SUMMARY
In horizontally-transmitted mutualisms between marine animals and their bacterial partners, the host environment promotes the initial colonization by specific symbionts that it harvests from the surrounding bacterioplankton. Subsequently, the host must develop long-term tolerance to immunogenic bacterial molecules, such as peptidoglycan and lipopolysaccaride derivatives. We describe the characterization of the activity of a host peptidoglycan-recognition protein (EsPGRP2) during establishment of the symbiosis between the squid Euprymna scolopes and its luminous bacterial symbiont Vibrio fischeri. Using confocal immunocytochemistry, we localized EsPGRP2 to all epithelial surfaces of the animal, and determined that it is exported in association with mucus shedding. Most notably, EsPGRP2 was released by the crypt epithelia into the extracellular spaces housing the symbionts. This translocation occurred only after the symbionts had triggered host morphogenesis, a process that is induced by exposure to the peptidoglycan monomer (TCT), a bacterial ‘toxin’ that is constitutively exported by V. fischeri. Enzymatic analyses demonstrated that, like many described PGRPs, EsPGRP2 has a TCT-degrading amidase activity. The timing of EsPGRP2 export into the crypts provides evidence that the host does not export this protein until after TCT induces morphogenesis, and thereafter EsPGRP2 is constantly present in the crypts ameliorating the effects of V. fischeri TCT.
INTRODUCTION
Environmentally transmitted mutualistic symbioses are widespread in the animal kingdom, often in the form of extracellular associations between the symbiont and host epithelial tissues. During the initial process of colonization, symbiotic partners undergo modifications that enable the transition from the non-symbiotic to the symbiotic state (Kelly and Coutts, 2000; Gage, 2004; J.E. Cooper, 2007). The microbial symbionts undergo cellular and molecular modifications that mediate their change from a free-living lifestyle, e.g. bacteria in the rhizosphere or in seawater, to that of an inhabitant of specific host tissues, e.g. symbionts in the root nodule of leguminous plants (Gage, 2004; J.E. Cooper, 2007) or in the trophosome of the hydrothermal-vent tube worm (Markert et al., 2007). Similarly, the environment of the host tissue that will house the symbiont must undergo a series of transitions: starting from a permissive state that allows sampling of the external environment, to a restrictive state that promotes colonization by the appropriate symbionts, and finally to a state of accommodation amenable to a long term association that benefits both partners. In this study, using the squid-vibrio symbiosis, we ask the question: Following colonization, how does a symbiotic host tolerate specific symbionts that release high levels of potentially toxic molecules into the tissue environments that support the association?
In the mutualistic association between the Hawaiian bobtail squid Euprymna scolopes and the luminous Gram-negative bacterium Vibrio fischeri, the host hatches without the symbiont and must acquire from the seawater V. fischeri cells, which then colonize deep epithelium-lined crypts of a specialized light organ (Fig. 1). As is characteristic of marine mollusks, many of the internal organs of the host squid, including the light organ, are exposed to the seawater containing a total of approximately 106 bacterial cells/ml, of which V. fischeri comprises about 0.01% (Lee and Ruby, 1994). Up to 1 h after hatching, the light organ is in a permissive state, in which environmental bacterial cells or even small inert particles can access the deep crypts (Nyholm et al., 2002). The light organ then enters a selective state. During this period, V. fischeri cells aggregate in host cell mucus, which has been shed from ciliated epithelial fields on the light organ surface (Fig. 1). Within a couple of hours, V. fischeri cells competitively dominate in the mucus, and then enter the light organ through three pores on each lateral face and slowly swim up the ducts, through the antechambers, and into the deep crypts (Nyholm et al., 2000).
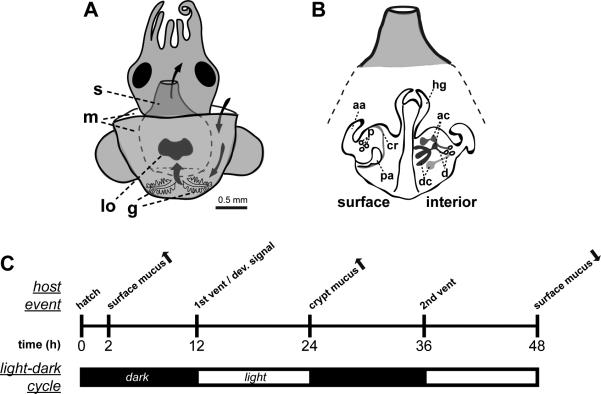
Figure 1. Early colonization of the E. scolopes light organ and host responses to colonization.
(A) E. scolopes organs of the mantle cavity are exposed to the bacterioplankton of the seawater environment by the process of ventilation (water flow depicted by curved arrows). g, gills; lo, light organ; m, mantle; and s, siphon. (B) Whereas the light organ surface epithelium (left) interacts transiently with V. fischeri during the initiation of symbiosis, the internal structures of the light organ (right) are the sites of persistent symbiotic association. aa, anterior appendage; ac, antechambers; cr, ciliated ridge; d, ducts; dc, deep crypts; p, pores; and pa, posterior appendage. (C) A timeline of some early host responses to the bacteria-rich environment, environmental light cues and the colonization by V. fischeri. Arrows indicate increase (up) or decrease (down) of activity.
V. fischeri cells then replicate and fill the deep crypts, until after about 12 h when the early morning light cues a venting of 90-95% of the symbionts back into the external environment (Nyholm and McFall-Ngai, 1998, 2004). Concurrent with this first venting, V. fischeri delivers a signal that irreversibly triggers a developmental loss, through apoptosis and epithelial cell shedding, of the ciliated fields on the light-organ surface, a process that occurs over the subsequent 4 – 5 d period (McFall-Ngai and Ruby, 1991; Doino and McFall-Ngai, 1995; Foster and McFall-Ngai, 1998). In addition to these morphogenic changes, the V. fischeri population in the crypt spaces induces reversible changes in the deep crypts, specifically increased density of the microvilli lining the apical surface of the epithelium (Lamarcq and McFall-Ngai, 1998) and swelling of the epithelia cells (Montgomery and McFall-Ngai, 1994; Visick et al., 2000). Curing the light organ bacterial population with antibiotic treatment results in a return of these two phenotypes to the non-symbiotic state. Patterns of light-organ mucus secretion are also changed during the onset of the symbiosis (Fig. 1). By 24 h, mucus has been secreted into the deep crypts colonized by V. fischeri and by 48 h mucus secretion from the light organ surface has ceased completely (Nyholm et al., 2002). In the 12 – 48 h timeframe, the light organ is beginning to establish the accommodation state that is required for life-long maintenance of the symbiosis. Each morning at dawn throughout the life of the host, the majority of symbionts are vented into the external environment (Nyholm and McFall-Ngai, 2004). The V. fischeri cells that remain after venting repopulate the deep crypts over the course of each day.
The interplay between microbe-associated molecular patterns (MAMPs) and the squid host is an important mediator of the transition from the free-living to the symbiotic state. Gram-negative or Gram-positive peptidoglycan (PGN), which has been shed by environmental bacteria, induces the secretion of mucus from the light organ surface (Nyholm et al., 2002). Upon colonization of the crypts, a tetrapeptide monomer of Gram-negative PGN secreted by V. fischeri, tracheal cytotoxin (TCT), at concentrations as low as 10 nM induces much of the light organ morphogenesis program (Koropatnick et al., 2004). The lipid A of V. fischeri lipopolysaccharide (LPS) induces early-stage apoptosis (Foster et al., 2000) and synergizes with TCT in the induction of light-organ development (Koropatnick et al., 2004; Troll et al., 2009). Furthermore, colonization of the light organ by a V. fischeri mutant deficient in the release of TCT (strain DMA388) results in a slowed light-organ morphogenesis response and reduces the number of apoptotic cells in the light-organ ciliated epithelia (Adin et al., 2008; Troll et al., 2009). These data indicate that V. fischeri releases TCT not only in vitro but also in the deep crypts of the squid light organ and that TCT is a key component of the 12-h irreversible morphogenetic signal.
Recent studies of the association have implicated a family of proteins, the peptidoglycan recognition proteins (PGRPs), in mediating responses to MAMPs during early symbiosis. Members of this protein family are mediators of innate immunity across the animal kingdom (Werner et al., 2000; Dziarski, 2004; Steiner, 2004; Goodson et al., 2005; Coteur et al., 2007). The PGRPs are known to have multiple functions within the host-immune response, including anti-microbial activity (Dziarski and Gupta, 2006), Gram-positive or Gram-negative peptidoglycan recognition leading to antimicrobial peptide induction (Michel et al., 2001; Kaneko et al., 2004), and degradation of PGN-ligands via N-acetylmuramyl-L-alanine amidase (NAMLAA) activity, which cleaves the lactyl-amide between the sugar backbone and the stem peptide (Gelius et al., 2003; Mellroth and Steiner, 2006). NAMLAA cleavage attenuates the cytopathological activities of TCT, which include damage/extrusion of ciliated tracheal or fallopian tube cells and inhibition of DNA synthesis, both mediated by induction of nitric oxide and interleukin-1α production (Melly et al., 1984; Luker et al., 1993; Luker et al., 1995; Cloud-Hansen et al., 2006). Thus, the activity of members of the PGRP family can result in a finely tuned innate immune response by activating antimicrobial peptide production, while also attenuating the effect of peptidoglycan ligands (Gottar et al., 2002; Mellroth et al., 2003; Bischoff et al., 2006; Zaidman-Remy et al., 2006).
Four homologs of the PGRP protein family, EsPGRP1-4, are expressed in the light organ of juvenile E. scolopes (Goodson et al., 2005). Analysis of the derived amino acid sequences of the EsPGRPs, in addition to revealing the PGN-binding domain, suggested cellular locations of each isoform -- EsPGRP1, intracellular; EsPGRP2, secreted; EsPGRP3, secreted and membrane-surface associated; EsPGRP4, integral membrane. In addition, the critical residues for NAMLAA activity are conserved in EsPGRPs 1-3 (Goodson et al., 2005). Thus far, only EsPGRP1 has been studied in depth; this isoform is involved with host-developmental responses to TCT, including apoptosis and tissue morphogenesis (Troll et al., 2009).
In this study, we characterized the role of EsPGRP2 in the dynamics of the symbiosis. We present evidence that EsPGRP2 is a secreted amidase involved in the maintenance of symbiosis with V. fischeri. Using a specific anti-EsPGRP2 antibody, we characterized the cellular and tissue locations of EsPGRP2 over time in both symbiotic and non-symbiotic E. scolopes. In addition, we tested EsPGRP2 for the ability to degrade the PGN-derived morphogen, TCT. Finally we sought to determine the effect of EsPGRP2-degradation on the activity of TCT in the squid light organ.
RESULTS
EsPGRP2 localizes to cytoplasmic stores in epithelial cells and is secreted into extracellular spaces
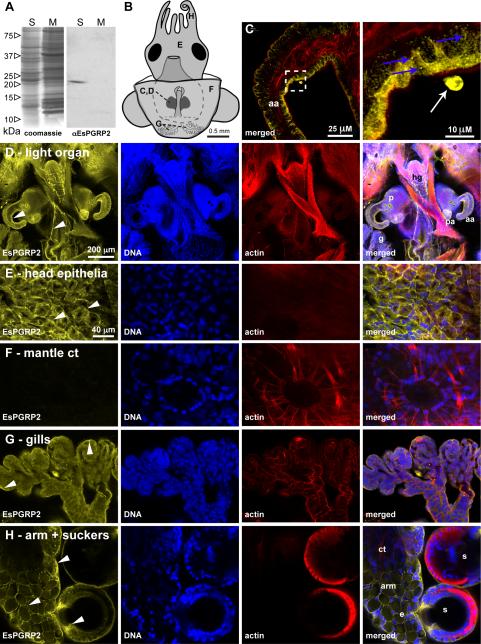
In western-blot analysis of homogenates of non-symbiotic whole juvenile E. scolopes, EsPGRP2 localized to the aqueous soluble fraction at a molecular weight of ~21 kDa (Fig. 2A). These size and localization results were consistent with predictions based on the derived amino-acid sequence from the EsPGRP2 cDNA (Goodson et al., 2005). We examined the localization of EsPGRP2 in a variety of tissues of the juvenile squid using immunocytochemistry visualized by confocal microscopy. The antibody recognized cytoplasmic sites in all epithelial tissues exposed to the seawater environment, including the gills, gut exterior, skin, and the light organ, but did not label most internal tissues, such as the connective tissues of the mantle associated with the chromatophores (Fig. 2 and Fig. S1). EsPGRP2 was observed in cytoplasmic striations associated with, but not colocalized with, striated actin filaments in some connective tissues (Fig. S1), demonstrating that the presence of EsPGRP2 is not restricted to the epithelia. Within the epithelial cells, EsPGRP2 localized to the apical cytoplasm or cell periphery in a granular or punctate form (Fig. 2C). These data, with the prediction of a signal sequence in the protein (Goodson et al., 2005), suggested that the protein is sequestered in vesicles for extracellular secretion. However, neither the presence of bacteria, such as the symbiont V. fischeri, nor developmental time, had any consistent effect on the localization of EsPGRP2 in any of these tissues (data not shown). We infrequently observed extracellular EsPGRP2 associated with the surface of the light organ in both symbiotic and non-symbiotic light organs (Fig. 2B and C). These data, coupled with the localization to the apical cytoplasm of cells known to secrete mucus and nitric oxide synthase (Nyholm et al., 2002; Davidson et al., 2004), led us to hypothesize that EsPGRP2 is secreted from E. scolopes epithelia exposed to seawater.
Figure 2. The localization of EsPGRP2 in E. scolopes tissues.
(A) The anti-EsPGRP2 antibody reacts with a single major protein species in the total soluble fraction of extracts from whole juvenile E. scolopes. The estimated molecular mass of the band was 20.5 kDa, consistent with the predicted mass of 20.8 kDa for the mature form of EsPGRP2. S, soluble fraction; M, membrane fraction. (B) Multiple tissues of juvenile E. scolopes were examined for expression of the EsPGRP2 protein by confocal immunocytochemistry, including the anterior appendage (C) of the light organ (D), the head epithelia (E), connective tissue of the mantle, including a chromatophore (F), the gills (G), and the arm and suckers (H). Juvenile E. scolopes were probed with the anti-EsPGRP2 antibody (yellow), and counterstained for actin-cytoskeleton (rhodamine-phalloidin) and DNA (TOTO-3). The EsPGRP2-antibody stained the apical portion of epithelial cells with the greatest intensity (arrowheads) and the staining appeared granular (blue arrows). Dashed box in left panel of C indicates area of the light-organ anterior appendage shown in higher magnification in right panel. Occasional extracellular protein staining was observed (white arrow). aa, anterior appendage; ct, connective tissue; e, epithelial cells of the arm; g, gills; hg, hindgut; p, pores; pa, posterior appendage; s, suckers.
To examine secretion from epithelia more closely, we performed confocal immunocytochemistry to assay for EsPGRP2 secretion from organ surfaces using a glyoxal-based fixative (see Material and Methods). We observed plumes of EsPGRP2 associated with mucus shedding from the light organ epithelia (Fig. S2).
Secretion of EsPGRP2 from deep crypt epithelia is temporally restricted
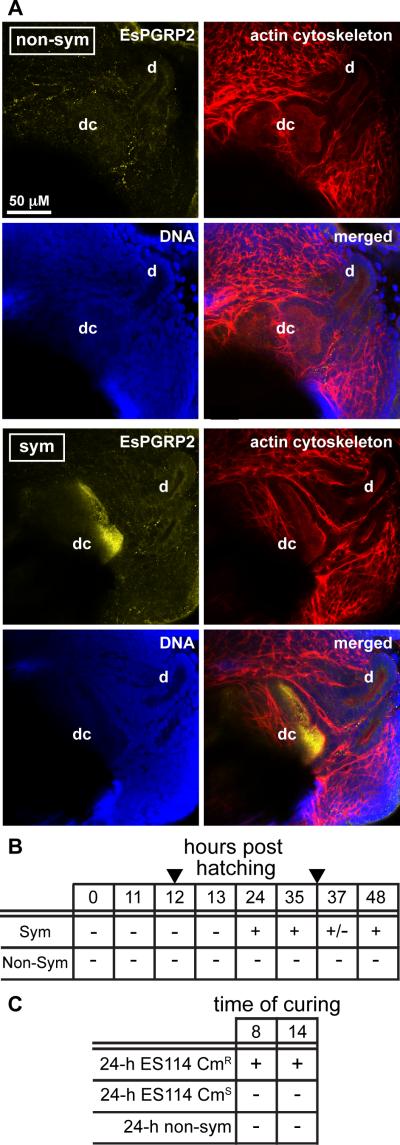
Whereas the light organ surface epithelia fleetingly interact with V. fischeri during the first few hours of the colonization process, the deep crypt epithelia are sites of persistent association with the symbiont. To determine whether the colonization of the deep crypts influences secretion of EsPGRP2, we used confocal immunocytochemistry to examine the internal epithelial structures of the light organ at approximately 12 h intervals through 48 h in non-symbiotic and symbiotic animals. At no time point was EsPGRP2 detected in the deep crypts of non-symbiotic animals (Fig. 3A and B). The lumina of fully colonized deep crypts, at 24 and 48 h, had abundant EsPGRP2 (Fig. 3B and C). However, in the 1 h interval around the critical 12 h time point (at 11, 12 and 13 h), which is coincident with the first vent (Nyholm and McFall-Ngai, 2004) and the delivery of the irreversible morphogenic signal (Doino and McFall-Ngai, 1995), EsPGRP2 was not detected in the deep crypts. Just before the second vent, at 35 h, EsPGRP2 was abundant in the deep crypts, suggesting that the lack of EsPGRP2 in the deep crypts at 12 h is not a result of down-regulation prior to venting events. The presence of EsPGRP2 in the deep crypts of 37-h symbiotic animals was variable. Because the amount of crypt contents vented and the time over which venting occurs can vary between individual animals, this variability of EsPGRP2 in the deep crypts at this time point is consistent with the diel rhythm (Graf and Ruby, 1998; Nyholm and McFall-Ngai, 1998). However, the reduction of EsPGRP2 in the deep crypts at time points associated with reduced symbiont number, e.g. at 37 h or in non-symbiotic compared to symbiotic, suggested that deep-crypt staining by the anti-EsPGRP2 antibody could result from non-specific cross-reactivity with V. fischeri. To eliminate this possibility, we incubated V. fischeri with the anti-EsPGRP2 antibody in vitro and found no fluorescence above background (data not shown). Thus, we concluded that EsPGRP2 is secreted into the luminal spaces housing the symbiotic bacteria beginning sometime between 13 and 24 h.
Figure 3. EsPGRP2 was secreted into the deep crypts of symbiotic light organs.
(A) The anti-EsPGRP2 antibody (yellow) did not label the deep crypts of 24-h non-symbiotic light organs, but intensely stained the lumina of the deep crypts of 24-h symbiotic juvenile E. scolopes. The light organs were counterstained for actin cytoskeleton (rhodamine-phalloidin, red) and DNA (TOTO-3, blue). d, duct; dc, deep crypt. (B) The deep crypts of both symbiotic and non-symbiotic light organs were assayed for the presence of EsPGRP2 over the first 2 days of the symbiosis. Arrowheads indicate venting time points, which occur at 12 h and 36 h. (C) Secretion of EsPGRP2 into the deep crypts is reversible, i.e. not triggered by the irreversible morphogenic signal at 12 h. Symbiotic juveniles were cured of their V. fischeri at either 8 or 14 h with 20 μg/ml chloramphenicol (CM) and then scored for EsPGRP2 secretion into the deep crypts at 24 h. Animals colonized with CM-resistant strains were used to control for any possible effects of the antibiotic on secretion of EsPGRP2. CMR, CM-resistant; CMS, CM-sensitive. For both B and C: -, no EsPGRP2 in the deep crypts of any individual; +/−, some individuals contain EsPGRP2 in their deep crypts; +, all individuals contain EsPGRP2 in deep crypts.
To determine whether the secretion of EsPGRP2 into the deep crypts was irreversible, i.e., similar to other deep crypt phenotype, we performed ‘curing’ experiments. In these experiments, symbiotic animals were treated with the antibiotic chloramphenicol before or after the delivery of the morphogenic signal, at 8 or 14 h respectively, and then scored for EsPGRP2 secretion at 24 h. Antibiotic clearing of the symbionts from the deep crypts at either time point resulted in a lack of detection of EsPGRP2 staining in the deep crypts at 24 h (Fig. 3C). This effect could not be attributed to chloramphenicol acting directly on the squid, because animals colonized with chloramphenicol-resistant V. fischeri secreted EsPGRP2 into their deep crypts at 24 h, regardless of exposure to the antibiotic. These data suggested that the secretion of EsPGRP2 into the deep crypts is dependent on the continuing presence of the symbiont.
EsPGRP2 degrades PGN
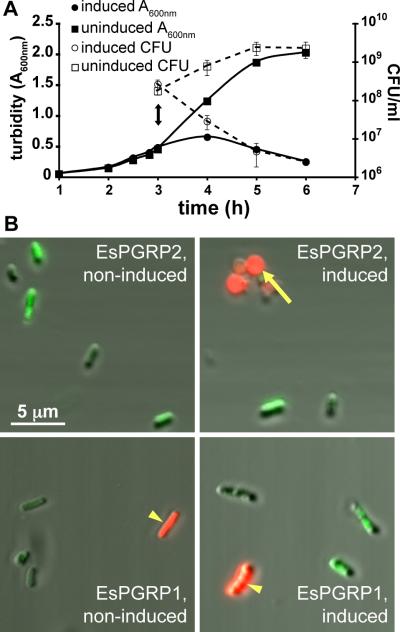
Several lines of evidence suggest that degradation of TCT by PGRPs is a protective host response against this bacterial PGN-derived toxin (Luker et al., 1995; Gelius et al., 2003; Mellroth et al., 2003; Bischoff et al., 2006). Up through the delivery of the irreversible morphogenic signal, TCT behaves as a morphogen in the squid (Koropatnick et al., 2004; Troll et al., 2009). Because TCT appears to be constitutively produced by V. fischeri and is known to be perturbing to animal cells, mechanisms likely exist to protect the host squid tissues from the potential accumulation and resultant toxicity of TCT over the lifetime of the symbiosis. Thus, we performed experiments to determine whether EsPGRP2 is capable of degrading TCT. The first suggestion that EsPGRP2 affects PGN came with our attempts to express and purify recombinant EsPGRP2 in E. coli. Upon induction of EsPGRP2 protein expression, the E. coli cells transitioned to spheroplast morphologies and rapidly lost viability (Fig. 4). Because an intact PGN layer maintains the rod shape of E. coli cells (Kaback and William, 1971), these data were consistent with EsPGRP2 gaining access to the PGN in E. coli and compromising the integrity of the bacterial cell wall.
Figure 4. E. coli-expressed EsPGRP2 degraded the peptidoglycan cell wall.
(A) Production of EsPGRP2 in E. coli cells caused a rapid clearing of cultures and resulted in a loss of cell viability. Double-headed arrow, time of addition of 1 mM IPTG. Error bars represent standard deviation among 3 replicate plates. (B) At 1 h post induction of EsPGRP2 gene expression, cells were live/dead stained and wet mounts were viewed by confocal fluorescence and DIC microscopy. Non-viable (red) cells were common upon expression of the EsPGRP2 gene and these cells were often spherical (arrow), whereas the majority of the cells not-induced for EsPGRP2 expression cells appeared to be viable (green) and rod shaped. Expression of the EsPGRP1 gene from the same vector had no effect on morphology or overall viability of the E. coli cells. Note that occasional non-viable cells (arrowhead) have not lost rod morphology.
To perform in vitro biochemical analyses on EsPGRP2, we expressed and purified the protein from Drosophila S2* cells (Fig 5A), as expression in E. coli was not feasible. One ng of purified EsPGRP2 quantitatively degraded ~86 ng of the TCT within 18 h at room temperature (Fig. 5B), in a manner consistent with the amidase activity of other PGRPs (Gelius et al., 2003; Mellroth et al., 2003; Mellroth and Steiner, 2006; Coteur et al., 2007). We confirmed that EsPGRP2 degraded TCT via NAMLAA activity using coupled liquid chromatography/mass spectroscopy. The undigested TCT showed a 922 Da peak in the positive ion spectrum, corresponding to the known molecular weight of 921 Da for TCT (data not shown). The products of the EsPGRP2/TCT reaction contained a molecular species of 462 Da, precisely corresponding to the expected mass of 461 Da for the tetrapeptide stem of TCT.
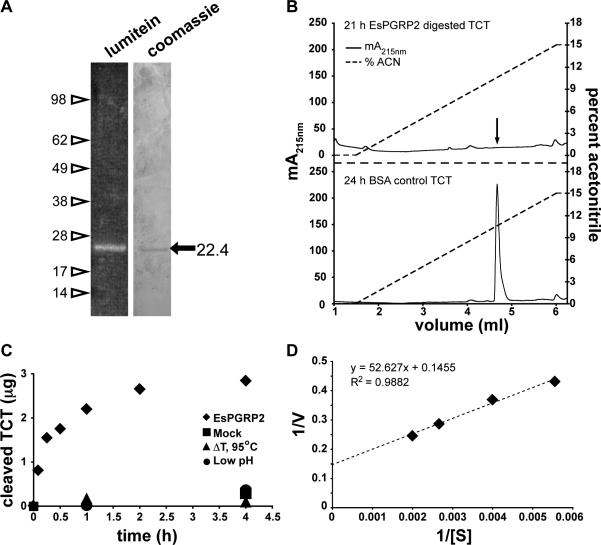
Figure 5. EsPGRP2 is an N-acetyl-muramoyl-L-alanine amidase.
(A) A single protein species at approximately the expected molecular mass (22.7 kDa) was detected in the preparation of EsPGRP2 from transfected S2* cells. One hundred and twenty ng of purified recombinant EsPGRP2 protein was visualized on a polyacrylamide gel with either the fluorescent protein-stain lumitein, or Coomassie. (B) EsPGRP2 NAMLAA activity on TCT, confirmed by LC-MS (data not shown), was visualized by resolution on reverse-phase HPLC. Arrow in top panel indicates location of TCT peak in bottom panel. (C) The kinetics of recombinant EsPGRP2 activity on TCT showed a typical progress curve for enzymatic reactions. The three alternate conditions, including a mock reaction (buffer only, no enzyme), heat-killed enzyme or the reaction carried out at pH 5, had negligible effect on the time-dependent loss of TCT. (D) The Km and Vmax of the recombinant EsPGRP2 amidase activity were determined using a Lineweaver-Burk plot. The units of [S] are μM TCT and V are μg TCT h−1.
To determine the kinetics of EsPGRP2 catalytic cleavage of TCT, we followed the EsPGRP2/TCT reaction over time. One ng of EsPGRP2 cleaved 39 ng of TCT within 1 h (Fig. 5C), degraded 51 ng within 4 h and showed a reduction in velocity over time, typical of many enzymatic reactions (Dixon and Webb, 1964). The amidase activity of EsPGRP2 against TCT was attenuated by heat treatment of the enzyme or by carrying out the reaction at pH 5. Using a standard Michaelis-Menten kinetic analysis including a Lineweaver-Burk plot, we found that EsPGRP2 has an apparent Km of approximately 360 μM and a Vmax of ~2200 moles TCT/ mole EsPGRP2 / h at 25°C (Fig. 5D). Together, these data demonstrate that EsPGRP2 is capable of degrading PGN through the NAMLAA activity conserved in many members of the PGRP protein family.
Next we sought to determine whether the in vitro biochemical activity of EsPGRP2 could be observed in vivo. We examined this question in two independent experiments: by analysis of either whole juvenile animals or adult light-organ central cores – the portion of the light organ that contains the epithelium closely associated with the symbionts. In experiments with juvenile squid, we hypothesized that EsPGRP2 secreted from epithelial surfaces exposed to the seawater milieu (Figs. 2 and S2) would be capable of breaking down TCT in the animal's environment. We exposed juvenile E. scolopes to TCT in filter-sterilized Instant Ocean (FSIO) medium and followed TCT levels in the FSIO over 4 d. Juvenile E. scolopes removed 30 μM, or ~1.5 μg/ml, of the TCT in about 72 h and over 50% of that loss of TCT occurs within the first 24 h (Fig. 6A). It is possible that some of this in vivo activity is due to EsPGRP3, because this member of the PGRP family is predicted to be secreted and to have NAMLAA activity. In mammals, PGRP-L is the only protein known to have NAMLAA activity (Gelius et al., 2003; Wang et al., 2003), so it is plausible that all the activity observed in live E. scolopes could be attributed to the combination of these two EsPGRPs. In the central cores of adult light organs, the EsPGRP2 protein was detected in the aqueous soluble fraction by western blot. In these central-core extracts, the rates of amidase activity were measured at up to 6 μg TCT / h / central core at room temperature (data not shown). In the adult central core experiments, some of the NAMLAA activity may be attributable to not only EsPGRP3, but also any amidases that may be released by the symbionts into the deep crypt lumina. These data suggested that the animal actively degrades PGN and possibly attenuates the activity of TCT. This hypothesis is consistent with the finding that PGRP-amidase cleavage of PGN reduces the immunogenicity of PGN in Drosophila (Mellroth et al., 2003).
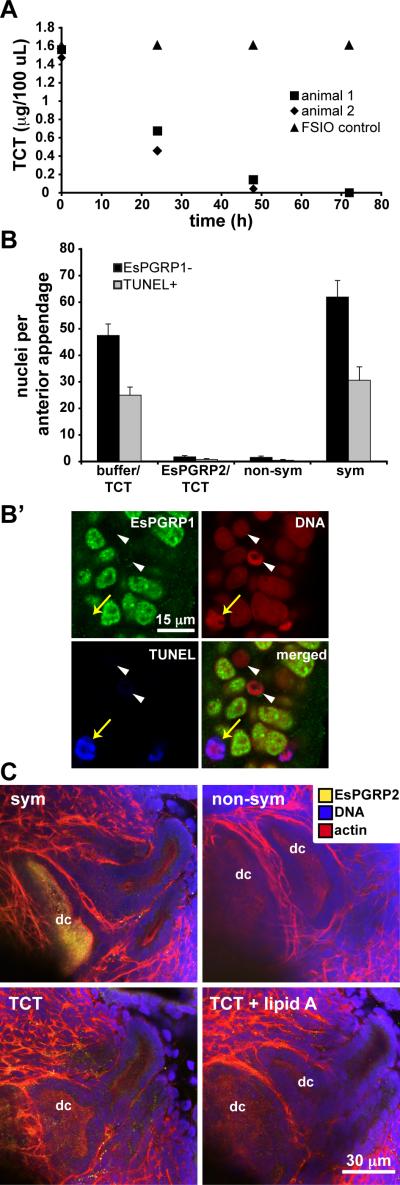
Figure 6. EsPGRP2 attenuates the morphogenic activity of TCT in vivo, but TCT does not induce the secretion of EsPGRP2 into the deep crypts.
(A) The presence of E. scolopes led to the disappearance of TCT from the seawater (representative experiment shown; n = 3, 2 animals per experiment). (B, B’) EsPGRP2-cleaved TCT failed to induce late-stage apoptosis and morphogenesis. (B) The morphogenic activity of EsPGRP2-cleaved TCT was examined by making direct-count estimates of EsPGRP1-negative and TUNEL-positive nuclei in the light organ anterior appendages. Hatchlings were treated with EsPGRP2-degraded TCT (EsPGRP2/TCT) or TCT incubated in a mock reaction (buffer/TCT) for 24 h and compared to 24-h symbiotic (sym) or non-symbiotic (non-sym) juveniles. n = 16, mean ± SEM. (B’) The nuclear protein EsPGRP1 is lost from light-organ epithelial nuclei (white arrowheads) upon induction of light organ morphogenesis. EsPGRP1-negative nuclei also become TUNEL-positive (yellow arrows) (Troll et al., 2009). (C) Pharmacological treatment of hatchling E. scolopes with either 10 μM TCT or 10 μM TCT + 10 ng/ml lipid A does not induce levels of secretion of EsPGRP2 into the deep crypts (dc) at 24 h comparable to that observed in symbiotic animals. non-sym, non-symbiotic; sym, symbiotic.
Degradation by EsPGRP2 abolishes the morphogenic activities of TCT
Previous studies of the symbiosis have shown that TCT must be present and active at the 12 h time point to induce morphogenesis (Doino and McFall-Ngai, 1995; Koropatnick et al., 2004). This observation, coupled with the finding that EsPGRP2 was not detected in the deep crypts at 12 h, led us to question whether EsPGRP2-treatment of TCT in vitro would abrogate its activity in vivo. We exposed juvenile E. scolopes to intact or EsPGRP2-cleaved TCT and assayed for known host responses: 1) TCT induced loss of EsPGRP1 from light-organ nuclei and DNA degradation associated with late-stage apoptosis (Troll et al., 2009) and, 2) in synergy with LPS, TCT induced chromatin condensation in the light organ epithelia (Koropatnick et al., 2004). TCT incubated in reaction buffer induced nuclear loss of EsPGRP1 and late-stage apoptosis at levels comparable to symbiosis, whereas EsPGRP2-digested TCT did not (Fig. 6B and B’). Further, EsPGRP2-degraded TCT failed to induce chromatin condensation in the light-organ epithelia, even in the presence of LPS (data not shown). The attenuation of TCT activity by EsPGRP2 suggested that secretion of EsPGRP2 into the deep crypts might be a direct response to continued release of TCT by V. fischeri after the delivery of the irreversible developmental signal at 12 h.
To determine whether secretion of EsPGRP2 into the deep crypts is triggered by TCT, we exposed juvenile E. scolopes to either 10 μM TCT or 10 μM TCT + 10 ng/ml lipid A at 21 h. Pharmacological treatment with TCT and/or lipid A at this time failed to induce secretion of EsPGRP2 into the deep crypts by 24 h (Figure 6C). In addition, treatment with TCT and/or lipid A for a full 24 h did not result in the localization of EsPGRP2 in the deep crypts (data not shown). Finally, colonization of the deep crypts by the V. fischeri strain DMA388 (Adin et al., 2008), a triple lytic-transglycosylase mutant defective in the release of TCT, induced secretion of EsPGRP2 into the deep crypts (data not shown). Taken together with the finding that the secretion of EsPGRP2 is reversible after 12 h (Fig. 3C), these data demonstrate that, in addition to TCT and lipid A, other factors are required for the induction of secretion of EsPGRP2 into the deep crypts.
DISCUSSION
In this study we characterized the spatial, temporal and biochemical activities of EsPGRP2 in the establishment of the persistent association between E. scolopes and V. fischeri. We provide evidence that EsPGRP2: i) localizes to environmentally-exposed epithelia, including those of the light organ, and is associated with mucus secreted from these tissues; ii) is secreted by the deep-crypt epithelia of the colonized light organ only after the symbiont has induced morphogenesis, a developmental process that is triggered by a PGN monomer released by the symbiont; iii) has a conserved PGRP amidase activity for the degradation of this PGN monomer. These data suggest that EsPGRP2 has a general role of defense and/or modulation of PGN-recognition in E. scolopes tissues exposed to the seawater environment, as well as a more specific role in modulating partner interactions in tissues colonized by symbionts.
The apparent roles for EsPGRP2 are striking in contrast to those of EsPGRP1, another member of this protein family that we characterized in a previous study (Troll et al., 2009). These two proteins share 74% identity and 84% similarity at the amino acid level (Goodson et al., 2005) and both are capable of degrading PGN via NAMLAA activity. However, EsPGRP1 is an intranuclear response element to the 12-h irreversible morphogenetic signal, intimately involved in the apoptosis program during the morphogenesis of the light organ (Troll et al., 2009). Thus, small differences in the primary structure of these proteins results in biologically divergent functions: one plays a role in responding to symbiont-derived PGN-signals while the other directly attenuates the activity of these very same compounds. These studies are beginning to reveal some of the mechanisms through which EsPGRPs mediate its mutualistic association with V. fischeri.
Implications of EsPGRP2 localization in squid tissues
EsPGRP2 is similar to certain members of the PGRP family in both vertebrates and invertebrates. All known vertebrate PGRPs are secreted and involved in defense against microbial infection. In invertebrates, studies of PGRP-SC1 (an amidase) and –LE (no amidase activity) in Drosophila suggest that they are secreted into the hemolymph and gut lumen, respectively, where they also serve in defense against pathogens (Aggrawal and Silverman, 2007; Charroux et al., 2009). One principal function of some PGRPs at these sites is amidase activity (Mellroth et al., 2003; Bischoff et al., 2006; Mellroth and Steiner, 2006; Zaidman-Remy et al., 2006; Troll et al., 2009), which abrogates the toxicity of the PGN monomer (Luker et al., 1995). In terrestrial animals, such as flies and mammals, PGRPs are principally associated with internal regions of the body, whereas the mucosal surfaces of many aquatic invertebrates are superficial and bathed in microbe-rich water. Thus, it would not be surprising to find that PGRPs are produced along all surface epithelia of animals in aquatic habitats, as they are in E. scolopes, and that these surfaces would thus have a higher threshold to become responsive to environmental PGN.
The lack of secretion of EsPGRP2 in deep crypts during the first few hours of colonization (Fig. 3) suggests that the host initially provides an environment in which symbiont-derived PGN molecules persist and are active in microbe to host signaling. Subsequent secretion of EsPGRP2 from the deep crypt epithelia following the 12-h irreversible morphogenetic signal suggests some sort of maturation of host tissues or symbiont cells, triggering secretion, i.e. host to microbe signaling. However, the secretion is reversible, which uncouples it from the morphogenetic signal. The timing of EsPGRP2 secretion was concomitant with the onset of mucus secretion into the crypts (Nyholm et al., 2002), suggesting that the secretion of these two products between 12 and 24 h is coupled. The host-symbiont interplay that results in the maturation of the deep-crypt environment and secretion of mucus and EsPGRP2 into the crypt spaces remains unknown. Although cell-surface molecules, such as LPS and TCT, were obvious candidates, they were not sufficient to induce these phenotypes.
The analyses of adult light organs provided evidence that the presence of EsPGRP2 in the deep crypts is constitutive throughout the animal's life. However, diel changes in symbiont population density and metabolism (Boettcher et al., 1996; Graf and Ruby, 1998; Nyholm and McFall-Ngai, 1998) could modulate activity of this protein. Metabolic profiles of symbiotic V. fischeri over the day/night cycle indicate a shift from anaerobic respiration in the morning to anaerobic fermentation in the late afternoon through morning (McFall-Ngai, unpublished data). When grown in vitro under conditions for anaerobic fermentation, V. fischeri acidifies the media to around pH 5.5 (McFall-Ngai, unpublished data); and the in vitro activity of EsPGRP2 is attenuated by low pH (Fig. 5C). Thus, whereas EsPGRP2 protein levels may be constant in symbiotic-tissue over the day-night cycle, degradative activity may vary with the chemistry of the crypt environmenta. Repression of EsPGRP2 activity would be expected to result in the accumulation of TCT to concentrations where it could once again act as a signaling molecule or toxin.
The in vitro biochemistry of EsPGRP2
To our knowledge, this is the first report of the determination of kinetic parameters for the NAMLAA activity of a PGRP. However, binding affinities (Kd) have been determined for several members of this protein family, with Kds for Gram-negative PGN derivatives in the 90-330 nM range (Kumar et al., 2005) and 55 nM - 60 μM range for Gram-positive PGN (Cho et al., 2005; Kumar et al., 2005). While the Km and Kd are not interchangeable measures, they both provide insight into the interactions between enzyme and substrate. Because the apparent Km for EsPGRP2 was 360 μM, we would expect that its Kd for Gram-negative PGN would be higher than that of other PGRPs. A lower efficiency of EsPGRP2 amidase activity may represent a tuning of the enzyme to the conditions presented by the crypt environment. Alternatively, TCT may not be the preferred substrate for EsPGRP2; removal of a single amino acid from the stem peptide resulted in a 67-fold increase in the affinity of human PGRP-S (Cho et al., 2005). Although Kms are not available for other PGRPs, a Km of ~ 300 μM has been reported for Staphylococcus epidermis autolysin E NAMLAA activity against a fluorescently labeled analog of a Gram-positive-type pentapeptide PGN monomer (Lützner et al., 2009). This value is similar to that of EsPGRP2, suggesting that the kinetics parameters of EsPGRP2 for TCT may be typical for NAMLAA cleavage of PGN-monomers.
Although it is technically unfeasible to measure TCT concentrations in the crypts, theoretical calculations suggest that the levels of TCT are at concentrations consistent with the kinetic parameters of EsPGRP2 amidase activity. V. fischeri releases as much as 220 nM TCT in a turbid culture with an A600nm of 0.9 (Koropatnick et al., 2004); these values correspond to ~1.2 × 10−18 moles TCT released per cell. A 24-h symbiotic light organ will typically contain 5 × 105 cells of V. fischeri in a volume of about 750 pL (McFall-Ngai and Ruby, 1991; Montgomery and McFall-Ngai, 1993; Ruby and Asato, 1993). Thus, if the levels of TCT released by V. fischeri in vivo are similar to those in liquid culture, the concentration of TCT in the deep crypts would be ~800 μM, well above the apparent Km of EsPGRP2. Based on these estimates, it is likely that the rate of TCT cleavage by EsPGRP2 approaches Vmax in the deep crypts, suggesting that the squid tolerates high rates of TCT release by V. fischeri.
EsPGRP2 in the squid/vibrio symbiosis
In summary, the data provide support for a model in which a single PGRP, EsPGRP2, plays two distinct roles in the squid E. scolopes depending on temporal and spatial regulation of the protein in the tissues. EsPGRP2 plays both a broad role in host immunity as a constitutively produced protein of the squid's surface-exposed epithelia and a specific role in modulating host-response to a toxic signaling molecule in the deep crypts of the light organ. Several host-derived enzymes that produce signaling molecules are down-regulated in response to symbiotic colonization by V. fischeri, including halide peroxidase and nitric oxide synthase (Small and McFall-Ngai, 1999; Davidson et al., 2004). The temporal regulation of TCT in the deep crypts by EsPGRP2 is the first example of E. scolopes regulating the levels of a V. fischeri-derived signaling molecule. The dynamics of EsPGRP2 provide another example of pathogenic and mutualistic symbioses using the same molecular language, but with alternate outcomes. Our findings with EsPGRP2, coupled with our previous study of EsPGRP1 (Troll et al., 2009), are defining the complex roles that this protein family can have in mediating mutualistic associations.
EXPERIMENTAL PROCEDURES
General Procedures
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Adult E. scolopes were collected from the near-shore waters of Oahu and maintained as previously described (Montgomery and McFall-Ngai, 1993). Within 10 min of hatching, juvenile squid were collected and washed 3 × with filter-sterilized Instant Ocean (FSIO) (Spectrum Brands, Atlanta, GA). Individual hatchling E. scolopes were placed in scintillation vials with 5 mls of FSIO (non-sym) or FSIO + 5 × 103 CFU/ml V. fischeri ES114 (sym). Colonization by V. fischeri was monitored by measuring luminescence in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) (Ruby and Asato, 1993). Animals were kept in a 12 h light/dark cycle throughout each experiment. In curing experiments, chloramphenicol was added directly to the FSIO to a final concentration of 20 μg/ml at either 8 h or 14 h. Clearing of symbionts was confirmed by monitoring luminescence. ES114 carrying the plasmid pVSV208 (Dunn et al., 2006) was used as chloramphenicol resistant strain to control for the effects of the antibiotic on the host.
Antibody Generation and Immunoblots
Rabbit polyclonal antibodies were raised against a synthetic peptide generated from the EsPGRP2 epitope, NISTCSEQMRKI, conjugated to ovalbumin (Harlan Biosciences, Indianapolis, IN). Aqueous soluble proteins were extracted from whole juvenile E. scolopes by homogenizing in PBS (50 mM sodium phosphate, pH 7.4, 150 mM NaCl) with a ground glass mortar and pestle, a process that would release all soluble cell and cell vesicle contents, then centrifuging at 20,000 × g for 20 min to pellet the insoluble material. The membrane fraction was generated by extracting the aqueous insoluble pellet with 1% SDS for 10 min, followed by centrifugation. Both fractions were run on a 12.5% SDS-PAGE gel and blotted onto PVDF using a Semi-Dry Trans-blot apparatus (Bio-Rad, Hercules, CA). Blots were briefly washed in TBS (20 mM Tris, pH 7.4, 150 mM NaCl) and then blocked in TBS + 1% goat serum + 0.5% BSA for 12 h. Blots were exposed to a 1:200 dilution of the anti-EsPGRP2 antibody in block solution for 24 h at room temperature and then rinsed twice with diH20, followed by 3 × 10 min washes with TBS. A goat anti-rabbit HRP-conjugated 2° antibody (Jackson ImmunolResearch labs, West Grove, PA) was diluted 1:3000 in block solution and applied to the blot for 1 h. Blots were rinsed in deionized water (diH2O) and washed in TBS as above and the blots were developed with ECL chemiluminescent substrates and exposed to film (Thermo Fisher Scientific, Waltham, MA).
Recombinant Protein Expression and Purification
The EsPGRP2 coding sequence was PCR amplified from E. scolopes cDNA using the following primers: JT69, CACCATGATGTTCCATATTTTATGCTTGG and JT70, GAATTCTTCGAGTTTAACCAATAATG. The EsPGRP2 product was TOPO cloned into the Gateway cloning system entry vector, pENTR D-TOPO (Invitrogen, Carlsbad, CA). The EsPGRP2 coding sequence was subcloned into the IPTG-inducible expression plasmid, pDEST-17 (Invitrogen, Carlsbad, CA) by in vitro site-specific recombination according to the manufacturer's instructions. The resulting plasmid, pJT18, was transformed into BL21(DE3) pLysS E. coli cells (Promega, Madison, WI) and expression was induced with 1 mM IPTG during early-log phase growth in LB at 37° C and followed by absorbance at 600 nm. CFUs/ml were determined by plating on LB-agar. Viability of cells during expression of EsPGRP2 was assessed using the LIVE/DEAD BacLight kit according to manufacturer's instructions (Invitrogen, Carlsbad, CA) and visualized on an LSM510 confocal microscope (Zeiss, Thornwood, NY).
To express and purify EsPGRP2 from Drosophila S2* cells, the EsPGRP2 coding sequence was subcloned into the pPAC-PL vector with a FLAG-tag by PCR amplification and subsequent restriction digest with the enzymes BamHI and NotI using the following primers: JT75, ACAGTGGATCCATGCACCATCACCATCACCATATGATGTTCCATATTTTATGCTTGGTGGC and JT76, CATGTGCGGCCGCTTATTTATCGTCATCGTCTTTGTAGTCACCAATAATGGTATCTACACC. The resulting plasmid was named pJT22. The EsPGRP2-FLAG (EsPGRP2-F) coding sequence was further subcloned from plasmid pJT22 into the expression vector, pRmHa-3, downstream of a metallothionine promoter, by restriction digestion with EcoRI and SalI of the PCR product of primers: JT131, TAGTTGAATTCATGCACCATCACCATCACCATAGTTTCGGAAATGGCACCTG and JT133, TACTTGAATTCATGCATCACCATCACCATCACGAACAGACTATCATGCAAAACTC. The resulting plasmid, pJT29, was stably transfected into Drosophila S2* cells as previously described (Silverman et al., 2000). EsPGRP1 protein was induced by addition of 500 μM CuSO4 into the media for 8-12 hours. Cells were then harvested, washed with PBS and frozen at −80°C. 100 ml cell cultures were induced with Cu2+ and pelleted by centrifugation. S2* cell pellets were resuspended in 5 ml of lysis buffer (50 mM Tris, pH 7.4, protease inhibitor cocktail III, 1 mM PMSF) and incubated on ice for 10 min. NaCl was added to a final concentration of 150 mM and the lysate was centrifuged at 20,000 × g for 20 min at 4°C. The supernatant was applied to an anti-FLAG affinity column 3 × by gravity flow at 4°C and the column was then washed with 600 column volumes of TBS by gravity flow at 4°C. Bound proteins were eluted with 100 μM FLAG peptide and fractions were assessed for EsPGRP2 concentration by anti-FLAG immunoblotting. Briefly, protein fractions were separated by 12.5% SDS-PAGE, transferred to PVDF as above, washed in TBS and blocked in TBS + 4% non-fat dry milk for 1 h at room temperature. Rabbit polyclonal anti-FLAG Ab was diluted 1:1000 in block solution and applied to the blot for 1 h at room temperature. The blot was washed and developed as above, except using 4% non-fat dry milk as a blocking agent. Fractions containing the highest levels of EsPGRP2 were pooled and dialyzed against two 0.5 L changes of storage buffer (20 mM Tris, pH7.4, 300 mM NaCl, 1mM DTT and 50% glycerol) and stored at −20°C. To determine the purity of the EsPGRP2-F protein, 120 ng of the active fractions were resolved on a 4-12% NuPAGE gradient gel (Invitrogen, Carlsbad, CA) and visualized with either lumitein according to manufacturer's instructions (Biotium, Berkeley, Ca) or Coomassie stain.
Immunocytochemistry
Juvenile E. scolopes were anaesthetized in FSIO + 2% ethanol and fixed in FSIO + 4% paraformaldehyde for 18 h at 4°C. All steps following fixation were carried out at 4°C. Samples were washed 4 × with 5 ml of mPBS (50 mM sodium phosphate pH 7.4, 0.4 M NaCl) for 30 min each, light organs were dissected out and permeabolized in mPBS + 1% Triton-X-100 (mPBST) for 48 h. The light organ samples were then placed in block solution (mPBST, 1% goat serum and 0.5% BSA) for 24 h and then exposed to 1:1000 anti-EsPGRP2 antibody in block solution for 7 days. The light organs were then washed 4 × with mPBST for 1 h, reblocked for 24 h, exposed to 1:25 FITC-conjugated goat anti-rabbit 2° antibody for 18 h. The 2° antibody was washed out with 4 × 30 min washes with mPBST, followed by 2 × 15 min washes with mPBS. To counterstain the actin cytoskeleton, 6.6 μM rhodamine-phalloidin was applied to the light organs for 18 h. RNA was degraded by equilibrating samples in 2 changes of 2 × SSC (0.3 M Sodium Chloride, 0.03 M Sodium Citrate, pH 7.0) for 10 min each, add RNaseA to a final concentration of 100 μM for 30 min at 37°C. The DNA was then stained with 1 μM TOTO-3, a nucleic acid specific dye, for 30 min at room temperature.
For immunocytochemistry experiments involving mucus staining, juvenile E. scolopes were fixed in Prefer (Anatech, Battle Creek, MI) for 12 h and then were permeabolized for 2 h. Squid mucus was counterstained with TRITC-conjugated wheat germ agglutinin (TRITC-WGA).
TCT degradation assays
For studies of the ability of EsPGRP2 to degrade TCT, the protein needed to be transferred from the storage buffer into the reaction buffer. Buffer exchange was performed by transferring 300 ng of purified EsPGRP2 into 500 μL reaction buffer (50 mM ammonium acetate, pH7, 4 μM ZnSO4, 550 mM NaCl, and 1% BSA) and then concentrating to 20 ng/μL by centrifugation through a 30-kDa cutoff microcon column (Millipore, Billerica, MA). Seventy ng of EsPGRP2 was diluted up to 7 μL with additional reaction buffer and added to 7 μL of TCT, then incubated for varying times at 25°C. To stop the reactions and store until they could be analyzed, reactions were diluted with 86 μL HPLC-grade H2O and snap-frozen in an ethanol/dry ice mixture. To determine TCT levels in each reaction, samples were thawed and centrifuged at 15,000 × g for 1 min, then loaded onto a reverse-phase Betasil C18 HPLC column (Thermo Hypersil-Keystone, Bellefonte, PA). TCT was eluted from the column using a 0-15% acetonitrile gradient and detected by monitoring absorbance at 215 nm. To compensate for variations in sample loading, the area of the TCT peak for each reaction was normalized to the volume loaded onto the HPLC.
To confirm amidase activity, EsPGRP2/TCT and mock/TCT reactions were fractionated through a 3-kDa cutoff microcon column (Millipore, Billerica, MA). The low molecular weight fractions were run on a Zorbax SB-C18 2.1 × 50 mm column (Agilent, Santa Clara, CA), 1.8um particle size, on an Agilent 1200 HPLC with a linear gradient of 98% Water: 2% acetonitrile 0.1% Formic acid to 90% Acetonitrile 10% water 0.1% Formic acid over 25 minutes at 0.25 ml/min. HPLC peaks were directly run on an Agilent LC/MSD TOF, using electrospray ionization in positive ion mode. Two lock masses were used to internally calibrate mass readings, purine (mass 121.050873) and HP-0921 (mass 922.009798).
To determine kinetic parameters of EsPGRP2 degradation of TCT, 100 μL reactions were set up with [TCT] of 500 μM, 375 μM, 250 μM and 180 μM. Every minute from 2-8 minutes, 10 μL were removed, added to 90 μL ddH2O and flash-frozen on dry-ice ethanol. The mass of TCT in each sample was determined, as above, and linear regressions were fit to the data to determine initial reaction rates. Data were then combined in a double reciprocal plot (Lineweaver-Burk) of reaction rate, V, vs. substrate concentration [S].
Whole animal TCT degradation
Hatchling E. scolopes were placed in 0.6 ml FSIO containing 15 μg TCT + 20 μg/ml chloramphenicol. One hundred μL of the FSIO media was sampled at 0, 24, 48 and 72 hours, flash frozen and stored at −20°C. Samples were thawed, centrifuged for 3 min at 13,000 RPMs and loaded onto the HPLC.
Morphogenic activity of degraded TCT
TCT was incubated with EsPGRP2, as above or with reaction buffer alone. The reaction products were separated from EsPGRP2 by spinning the reaction through a 3 kDa-cutoff microcon column. The filtrate was applied to hatchling E. scolopes at concentrations equivalent to 1 μM TCT for 24 h. Animals were fixed and assayed for nuclear loss of EsPGRP1, and late-stage apoptosis (TUNEL) as previously described (Troll et al., 2009).
Supplementary Material
Figure S1. EsPGRP2 was present in squid muscle tissues. Confocal immunocytochemistry of various tissues from E. scolopes juveniles revealed that unlike other types of connective tissue, those containing actin filaments (rhodamine-phalloidin, red) also contained EsPGRP2 protein (yellow) in cytoplasmic striations. The actin filaments and the EsPGRP2 striations did not colocalize, suggesting that the two do not directly associate. Epithelial tissues in close proximity to the striated connective tissues, such as the gut epithelia, maintained a more typical apical cytoplasmic localization. (A) A schematic of the juvenile squid. (B) Head muscle tissue, (C) outer epithelia of the gut and (C’) muscle tissue of the gut. mt, muscle tissue. Nuclei with TOTO-3 (blue).
Figure S2. EsPGRP2 was secreted from mucosal surfaces, including the light organ. (A) Merged immunocytochemistry image of juvenile light organ stained with WGA-Oregon green (B, blue) or with the anti-EsPGRP2 antibody (C, yellow). Dashed box represents area shown in higher magnification in (D). EsPGRP2 was associated with shed mucus (arrow heads) and was also clearly visible in vesicle-like structures present in epithelial tissues (arrows).
ACKNOWLEDGEMENTS
We are grateful to Michael Apicella for kindly providing the V. fischeri LPS and lipid A used in this study. We thank C. Brennan, J. Dillard, H. Goodrich-Blair, E. Heath-Heckman, L. Knoll, M. Mandel, B. Rader and E. Ruby for helpful comments on the manuscript. This work was funded by NIH RO1-AI50661 to M.M.N., NSF IOS 0817232 to M.M.N. and E.G. Ruby, NIH RR R01-12294 to E.G. Ruby, and by NIH NRSA AI55397 to J.V.T. through the Microbial Pathogenesis and Host Responses Training Program. Additional support for E.H.B. was provided by NSF Research Experience for Undergraduates award No. 0552809.
REFERENCES
- Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2008;191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggrawal K, Silverman N. Peptidoglycan recognition in Drosophila. Biochem Soc Trans. 2007;35:1496–1500. doi: 10.1042/BST0351496. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG, McFall-Ngai MJ. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. Journal of Comparative Physiology A Sensory Neural and Behavioral Physiology. 1996;179:65–73. [Google Scholar]
- Charroux B, Rival T, Narbonne-Reveau K, Royet J. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 2009 doi: 10.1016/j.micinf.2009.03.004. in press. [DOI] [PubMed] [Google Scholar]
- Cho JH, Fraser IP, Fukase K, Kusumoto S, Fujimoto Y, Stahl GL, Ezekowitz RA. Human peptidoglycan recognition protein S is an effector of neutrophil-mediated innate immunity. Blood. 2005;106:2551–2558. doi: 10.1182/blood-2005-02-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- Coteur G, Mellroth P, De Lefortery C, Gillan D, Dubois P, Communi D, Steiner H. Peptidoglycan recognition proteins with amidase activity in early deuterostomes (Echinodermata). Dev Comp Immunol. 2007;31:790–804. doi: 10.1016/j.dci.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Dixon M, Webb EC. Enzymes. Academic Press; New York: 1964. [Google Scholar]
- Doino JA, McFall-Ngai MJ. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull. 1995;189:347–355. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R. Peptidoglycan recognition proteins (PGRPs). Mol Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell Microbiol. 2006;8:1059–1069. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- Foster JS, McFall-Ngai MJ. Induction of apoptosis by cooperative bacteria in the morphogenesis of host epithelial tissues. Dev Genes Evol. 1998;208:295–303. doi: 10.1007/s004270050185. [DOI] [PubMed] [Google Scholar]
- Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem Biophys Res Commun. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- Goodson MS, Kojadinovic M, Troll JV, Scheetz TE, Casavant TL, Soares MB, McFall-Ngai MJ. Identifying components of the NF-κB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl Environ Microbiol. 2005;71:6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci U S A. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JE. Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. Journal of Applied Microbiology. 2007;103:1355–1365. doi: 10.1111/j.1365-2672.2007.03366.x. [DOI] [PubMed] [Google Scholar]
- Kaback HR, William BJ. Methods in Enzymology. Academic Press; 1971. Bacterial membranes. pp. 99–120. [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- Kelly D, Coutts AG. Early nutrition and the development of immune function in the neonate. Proc Nutr Soc. 2000;59:177–185. doi: 10.1017/s0029665100000197. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Kumar S, Roychowdhury A, Ember B, Wang Q, Guan R, Mariuzza RA, Boons GJ. Selective recognition of synthetic lysine and meso-diaminopimelic acid-type peptidoglycan fragments by human peptidoglycan recognition proteins Iα and S. J Biol Chem. 2005;280:37005–37012. doi: 10.1074/jbc.M506385200. [DOI] [PubMed] [Google Scholar]
- Lamarcq LH, McFall-Ngai MJ. Induction of a gradual, reversible morphogenesis of its host's epithelial brush border by Vibrio fischeri. Infect Immun. 1998;66:777–785. doi: 10.1128/iai.66.2.777-785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Tyler AN, Marshall GR, Goldman WE. Tracheal cytotoxin structural requirements for respiratory epithelial damage in pertussis. Mol Microbiol. 1995;16:733–743. doi: 10.1111/j.1365-2958.1995.tb02434.x. [DOI] [PubMed] [Google Scholar]
- Luker KE, Collier JL, Kolodziej EW, Marshall GR, Goldman WE. Bordetella pertussis tracheal cytotoxin and other muramyl peptides: distinct structure-activity relationships for respiratory epithelial cytopathology. Proc Natl Acad Sci U S A. 1993;90:2365–2369. doi: 10.1073/pnas.90.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lützner N, Pätzold B, Zoll S, Stehle T, Kalbacher H. Development of a novel fluorescent substrate for Autolysin E, a bacterial type II amidase. Biochemical and Biophysical Research Communications. 2009;380:554–558. doi: 10.1016/j.bbrc.2009.01.140. [DOI] [PubMed] [Google Scholar]
- Markert S, Arndt C, Felbeck H, Becher D, Sievert SM, Hugler M, et al. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science. 2007;315:247–250. doi: 10.1126/science.1132913. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem Biophys Res Commun. 2006;350:994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. 2003;278:7059–7064. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- Melly MA, McGee ZA, Rosenthal RS. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeae to damage human fallopian-tube mucosa. J Infect Dis. 1984;149:378–386. doi: 10.1093/infdis/149.3.378. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai M. Embryonic-development of the light organ of the sepiolid squid Euprymna-scolopes berry. Biological Bulletin. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai M. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development. 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;68:5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small AL, McFall-Ngai MJ. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem. 1999;72:445–457. [PubMed] [Google Scholar]
- Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev. 2004;198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x. [DOI] [PubMed] [Google Scholar]
- Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, et al. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cellular Microbiology. 2009;11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZM, Li X, Cocklin RR, Wang M, Fukase K, Inamura S, et al. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J Biol Chem. 2003;278:49044–49052. doi: 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. EsPGRP2 was present in squid muscle tissues. Confocal immunocytochemistry of various tissues from E. scolopes juveniles revealed that unlike other types of connective tissue, those containing actin filaments (rhodamine-phalloidin, red) also contained EsPGRP2 protein (yellow) in cytoplasmic striations. The actin filaments and the EsPGRP2 striations did not colocalize, suggesting that the two do not directly associate. Epithelial tissues in close proximity to the striated connective tissues, such as the gut epithelia, maintained a more typical apical cytoplasmic localization. (A) A schematic of the juvenile squid. (B) Head muscle tissue, (C) outer epithelia of the gut and (C’) muscle tissue of the gut. mt, muscle tissue. Nuclei with TOTO-3 (blue).
Figure S2. EsPGRP2 was secreted from mucosal surfaces, including the light organ. (A) Merged immunocytochemistry image of juvenile light organ stained with WGA-Oregon green (B, blue) or with the anti-EsPGRP2 antibody (C, yellow). Dashed box represents area shown in higher magnification in (D). EsPGRP2 was associated with shed mucus (arrow heads) and was also clearly visible in vesicle-like structures present in epithelial tissues (arrows).