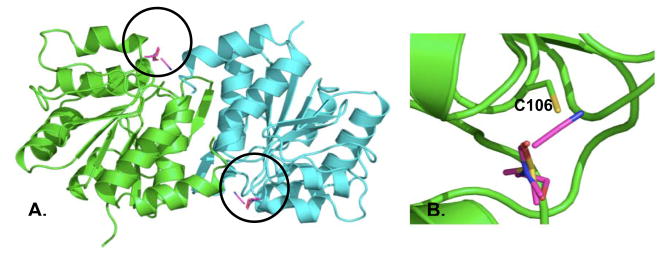
Experimental solvent mapping of DJ-1 revealed a single hot spot bound by all solvents on each monomer (Fig 2A, circled). This hot spot emerges in the region containing Cys 106 and is shown in close-up in Fig 2B. Published studies have shown that xxidation of Cys 106 results in the localization of DJ-1 to the mitochondria, where it is has a demonstrated neuroprotective effect [41]. Independent of the MSCS experiments, FTMap simulations (see Methods) were conducted on a dimeric crystal structure of apo DJ-1 (PDB entry 1SOA, [8]). Several consensus regions for fragment binding are detected using FTMap (magenta, Fig 3, Table 1). In excellent agreement with the MSCS data, the most highly populated consensus site resulting from FTMap coincides with the region surrounding Cys 106 (Fig 3B). In addition to reproducing the experimental solvent mapping data, multiple consensus sites unique to the FTMap simulations are formed near the dimer interface (magenta sticks, Figs 3A and 3B). These hot spots may provide a novel starting point for the development of stabilizing agents of DJ-1. In Table 3 the ranks of the consensus sites are given with their size, i.e. the number of fragment clusters comprising each site. Interacting residues are determined as those residues located within 5Å of a fragment cluster and were calculated using Pymol version 0.99 ([42], http://pymol.sourceforge.net).
Fig 2.
Results of the DJ-1 MSCS experiments. (A) Solvents (magenta sticks, circled) bind to both monomers (green and blue cartoon) of DJ-1 in the region of cysteine 106. (B) Close-up view of the region surrounding the bound solvents. Cys106 is shown in stick to illustrate its proximity to the bound solvents.
Fig 3.
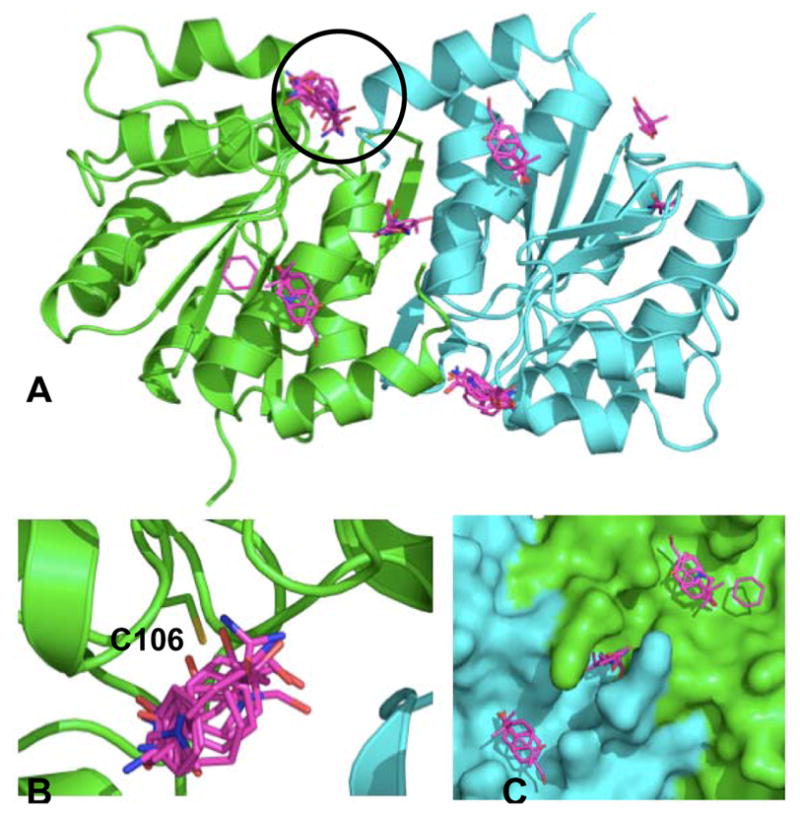
Hot spots detected for DJ-1 via FTMap simulations. (A) Fragment clusters (magenta sticks) populating the top six consensus sites resulting from the simulations are superimposed on dimeric DJ-1. The largest consensus site (circled) is located in the region also detected via MSCS. (B) Close-up view of circled region in (A). C106 is labeled. (C) Close-up view of additional hot spots formed close to the dimer interface. Based on these data, we hypothesize that a pharmacological chaperone could be targeted to this region.
Table 3.
Summary of FTMap-derived hot spots for DJ-1. All results listed are for a single chain unless otherwise denoted.
| CS ranks | CS size | Residues |
|---|---|---|
| 1, 2(B) | 23, 18(B) | E15, E18, R48, G74, N76, C106, A107, H126 |
| 3, 4(B), 5 | 13, 8(B), 5 | R145, G159, F162, E163, L187, K188, R145(B), R158(B), G159(B), F162(B), E163(B), L163(B), L187(B), K188(B) |
CS = consensus site, (B) denotes chain B
Detection of novel hot spots for GCase via MSCS and FTMap
Structures of cross-linked GCase soaked with methanol (Structure A) and phenol (Structure B) are shown here as Figs 4 and 5 respectively. Four molecules are present in the asymmetric units of both structures, where the individual monomers are nearly identical to previously solved structures ([22], not shown). Fourteen sulfate, and 12 methanol molecules were modeled in structure A, whereas nine sulfates and 18 phenol molecules were modeled in structure B. Four glycerol molecules, used as a cryoprotectant, were bound in the active site of structure A, a result we obtained previously and will not be discussed further in this analysis [22]. The modeled sulfate, phenol and methanol molecules are located throughout the surface of GCase (Figs 4–6), and represent sites previously modeled with water and sulfates only. Clusters described in detail below are limited to those regions where a heteroatom of interest was modeled in at least two of the four monomers in the asymmetric unit of GCase. Inspection of the regions bound by the solvents yields several points are noteworthy. First, two tight-binding anionic sites are found in the structure A (Fig 6). These clusters have been described previously [12,43,44], occupied by sulfate or phosphate anions in numerous other GCase crystal structures (not shown). Cluster A1 (Fig. 6B) resides on the same face of the TIM barrel domain as the active site, and sits at the end of the α-helix harboring N370, the site of the most common mutation among Ashkenazi Jews [20]. The sulfate anion forms electrostatic/hydrogen bonding interactions with Arg 353 and Ser 12. Cluster A2 (Fig. 6C) is located at the opposite end of the TIM barrel from the active site, and is stabilized by interactions with Arg 277, Trp 228 and His 306. The analogous site in structure B (see Fig. 5A) site is modeled as a mixture of sulfate and phenol. The observation that cluster A1 is best modeled with sulfate anion in both structures, even after cross-linking and soaking with organic solvents, provides experimental evidence for cluster A1 as a phospholipid binding site. This hypothesis has been suggested previously [22], and is consistent with the notion that a destabilized helix harboring the N370S mutation would impair phospholipid binding [45].
Fig 4.
Bound methanols in GCase (Structure A) (A) Overall view of methanol clusters in GCase relative to active site. (B) Cluster A3, which is equivalent to cluster B in Fig 6. (C) Cluster A4. Stabilizing interactions (< 3.7 Å) between sulfate and side chains are shown with dashed lines.
Fig 5.
Bound phenols in GCase (Structure B) (A) Overall view of modeled phenols after superposition of all monomers in asymmetric unit. (B) Cluster B at interface of three secondary structural elements. Top, ball-and-stick view. Bottom, surface representation. (C) Cluster C at antiparallel β-strand domain. Top, ball-and-stick view. Bottom, surface representation. Stabilizing interactions (< 3.7 Å) between sulfate and side chains are shown with dashed lines.
Fig 6.
Bound sulfates in methanol-bound GCase (Structure A) (A) Overall view of the location of the clusters relative to the active site after superposition of all four monomers in the asymmetric unit. (B) Cluster A1 (C) Cluster A2. Stabilizing interactions (< 3.7 Å) between sulfate and side chains are shown with dashed lines.
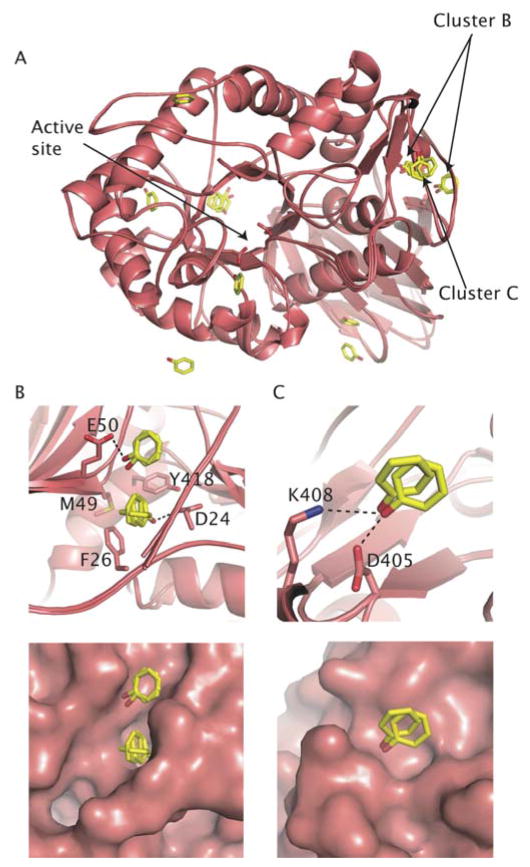
One cluster of interest is located at the interface of all three secondary structure elements in GCase, a groove shaped by amino acid side chains derived from the TIM barrel, β-barrel and anti-parallel β-sheet. In this site we have found phenol (Fig. 5B, cluster B), methanol (Fig 4B, cluster A3), and previously, glycerol, which was introduced as a cryoprotectant (see for example PDB entry 2NT0). Another phenol appears bound close by in two of the four crystallographic monomers (Fig. 5B). A second site (Fig. 5C, cluster C) found on the anti-parallel β-sheet, is separated from the previous site by the N-terminus (residues 1–10). The functional significance of these sites is not yet clear, but it is possible to envision a scenario in which a signal triggers a conformational change in the N-terminus to expose this site for binding to a protein partner. Two partners are currently known: LIMP-II [46], which GCase is proposed to traffic to the lysosome, and saposin C, an activator protein [47–49]. However, the precise binding site for either of these proteins is not currently known. Finally, we have identified a cluster of methanol molecules in structure A (Fig. 4C, cluster A4) at the edge of the β-barrel that abuts the TIM barrel, particularly at the N370-containing helix (see above). N370 has been shown to be critical in stabilizing an active site loop in a conformation of GCase that substrate-ready [22]. It may be possible to develop a small molecule stabilizer for N370S GCase at this interface, which would both improve cellular trafficking as well as improve lysosomal catalysis. This region is also a candidate for a binding site for saposin C, as N370S mutant GCase exhibits poor binding to this activator protein [45]. Further functional studies will be required to determine the binding surface on GCase for both Sap C and LIMP-II, but these experimental methods provide clues to sites of interest.
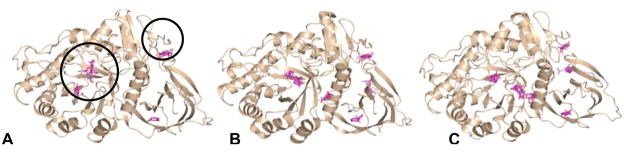
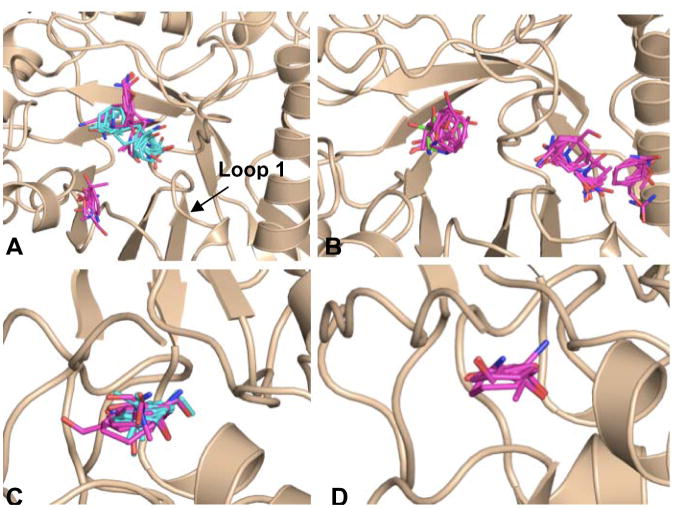
Computational mapping simulations using FTMap were performed on three GCase monomers (Fig 7, Table 4). These monomers were chosen to ensure that all published conformations of GCase were sampled for this analysis. For this study two monomers of PDB entry 2NSX [22] were utilized: chain B, to which IFG is bound, and chain A, to which only glycerol is bound. When IFG is bound, a conformational change in loop 1 occurs, resulting in a shallower surface topology as compared to the apo structure [22]. The third structure utilized was the A chain of PDB entry 2NTl, a neutral pH apo structure of GCase where no bound ligands are present. Two regions containing well-populated consensus sites are found in all three structures (Fig 7, circled in (A)). The largest concentration of fragment clusters contains the catalytic center and proposed hydrophobic subsites for binding of the substrate, glucosylceramide (large circle in Fig 7A, Fig 8). A comparison of the consensus sites for glycerol-bound GCase (2NSX chain A) and apo GCase (2NT1 chain A) shows high overlap at the site of catalysis (Fig 8A). In addition to this region, a second cluster is observed only in glycerol-bound GCase (Fig 8A). Some of these regions appear to clash with the protein side chains in GCase, but are clarified in a comparison with the IFG-bound structure (Fig 8A). Overlay of all hot spots formed in the catalytic region point to a binding region that encompasses both the site of catalysis and the two emerging troughs; these data lend further credibility to the hypothesis that the induced fit conformation adopted by bound IFG is the substrate-ready form of the enzyme. Further, the hot spots that clash with GCase in the non-IFG bound structures (Fig 8A) appear to predict the conformational change observed experimentally with IFG-bound GCase. In terms of therapeutics, these data suggest that a larger molecule that mimics not only the substrate intermediate but can bind to surrounding hydrophobic subsites may be a suitable chaperone.
Fig 7.
Hot spots detected on the surface of GCase using FTMap. Simulations were performed on (A) apo GCase (2NT1) (B) glycerol-bound GCase (2NSX chain A) and (C) IFG-bound GCase (2NSX chain B). Although differences in the location of consensus sites are evident (cluster representatives colored in cyan) two hot spot regions, circled in (A) are conserved in all three structures.
Table 4.
FTMap results for glucocerebrosidase.
| Region | CS ranks (size) | Residues | ||
|---|---|---|---|---|
| 2nsxA | 2nsxB | 2nt1 | ||
| 1 | 2(17) | 1(26) | 2(15) | D127, F128, E235, Y244, F246, Y213, W312, Y313, D315, F316, E340, C342, V343, S345, E349, Q362, W381, N396, V398 |
| 3 (13) | 2(19) | 3(12) | ||
| 5(12) | 4(10) | 6(5) | ||
| 2 | 1(20) | 3(14) | 1(20) | A1, R2, P3, C4, D24, S25, F26, R48, M49, Y418 |
| 3 | 6(10) | 6(8) | 6(6) | L67, L372, L436, V437, A438, N442, L444, D445, A446, V446 |
CS = consensus site
Fig 8.
Close-up view of hot spots formed in the catalytic center (Figure 7A, large circle) and N terminal region (Figure 7A, small circle) of GCase from FTMap simulations (A) Fragment clusters formed in catalytic center of the apo (magenta sticks) and glycerol-bound (cyan sticks) conformations of GCase. (B) Binding of IFG results in a conformational change that allows for the formation of additional hot spots in the active site region (C) The largest number of fragment clusters is found in the N-terminal region of GCase for both the apo (fragment clusters colored in magenta) and glycerol-bound structures of GCase (fragment clusters colored in cyan) (D) Although smaller, a hot spot is also observed in the N-terminal region of the IFM-bound structure (fragment clusters colored in magenta).
Although the catalytic region contains the highest number of consensus sites, the mostly highly populated consensus sites for two of the three structures is located near the N terminus of GCase (smaller circle in Fig 7A, Fig 8C–D), in a groove at the interface between the β-sheet and β-barrel domains. This region coincides with cluster B identified in the MSCS experiments described above (Fig 4B, Fig 5B), in addition to glycerol observed previously [22]. Although not present in the apo structure (2NT1), fragment clusters are found located in a site proximal to region 2 for both glycerol- and IFG-bound GCase; based on this data, we hypothesize that a drug-sized molecule could be accommodated in this region. Overall, we observe considerable consistency of mapping data across the three structures, in terms of the size and rank of consensus sites, e.g. hot spots, determined for each region (Table 4). These new regions can now be targeted for the discovery of new remote-binding chaperones.
Conclusions
In this study we successfully applied MSCS and FTMap to the identification of novel hot spots for DJ-1 and GCase, two pharmaceutical targets for the development of treatments for neurological disorders. Novel hot spots were identified on the surface of DJ-1 in two regions. One hot spot was found in the region containing a residue whose oxidation may protect against PD and the other in the dimer interface, where a pharmacological chaperone could be bound to increase the stability of the dimeric structure. Three regions of interest were identified for GCase, with multiple hot spots emerging in the catalytic region. While the catalytic function of GCase is known, interactions with substrates and other proteins are poorly understood on a structural level. Results of the MSCS experiments and FTMap simulations corroborate the location of the catalytic center, and substantiate claims of hydrophobic binding sites for the substrate ceramide. Additional hot spots found on the surface of GCase provide new hypotheses for the binding sites of key players in trafficking and catalysis, both of which are disrupted in patients with Gaucher’s Disease. These hot spots also provide starting points for drug discovery efforts.
In addition to these findings, the excellent agreement observed between hot spots derived from the two methods supports the use of computational simulations in lieu of expensive and time-consuming experiments. While MSCS is a powerful approach, often the resolution of structures derived is too low to accurately determine solvent binding positions, in addition to the method itself being costly and difficult to perform. For example, higher resolution structures will be required to more confidently model organic heteroatoms in the GCase structure, a goal that will continue to be challenged by the somewhat lipophilic nature of GCase and subsequent tendency of cross-linked GCase crystals to disintegrate when exposed to organic solvents. In addition to successfully uncovering hot spots found using MSCS, we were able to uncover additional hot spots using that were not observed in the MSCS experiments. Although one caveat of using FTMap is the conformational dependence of the results, as illustrated by the analysis of GCase, consensus information gained from the use of multiple structures as input for FTMap simulations increases the robustness of the results. FTMap is available free to the academic community at http://ftmap.bu.edu/~ftmap/.
Acknowledgments
M.R.L was supported by grant F32NS061415 from the National Institute of Neurological Disorders and Stroke. Research performed in the laboratory of S.V. was supported by grant GM064700 from the National Institutes of Health (NIH). R.L.L. was supported by fellowship F32AG027647 from the National Institutes of Health. G.A.P. is a Duvoisin fellow of the American Parkinson’s Disease Association. G.A.P. and D.R. are recipients of an award from the McKnight Endowment Fund for Neuroscience. Parkinson’s Disease work at Brandeis University was initiated with generous support from the Ellison Medical Foundation. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL) and the Advanced Photo Source (APS), national user facilities operated on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. Work performed at FM/CA CAT at APS has been funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). We would also like to thank Amicus Therapeutics for their generous support.
References
- 1.Bembenek SD, Tounge BA, Reynolds CH. Drug Discov Today. 2009 doi: 10.1016/j.drudis.2008.11.007. In press. [DOI] [PubMed] [Google Scholar]
- 2.Ciulli A, Williams G, et al. J Med Chem. 2006;49:4992–5000. doi: 10.1021/jm060490r. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson J. An Essay on the Shaking Palsy. Whitingham and Rowland; London UK: [Google Scholar]
- 4.Jankovic J. J Neurol Neurosurg Psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 5.Bonifati V. Parkinsonism Relat Disord. 2007;13(Suppl 3):S233–S241. doi: 10.1016/S1353-8020(08)70008-7. [DOI] [PubMed] [Google Scholar]
- 6.da Costa CA. Curr Mol Med. 2007;7:650–657. doi: 10.2174/156652407782564426. [DOI] [PubMed] [Google Scholar]
- 7.Biskup S, Gerlach M, et al. J Neurol. 2008;255(Suppl 5):8–17. doi: 10.1007/s00415-008-5005-2. [DOI] [PubMed] [Google Scholar]
- 8.Wilson MA, Collins JL, et al. Proc Natl Acad Sci U S A. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore DJ, Zhang L, et al. J Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 10.Miller DW, Ahmad R, et al. J Biol Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- 11.Rohrbach M, Clarke JT. Drugs. 2007;67:2697–2716. doi: 10.2165/00003495-200767180-00005. [DOI] [PubMed] [Google Scholar]
- 12.Liou B, Kazimierczuk A, et al. J Biol Chem. 2006;281:4242–4253. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz M, Alfalah M, et al. Int J Biochem Cell Biol. 2005;37:2310–2320. doi: 10.1016/j.biocel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Grace ME, Newman KM, et al. J Biol Chem. 1994;269:2283–2291. [PubMed] [Google Scholar]
- 15.Yu Z, Sawkar AR, Kelly JW. FEBS. 2007;274:4944–4950. doi: 10.1111/j.1742-4658.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 16.Steet RA, Chung S, et al. Proc Natl Acad Sci U S A. 2006;103:13813–13818. doi: 10.1073/pnas.0605928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawkar AR, MS, Zimmer KD, et al. ACS Chemical Biology. 2006;1:235–251. doi: 10.1021/cb600187q. [DOI] [PubMed] [Google Scholar]
- 18.Compain P, Martin OR, et al. Chembiochem. 2006;7:1356–1359. doi: 10.1002/cbic.200600217. [DOI] [PubMed] [Google Scholar]
- 19.Sawkar AR, D’Haeze W, Kelley JW. Cell Mol Life Sci. 2006;63:1179–1192. doi: 10.1007/s00018-005-5437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawkar AR, Adamski-Werner SL, et al. Chem Biol. 2005;12:1235–1244. doi: 10.1016/j.chembiol.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Sawkar AR, Cheng WC, et al. Proc Natl Acad Sci U S A. 2002;99:15428–15433. doi: 10.1073/pnas.192582899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman RL, Wustman BA, et al. Nat Chem Biol. 2007;3:101–107. doi: 10.1038/nchembio850. [DOI] [PubMed] [Google Scholar]
- 23.Allen KN, Bellamacina CR, et al. J Phys Chem. 1996;100:2605–2611. [Google Scholar]
- 24.Mattos C, Bellamacina CR, et al. J Mol Biol. 2006;357:1471–1482. doi: 10.1016/j.jmb.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 25.Mattos C, Ringe D. Nat Biotechnol. 1996;14:595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 26.Brenke R, Kozakov D, et al. Bioinformatics. 2009 In Press. [Google Scholar]
- 27.Dennis S, Kortvelyesi T, Vajda S. Proc Natl Acad Sci U S A. 2002;99:4290–4295. doi: 10.1073/pnas.062398499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kortvelyesi T, Dennis S, et al. Proteins. 2003;51:340–351. doi: 10.1002/prot.10287. [DOI] [PubMed] [Google Scholar]
- 29.Silberstein M, Dennis S, et al. J Mol Biol. 2003;332:1095–1113. doi: 10.1016/j.jmb.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Landon MR, Amaro RE, et al. Chem Biol Drug Des. 2008;71:106–116. doi: 10.1111/j.1747-0285.2007.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landon MR, Lancia DR, et al. J Med Chem. 2007;50:1231–1240. doi: 10.1021/jm061134b. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. 1997;276:307–326. [Google Scholar]
- 33.Vagin A, Teplyakov A. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project, Number 4. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. [Google Scholar]
- 36.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Kozakov D, Brenke R, et al. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 38.Schaefer M, Karplus M. J Phys Chem. 1996;100:1578–1599. [Google Scholar]
- 39.Brooks BR, Bruccoleri RE, et al. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 40.Ruvinsky AM, Kozintsev AV. Proteins. 2006;62:202–208. doi: 10.1002/prot.20673. [DOI] [PubMed] [Google Scholar]
- 41.Blackinton R, Lakshminarasimhan M, et al. J Biol Chem. 2009 doi: 10.1074/jbc.M806599200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delano WL. The PyMol Molecular Graphics System. Delano Scientific; Palo Alto, California, USA: 2008. [Google Scholar]
- 43.Brumshtein B, Wormald MR, et al. Acta Crystallogr D Biol Crystallogr. 2006;62:1458–1465. doi: 10.1107/S0907444906038303. [DOI] [PubMed] [Google Scholar]
- 44.Kacher Y, Brumshtein B, et al. Biol Chem. 2008;389:1361–1369. doi: 10.1515/BC.2008.163. [DOI] [PubMed] [Google Scholar]
- 45.Salvioli R, Tatti M, et al. Biochem J. 2005;390:95–103. doi: 10.1042/BJ20050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reczek D, Schwake M, et al. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 47.de Alba E, Weiler S, Tjandra N. Biochemistry. 2003;42:14729–14740. doi: 10.1021/bi0301338. [DOI] [PubMed] [Google Scholar]
- 48.Hawkins CA, de Alba E, Tjandra N. J Mol Biol. 2005;346:1381–1392. doi: 10.1016/j.jmb.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 49.John M, Wendeler M, et al. Biochemistry. 2006;45:5206–5216. doi: 10.1021/bi051944+. [DOI] [PubMed] [Google Scholar]