Abstract
Most current genetic association studies, including genome-wide association studies, look for the single nucleotide polymorphisms (SNPs) with a relatively large minor allele frequency (MAF) (e.g. >5%) in the search for genetic loci underlying the susceptibility for complex diseases. The strategy of focusing on common SNPs in genetic association studies is very effective under the common-disease-common-variant (CDCV) hypothesis, which claims that common diseases are caused by common variants that have relatively small to moderate effects. Although the CDCV hypothesis has become the dogma guiding the conduct of association studies over the past decade, growing evidence from recent empirical data and simulations suggests that the causal genetic polymorphisms, including SNPs and copy number variants (CNVs), for common diseases have a wide spectrum of MAFs, ranging from rare to common. Unlike the analysis for common genetic variants, statistical approaches for the analysis of rare variants receive very little attention. Methods developed for common variants usually rely on their asymptotic properties, which can be inaccurate for the study of the rare variants with limited sample size. Although Fisher's exact test can be used for such a scenario, it is usually conservative and thus its usefulness is diminished to some extent. Here we propose two novel approaches for the analysis of rare genetic variants. Simulation studies and two real examples demonstrate the advantages of the proposed methods over the existing methods.
Key Words: Association test, CDRV, Rare polymorphisms
Introduction
Most current genetic association studies, including genome-wide association studies (GWAS), look for the single-nucleotide polymorphisms (SNPs) with relatively high minor allele frequencies (MAFs) (say, MAF >5%) [1,2,3,4] in the search for genetic loci underlying susceptibility to complex diseases. The strategy of focusing on common SNPs in genetic association studies is very effective under the common-disease-common-variant (CDCV) scenario, that is, when common diseases are caused by common variants with relatively small to moderate effects. GWAS based on a quarter of a million to one million common SNPs have been very successful in identifying disease-susceptibility regions through indirect linkage disequilibrium (LD) mapping [5]. Under the CDCV paradigm, the set of common SNPs (tagSNPs) provided by the existing high-throughout genotyping platforms can cover the genome well enough so that they can capture the relationship between the ‘common’ causal variants at unmeasured loci and the disease through their high LD with the functional loci.
Although the CDCV has been the dominant dogma guiding the conduct of association studies for the past decade, growing evidence from recent empirical and simulation studies [6,7,8,9,10,11,12,13,14,15,16] suggests that the causal variants for common diseases have a wide spectrum of MAFs, ranging from rare to common. For example, Gorlov et al. [14] found that functional SNPs tended to have low MAFs. A recent study by Need et al. [16] suggested that common genetic variants do not appear to have a major impact on predisposition to schizophrenia and that rare copy number variants (CNVs) may be more important in susceptibility to schizophrenia than common polymorphisms. Thus, in addition to the CDCV scenario, the common-disease-rare-variant (CDRV) hypothesis, which asserts that there are multiple rare variants underlying the susceptibility to a common disease, is a very plausible scenario for many complex diseases. Furthermore, some researchers believe that both CDCV and CDRV hypotheses could be true even within the same susceptibility gene for a complex disease [5,14].
Under the CDRV scenario, the population-based association studies that adopt the strategy of using common tagSNPs would be underpowered, as those common SNPs tend to have a low correlation with the unmeasured disease-causing (rare) variants, and thus are not very informative when used in indirect LD mapping [5]. Given the fact that the majority of SNPs in the human genome are rare [14] (MAF <5%) and that the CDRV scenario appears to be the norm instead of a rarity for complex disease, it would be beneficial to study rare SNPs in large-scale population-based association studies to enhance the chance of disease-gene detection.
There are a vast number of analytic approaches for studying the association between the disease and a genetic variant or set of variants. Most of them are designed for the analysis of common variants, relying on asymptotic distributions for their statistical significance evaluation. Their accuracy on rare variants could be suspected. Li and Leal [5] have recently proposed a method targeting the analysis of multiple rare variants within a candidate region. Their approach, called the collapsing method, tries to enrich the association signals and to reduce the degrees of freedom by collapsing genotypes at multiple rare SNPs into a univariate test.
In anticipating the agnostic screening for rare SNPs or CNVs in future studies, we focus on the single-marker analysis of rare variants. Fisher's exact test is the standard approach when the sample size is limited, although it is well known that it is conservative [17,18] and thus has its power diminished to some extent. The aim of this work is to develop more powerful single-marker tests for the analysis of rare variants (SNPs or CNVs) with MAFs below 5%.
Materials and Methods
Notation
We assume that there are cases and controls in a case-control genetic association study and that the genetic polymorphism under study is bi-allelic (e.g. SNPs). We denote two alleles at a bi-allelic marker as A and a. We represent the three genotypes as aa, Aa, and AA. We further assume that the high-risk allele A is a rare allele. When the sample size is limited, we expect the genotype count for AA is close to zero in both cases and controls. For the convenience of our analysis, we combine the genotypes Aa and AA into one type and denote the counts for the collapsed genotypes by (x, n1 – x) for cases where x is the number of cases having genotype Aa or AA, n1 – x is the number of cases having genotype aa. Similarly, we denote the genotype count for controls by (y, n2 – y).
Fisher's Exact Test
Fisher's exact test is the standard approach when the sample size is limited. For the sake of completeness, we describe it here briefly. Let θ1 and θ2 be the probabilities of the event that (Aa or AA) occurs in cases and controls, respectively. Denote the observed count of (Aa or AA) in cases by X and the count in controls by Y.
Under the null hypothesis, i.e. θ1 = θ2 = θ, b = x + y is the sufficient statistic for θ. The conditional probability of observing X = i and Y = j given b, the total sum of the events, follows a hyper-geometric distribution,
where the symbol ! denotes the factorial operator. The p value is then calculated by summing the probabilities of the more extreme (in term of the probability) events with X + Y = b than the observed one. There is a user friendly function in R (function fisher.test) to calculate Fisher's exact test.
AC-Test
Audic and Claverie [19] considered an alternative approach, assuming the unknown parameters θ1 and θ2 to be random, instead of fixed. In particular, they assumed θ1 = θ2 = θ under the null hypothesis, where θ followed a given random distribution. Their test built upon , the conditional probability under the null. They proposed to approximate the binomial distributions Prn1 (X = x∣θ) and Prn2 (Y = y∣θ) through the corresponding Poisson distributions. They obtained the following formula for calculating ,
If the is treated as the observed value of the test statistic, the p value (two-sided) could be written to be
where I(.) is the indictor function. This formula is different from equations 9a and 9b in Audic and Claverie's paper [19], which were used for calculating the confidence interval.
The Proposed Tests
In the AC-test described above, two binomial distributions, Prn1 (X = x∣θ) and Prn2 (Y = y∣θ), are approximated by Poisson distributions. This approximation might not be appropriate, especially when the observed X or Y are not too rare. Instead of relying on the Poisson approximation, we can actually calculate Prn1,n2 (X = x∣Y = y) exactly by assuming a suitable prior or posterior distribution for θ. Here we propose two alternative tests under such motivation.
Uniform-Test
For x ∈ {0, 1, …, n1}, and y ∈ {0, 1, …, n2}, we notice that
| (1) |
where f(θ) is the prior density function of θ under the null and f(θ∣Y = y) is the posterior density function of θ after observing the value of Y.
Since we have no knowledge on θ, the least constrained hypothesis on θ is that the prior of θ is uniform over (0, 1). Then f(θ∣Y = y) becomes the specified Beta distribution density
0 < θ < 1, with ξ = y + 1 and η = n2 – y + 1, and where
is the beta function. Therefore, we have, from (1),
Thus we can calculate numerically through the beta function. Similar to the AC-test, we can treat at the observed value as the test statistic and evaluate its significance (the p value). The p value can be exactly calculated as
| (2) |
This test, called the Uniform-test, is unconventional compared to the standard statistical testing procedure, as the probability used in (2) is not the standard one defined under the null hypothesis. A natural question is whether the test with its significance evaluated by (2) can maintain its type I error properly. From the Theorem in the Appendix, we can see that the type I error of the proposed test is well controlled for large n2. Simulation results shown later will demonstrate that the proposed test also has a satisfactory type I error rate for relatively small n2 (a few hundred).
Beta-Test
While deriving the Uniform-test, we showed that f(θ∣Y = y), the posterior distribution of θ given Y = y, is a beta distribution when we assume an uninformative prior for θ. Motivated by this, we could follow an alternative approach and make an assumption directly about f(θ∣Y = y) without specifying the prior distribution for θ. For convenience, we assume θ∣Y = y follows the Beta distribution Beta(θ; u, ν), θ ∈ (0, 1), but with u and ν being calibrated by the observed Y. Since the mean and variance of the Beta distribution Beta(θ; u, ν) are
respectively, and the moment estimate for θ, given that Y = y is
a natural choice for u and ν is to calibrate them according to the following two equations:
This gives u = y(n2 − 1)/n2 and ν = (n2 − 1)(n2 – y)/n2. They are slightly different from the ones derived for the Uniform-test. Once we have specified f(θ∣Y = y), we have
Then after observing X = x and Y = y, we can exactly calculate the two-sided p value as
Thus, similar to the Uniform-test, the above test, called the Beta-test, has its type I error well controlled when n2 is large. Simulation results shown later also demonstrate that the Beta-test has a satisfactory type I error rate for relatively small n2 (a few hundred). We pointed out that both proposed tests are Bayesian tests. For the second one, we used the Empirical Bayes approach where mean and variance are used to derive the parameters.
Simulation Design
Denote the three possible genotypes at the study SNP by G0 = aa, G1 = Aa, and G2 = AA, with ‘A’ being the minor allele and the high-risk allele in a case-control genetic association study. We assume that the Hardy-Weinberg equilibrium (HWE) holds in the study population. Thus, the genotype frequencies are given as Pr(G0) = (1 – f)2, Pr(G1) = 2f(1 – f), and Pr(G2) = f2, where f is the minor allele frequency (MAF). Let the odds ratio (OR) for having 1 copy or 2 copies of the high-risk allele ‘A’ be λ1 and λ2. We further assume that the disease under study is rare in the source population. Then the genotype frequencies in controls are (almost) the same as in the source population, and in cases they are given as
with K = (1 – f)2 + 2λ1f(1 – f) + λ2f2. Based on the above genotype frequencies in cases and controls, we can randomly generate the genotypes for a sample of cases and controls given specified values of (f, λ1, λ2).
We first generate the data under the null hypothesis that there is no association (λ1 = λ2 = 1) between the SNP and disease to assess the type I error rates of various considered tests, including Fisher's exact test, the AC-test, and the proposed two tests. Following Gorlov et al. [14], we consider MAFs with the following values: 0.05, 0.04, 0.03, 0.02, 0.01, 0.009, 0.008, 0.007, 0.006, and 0.005. The sample sizes are set to be n1 = n2 = 250 or 500. For each considered scenario, the type I error rate at the significance level of 0.05 is estimated based on 10,000 replicated datasets.
For the purpose of comparing power, similar to the above configurations, we generate the data under the following two disease models, the multiplicative model with λ1 = and λ2 = 4 and the dominant model with λ1 = λ2 = 2. Again, for each considered scenario, the power is estimated based on 10,000 datasets.
Results
Type I Error Rates
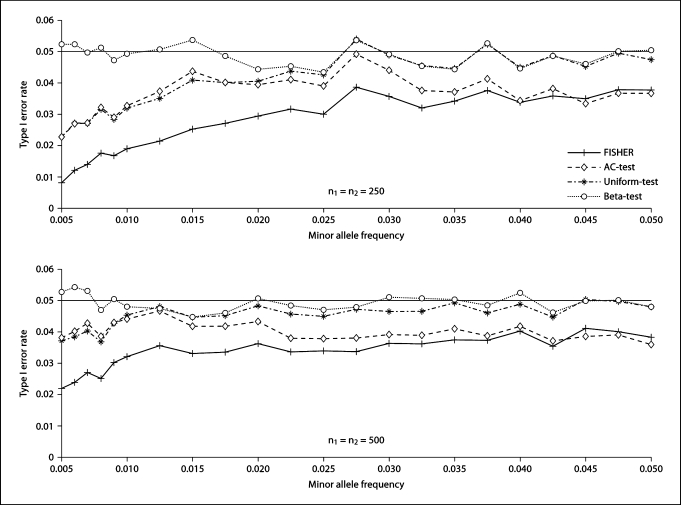
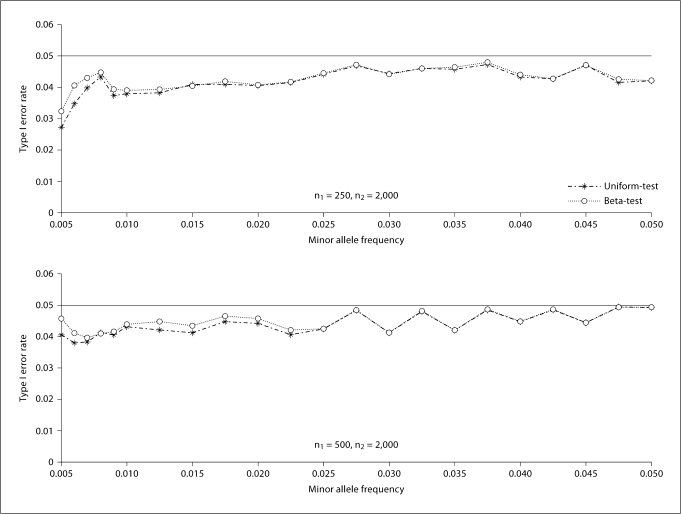
Figure 1 shows the empirical type I error rates of all considered tests. From figure 1, we find that all tests can control their type I error rates, with Fisher's exact test the most conservative, the AC-test the second most conservative. Both the Uniform-test and Beta-test have empirical type I error rates close to the nominal level, with the Uniform-test more conservative than the Beta-test. They are occasionally anti-conservative for the relatively small sample size. For large sample size, we have proved theoretically that both tests are conservative. This can be further verified by figure 2 for n2 being 2,000.
Fig. 1.
Empirical type I error rates of Fisher's exact test (FISHER), the Audic-Claverie test (AC-test), and the proposed two tests (the Uniform-test and the Beta-test). n1 is the number of cases and n2 is the number of controls. The number of replicates is 10,000.
Fig. 2.
Empirical type I error rates of the proposed two tests (the Uniform-test and the Beta-test). n1 is the number of cases and n2 is the number of controls. The number of replicates is 10,000.
Power
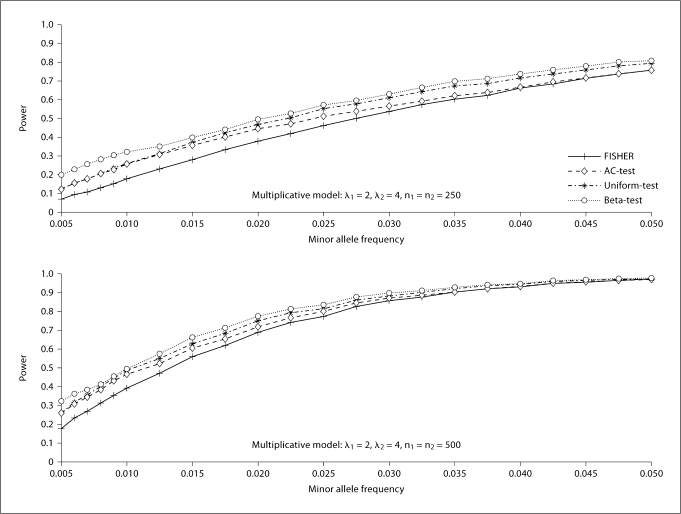
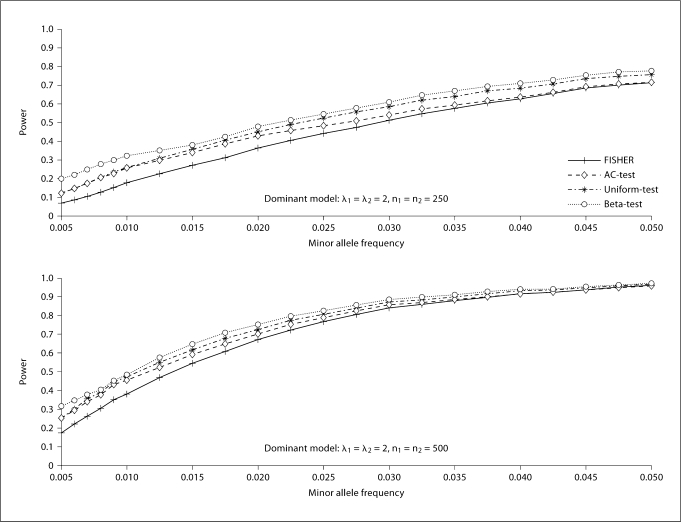
Figures 3 and 4 show the powers under the various considered scenarios at the significance level of 0.05. In general, the proposed two tests, especially the Beta-test, have a noticeable power advantage over Fisher's exact test and the AC-test. It is also seen that the Beta-test is always dominant over Fisher's exact test, the AC-test, and the Uniform-test in terms of power in the simulation situations. This could be due partially to the fact that the evaluation of the significance level for the Beta-test is less conservative than that of the other three tests. It is also seen that this trend does not alter for different choices of sample size (250 vs. 500). We also notice that the power is quite similar between the Uniform-test and the AC-test in situations where the risk allele frequency is small. This is to be expected, since the difference between the two tests stems from the way the conditional probability is calculated. The Uniform-test calculates the probability exactly, while the AC-test is based on the Poisson approximation, which is fairly accurate when the risk allele frequency is small. The AC-test is less powerful than the Uniform-test and Beta-test for relatively large MAF (>0.025). This is not surprising, since the Poisson approximation for the AC-test is accurate only when the MAF is small.
Fig. 3.
Power of Fisher's exact test (FISHER), the Audic-Claverie test (AC-test), and the proposed two tests (the Uniform-test and the Beta-test). n1 is the number of cases and n2 is the number of controls. The number of replicates is 10,000.
Fig. 4.
Power of Fisher's exact test (FISHER), the Audic-Claverie test (AC-test), and the proposed two tests (the Uniform-test and the Beta-test). n1 is the number of cases and n2 is the number of controls. The number of replicates is 10,000.
Real Examples
Example I: The Association between Copy Number Variants and Schizophrenia
Need et al. [16] investigated the association between the copy number variants with length greater than 2 Mb and schizophrenia, using a total of 1,013 cases and 1,084 controls. They observed 14 copy number variants in cases and 3 in controls. Using Fisher's exact test, they reported the p value of 0.006 based on the 2-sided test. The AC-test, Uniform-test, and Beta-test give p values of 0.0043, 0.0041, and 0.0024, respectively. Among the four considered tests, the Beta-test provides the strongest evidence for an association. This is consistent with the simulation results.
Example II: The Association between Microduplications and Schizophrenia
McCarthy et al. [20] investigated the association between microduplications of 16p11.2 and schizophrenia. In the replicated study, they observed 9 subjects with microduplications in 2,645 cases and only 1 in 2,420 controls. Using Fisher's exact test, they reported the p value of 0.022 based on the 2-sided test. The AC-test, Uniform-test and Beta-test give p values of 0.015, 0.015, and 0.0074, respectively. Again, among the four considered tests, the Beta-test provides the strongest evidence for association.
Discussion
Guided by the common-disease-common-variant (CDCV) assumption, common genetic polymorphisms (mainly SNPs) are the main focus of most genetic association studies in the pursuit of genetic loci underlying susceptibility to complex diseases, with rare genetic variants being mostly ignored. GWAS equipped with the state-of-the-art genotyping arrays have enjoyed tremendous success in finding common genetic variants for certain complex diseases. The strategy of focusing on common variants, however, might not be as effective for some other diseases, such as schizophrenia [16]. There is a growing consensus on seeking more rare genetic variants when examining genetic predisposition to disease. In contrast to the large number of statistical approaches available for the analysis of common variants, there are only a few options for the study of rare ones, mainly Fisher's exact test and the AC-test [19]. In this report we proposed two more powerful alternatives and recommend their usage for future studies of rare genetic polymorphisms, such as SNPs and CNVs.
By theoretical arguments and simulation studies, we have shown that both proposed tests can properly control their type I error rates for a wide range of sample sizes and are less conservative than Fisher's exact test and the AC-test. Unlike the AC-test [19], which relies on the Poisson approximation for the binominal probability calculation and thus is appropriate only for the study of rare variants, the two proposed tests do not involve any such approximation in their calculations. As a result, these two tests can be used to analyze not only rare genetic variants but also common ones. Thus, the two tests can be used as exact tests for the analysis of 2 by 2 tables with limited sample sizes, just as Fisher's exact test can. Among the two tests, the Beta-test appears to be slightly more powerful than the Uniform-test.
We chose three different prior distributions of uniform distribution for the Uniform-test to evaluate its power sensitivity. Results (data not shown here) show that there is no big difference for the choice of the prior of uniform distribution when the minor allele frequency is relatively small.
The Uniform-test we proposed above is based on . Due to the symmetric role played by X and Y, a different test can be derived based on , which is conditioned on X. The two tests are not equivalent. Based on the Theorem in the Appendix, we recommend using the test conditioning on the sample with the larger sample size to ensure that the type I error is properly controlled. If the number of cases and controls are equal, we recommend using the control as the condition since the minor allele frequency of control is more close to that of the source population for rare disease. The same argument also applies to the Beta-test.
The two new tests are derived under the setting of 2 by 2-table analysis. They can be extended to the study of genetic polymorphisms with more than 2 categories, where the priori distribution of the unknown parameters is the Dirichlet distribution. Finally, the proposed tests can be used together with the collapsing strategy of Li and Leal [5] to evaluate jointly the association between multiple rare genetic variants and the outcome.
Acknowledgements
We would like to thank the editor and two anonymous reviewers for their insightful comments. We thank B.J. Stone for her valuable suggestions. H. Zhang and K. Yu are supported by the Intramural Program of the National Institutes of Health. Q. Li is partially supported by the National Young Science Foundation of China. No. 10901155. The opinions expressed in the article are not necessarily those of the National Institutes of Health.
Appendix
Theorem
For any given significance level α and any n1,
Before proving the theorem, we provide a lemma.
Lemma 1
Suppose that θ1 = θ under the null hypothesis. Adopting the notation in the text, we have, for any given α (0 < α < 1),
Proof
For i ∈ {0, 1, …, n1], define
Then, for any given α (0 < α < 1),
Let t = max{k|k ≤ n1θ, k = 0, 1, … n1}, Ξα,1 = {k|V(k) ≤ α, k = 0, 1, …, t}, and Ξα,2 = {k|V(k) ≤ α, k = t, t + 1, …, n1}. Define
We now consider two cases:
-
(1)When ,
-
(2)When in the similar way, we have
The lemma is proved by (1), (2), and the definition of kα,1 and kα,2.
Proof of the Theorem
Suppose that θ1 = θ2 = θ under the null hypothesis. Denote the true probabilities of observing X = i and Y = j by πn1 (X = i|θ) and πn2 (Y = j|θ), respectively. Then, for any α and n1, the type I error rate of the Uniform-test is
Let
and
where
In view of lemma 1, it suffices to show that
converges to g(πn1 (X = 1|θ), …, n1 (X = n1|θ), i, α) as goes to infinity.
Let a random variable Z|Y n Beta(z; Y + 1, n2 + Y + 1), z ∈ (0, 1) with the density function f(z|Y), where Y n Binom (n2, θ). Then we have
as n2 goes to infinity, and
Therefore, Z converges to θ in probability. Hence,
converges to πn1 (X = i|θ) as n2 goes to infinity, and consequently converges to .
By now we have proved
similarly, we can prove
References
- 1.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 2.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 4.Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, Goldstein McHenry T, Cooper ME, Govil M, Daack-Hirsch S, Riley B, Jugessur A, Felix T, Moreno L, Mansilla MA, Vieira AR, Doheny K, Pugh E, Valencia-Ramirez C, Arcos-Burgos M. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Hum Hered. 2009;68:151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnett FC, Howard RF, Tan F, Moulds JM, Bias WB, Durban E, Cameron HD, Paxton G, Hodge TJ, Weathers PE, Reveille JD. Increased prevalence of systemic sclerosis in a Native American tribe in Oklahoma. Association with an Amerindian HLA haplotype. Arthritis Rheum. 1996;39:1362–1370. doi: 10.1002/art.1780390814. [DOI] [PubMed] [Google Scholar]
- 7.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 9.Fearnhead NS, Wilding JL, Winney B, Tonks S, Bartlett S, Bicknell DC, Tomlinson IP, Mortensen NJ, Bodmer WF. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci USA. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 11.Iyengar SK, Elston RC. The genetic basis of complex traits: Rare variants or ‘common gene, common disease'? Methods Mol Biol. 2007;376:71–84. doi: 10.1007/978-1-59745-389-9_6. [DOI] [PubMed] [Google Scholar]
- 12.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzopardi D, Dallosso AR, Eliason K, Hendrickson BC, Jones N, Rawstorne E, Colley J, Moskvina V, Frye C, Sampson JR, Wenstrup R, Scholl T, Cheadle JP. Multiple rare nonsynonymous variants in the adenomatous polyposis coli gene predispose to colorectal adenomas. Cancer Res. 2008;68:358–363. doi: 10.1158/0008-5472.CAN-07-5733. [DOI] [PubMed] [Google Scholar]
- 14.Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slatter TL, Jones GT, Williams MJ, van Rij AM, McCormick SP. Novel rare mutations and promoter haplotypes in ABCA1 contribute to low-HDL-C levels. Clin Genet. 2008;73:179–184. doi: 10.1111/j.1399-0004.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- 16.Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciūte D, Gennarelli M, Strittmatter WJ, Bonvicini C, Rossi G, Jayathilake K, Cola PA, McEvoy JP, Keefe RS, Fisher EM, St Jean PL, Giegling I, Hartmann AM, Möller HJ, Ruppert A, Fraser G, Crombie C, Middleton LT, St Clair D, Roses AD, Muglia P, Francks C, Rujescu D, Meltzer HY, Goldstein DB. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altham P. Exact Bayesian analysis of a 2 × 2 contingency table, and Fisher's ‘exact’ significance test. J R Stat Soc Series B Stat Methodol. 1969;31:261–269. [Google Scholar]
- 18.Howard JV. The 2 × 2 Table: A Discussion from a Bayesian Viewpoint. Stat Sci. 1998;13:351–367. [Google Scholar]
- 19.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, Krause V, Kumar RA, Grozeva D, Malhotra D, Walsh T, Zackai EH, Kaplan P, Ganesh J, Krantz ID, Spinner NB, Roccanova P, Bhandari A, Pavon K, Lakshmi B, Leotta A, Kendall J, Lee YH, Vacic V, Gary S, Iakoucheva LM, Crow TJ, Christian SL, Lieberman JA, Stroup TS, Lehtimäki T, Puura K, Haldeman-Englert C, Pearl J, Goodell M, Willour VL, Derosse P, Steele J, Kassem L, Wolff J, Chitkara N, McMahon FJ, Malhotra AK, Potash JB, Schulze TG, Nöthen MM, Cichon S, Rietschel M, Leibenluft E, Kustanovich V, Lajonchere CM, Sutcliffe JS, Skuse D, Gill M, Gallagher L, Mendell NR, Wellcome Trust Case Control Consortium, Craddock N, Owen MJ, O’Donovan MC, Shaikh TH, Susser E, Delisi LE, Sullivan PF, Deutsch CK, Rapoport J, Levy DL, King MC, Sebat J. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]