Abstract
KLF13 and FGFR3 have important cellular functions and each is believed to play a role in cancer. KLF13 is a transcription factor required for the expression of several oncogenes. FGFR3 is a fibroblast growth factor receptor that initiates a signaling cascade leading to the activation of numerous cellular pathways. Here we show that KLF13 and FGFR3 are overexpressed in oral cancer cells. We also show that artificially reducing cellular levels of KLF13 and FGFR3 decreases cell proliferation and increases sensitivity to ionizing radiation. These data suggest that KLF13 and FGFR3 contribute to malignancy in oral cancer cells and may be useful biomarkers for early detection and possible targets for therapy.
Key Words: FGFR3, KLF13, miRNA, Oral cancer
Introduction
Oral squamous cell carcinoma (OSCC) is the 8th most common cancer worldwide, and includes tumors within the oral cavity [Jemal et al., 2009]. Causes of OSCC include tobacco and alcohol use as well as human papillomavirus infection. Although diagnosis and treatment of OSCC have improved, the survival rate has not increased substantially in 40 years. Like other cancers, OSCC is characterized by genomic instability and altered gene expression. Recently, we examined how the altered expression of the microRNAs (miRNA) miR-125b and miR-100 contributes to malignancy in OSCC [Henson et al., 2009]. We found that expression levels of miR-125b and miR-100 are substantially lower in OSCC cell lines and tumors than in normal human oral keratinocyte (NHOK) controls. We also found that increasing cellular levels of miR-125b and miR-100 affected the expression of both predicted target genes and non-target genes. This included KLF13 and FGFR3, both of which had reduced expression levels in response to increased levels of miR-125b and miR-100, respectively. Public databases predict that KLF13 and FGFR3 are targets of miR-125b and miR-100, respectively. Thus, the reduced expression of KLF13 and FGFR3 was presumably due to them being targets of miR-125b and miR-100; however, this has yet to be validated experimentally.
The altered expression of KLF13 and FGFR3 is intriguing since they are involved in a number of important biological processes. KLF13 (Krüppel-like factor 13) is a zinc finger transcription factor known to play a role in proliferation, differentiation, cell cycle progression, and apoptosis [Chen et al., 2001; Kaczynski et al., 2003; Nemer and Horb, 2007]. KLF13 is also involved in T and B lymphocyte development [Outram et al., 2008], and in proliferation and differentiation of the heart [Lavallee et al., 2006; Nemer and Horb, 2007]. FGFR3 (fibroblast growth factor receptor 3) is a member of the fibroblast growth factor receptor family of tyrosine kinases. It is a transmembrane protein that binds fibroblast growth factors, initiating a signaling cascade that leads to the activation of a number of cellular pathways, including the PI3K, MAPK, p38, STAT1, STAT3, and STAT5 pathways [Eswarakumar et al., 2005; Toll and Real, 2008; Matsushita et al., 2009].
In addition, KLF13 and FGFR3 have been implicated in a number of malignancies. For instance, altered expression of Krüppel-like transcription factors is believed to contribute to tumorigenesis [Black et al., 2001; Dang et al., 2001; Miller et al., 2001; Martin et al., 2003]. KLF13 can play the role of an activator, as it does for CCL5 (RANTES) [Song et al., 1999], or that of a repressor, as it does for CYP1A1 [Kaczynski et al., 2003]. This is of particular importance, since CCL5 is a chemotactic chemokine in inflammatory cells and is overexpressed in lung cancer, breast cancer, prostate cancer, melanoma, T-cell leukemia, ovarian cancer, and oral cancer [Negus et al., 1997; Robinson et al., 2003; Mori et al., 2004; Vaday et al., 2006; Karnoub et al., 2007; Borczuk et al., 2008; Chuang et al., 2009], and CYP1A1 plays a role in head and neck and lung cancers [Agundez, 2004; Li et al., 2004; Hiyama et al., 2008]. KLF13 is also required for the expression of cyclin D1, which is a known oncogene in OSCC [Nemer and Horb, 2007]. A mutated form of FGFR3 is overexpressed in some bladder cancers, urothelial cancer, multiple myeloma, and cervical cancer [Dailey et al., 2005; Knowles, 2007, 2008; Tomlinson et al., 2007b], whereas the wild-type FGFR3 is overexpressed in some bladder cancers [Tomlinson et al., 2007b] and multiple myeloma [Chesi et al., 2001]. Reducing FGFR3 levels has been shown to significantly decrease cell proliferation and clonogenicity in bladder cancer [Tomlinson et al., 2007a; Martinez-Torrecuadrada et al., 2008]. Karoui et al. [2001] showed that oral cancer cells do not contain mutations in FGFR3; however, expression levels were not examined.
Although KLF13 and FGFR3 are believed to be involved in other cancers, it is not known whether they contribute to malignancy in oral cancer cells. Our objective was to examine the expression of KLF13 and FGFR3 in OSCC cell lines and tumors, and determine what, if any, role they play in cell proliferation and radiosensitivity in oral cancer cells. We hypothesize that KLF13 and FGFR3 are overexpressed in oral cancer cells and that they contribute to malignancy.
Materials and Methods
Cell Culture
Fourteen OSCC cell lines were chosen from our collection [White et al., 2007; Martin et al., 2008] and cultured in Minimal Essential Medium (MEM) (Invitrogen, Carlsbad, Calif., USA) supplemented with gentamicin, L-glutamine, nonessential amino acids, and 10% fetal bovine serum. As a control, we used 2 independent NHOK samples, which were derived from fresh uvulopalatopharyngoplasty (UP3) specimens from consenting individuals undergoing corrective surgery for sleep apnea. NHOK were cultured in keratinocyte serum-free medium (Ker-sfm) (Invitrogen) supplemented with hEGF in 0.1% BSA, Penicillin/Streptomycin, L-Glutamine, and bovine pituitary extract. OSCC primary tumor samples and NHOK controls were obtained from the University of Pittsburgh Head & Neck SPORE Tissue Bank under our consolidated IRB approval. Table 1 lists the OSCC cell lines, tumors, and NHOK samples used in this study, along with demographic information.
Table 1.
Characteristics of the OSCC cell lines, OSCC tumors, and NHOK samples used in this study
| Sex | Race | Age | Smoking | Alcohol | Family history | Site | Stage | |
|---|---|---|---|---|---|---|---|---|
| Cell line | ||||||||
| UPŒSCC029 | M | Cau | 85 | Yes | Yes | No | Buc | T4N2 |
| UPŒSCC032 | M | Cau | 60 | Yes | Yes | Yes | RMT | T2N2B |
| UPŒSCC040 | M | Cau | 51 | No | Yes | Yes | Tong | T2N2 |
| UPŒSCC066 | F | Cau | 75 | Yes | Yes | Yes | Alv | TINO |
| UPŒSCC070 | F | Cau | 34 | Yes | Yes | Yes | RMT | T3N1 |
| UPŒSCC078 | M | Cau | 60 | No | Yes | Yes | FOM | T2N0 |
| UPCI:SCC084 | M | Cau | 53 | Yes | Yes | No | RMT | T2N2B |
| UPŒSCC103 | F | Cau | 27 | Yes | No | No | Tong | TINO |
| UPCI:SCC104 | M | Cau | 58 | No | No | No | FOM | T4NX |
| UPCI:SCC116 | M | Cau | 58 | Yes | Yes | Yes | Alv | T2N0 |
| UPCI:SCC122 | M | Cau | 63 | Yes | Yes | No | Tong | T1N1 |
| UPCI:SCC131 | M | Cau | 73 | No | No | No | FOM | T2N2 |
| UPŒSCC142 | M | Cau | 58 | No | No | No | FOM | T4NX |
| UPCI:SCC182 | M | Cau | 71 | Yes | Yes | No | RMT | T2N1 |
| Tumor | ||||||||
| 04-1555 | M | Cau | 65 | Yes | Yes | Yes | Tong | T2N1 |
| 05-1797 | M | Cau | 42 | Yes | Yes | Yes | Buc | T2N0 |
| 05-1872 | M | Cau | 79 | Yes | No | No | Buc | T4N0 |
| 05-1902 | M | Cau | 68 | Yes | No | Yes | Tong | T3N2B |
| 05-1980 | M | Cau | 41 | No | Yes | Yes | Tong | T3N2 |
| NHOK | ||||||||
| 06-2665 | M | N/A | 35 | No | No | N/A | UP3 | ni |
| 07-2729 | F | Cau | 19 | No | No | N/A | UP3 | ni |
Included are demographic data as well as smoking history, alcohol use, family history of cancer, tumor stage, and tumor location.
RMT = Retromolar trigone; Tong = tongue; FOM = floor of mouth; Alv = alveolar ridge; Buc = buccal mucosa; N/A = data not available; nl = normal histopathology; Cau = Caucasian; UP3 = uvula, palate, and/or pharynx.
Transfections
For all transfections in this study, we used the OSCC cell line UPCI:SCC029. To establish optimal transfection efficiencies, UPCI:SCC029 cells were transfected with Alexa Fluor 488-labeled AllStars Negative Control siRNA (Qiagen, Valencia, Calif., USA) under a variety of experimental conditions followed by fluorescence microscopy. In 6-well plates, we seeded 1 × 106 cells, and in 96-well plates, we seeded 2.5 × 104 cells on the day before transfection in MEM without antibiotics. Transfections were carried out in Opti-MEM® I (Invitrogen) with Lipofectamine™ 2000 (Invitrogen) at a dilution of 1:100. Transfections were carried out for 24 h, after which time the cells were either harvested or the medium was replaced with antibiotic-free MEM. To decrease cellular levels of KLF13 and FGFR3, we transfected UPCI:SCC029 cells with KLF13 and FGFR3 siRNAs (Ambion, Austin, Tex., USA) at a final concentration of 200 μm. As a control, we transfected cells with the AllStars Negative Control siRNA (Qiagen) at a final concentration of 200 μm.
Cell Proliferation Assays
To assess the ability of KLF13 and FGFR3 to alter proliferation, we carried out cytotoxicity assays (MTT) using the MTT Cell Growth Assay Kit (Millipore, Billerica, Mass., USA). We transfected UPCI:SCC029 cells with the KLF13 siRNA, FGFR3 siRNA, or the negative control siRNA and examined them at 0 h, 24 h, 48 h, and 72 h post-transfection in 3 biological replicates. To eliminate any effect that Lipofectamine 2000 might have on proliferation, all experimental groups were normalized to the mock-transfection control at all time points. Our plates were read using a Spectramax M2/M2e Microplate Reader (Molecular Devices, Sunnyvale, Calif., USA).
Western Blotting
OSCC cells were collected and washed with cold PBS and lysed (on ice) with a solution containing 50 mM Tris, 1% Triton X-100, 10 μg/ml leupeptin, 150 mM NaCl, 1 mM DTT, 0.1% sodium dodecyl sulfate, 10 μg/ml pepstatin, and 1 nM PMSF. Protein concentrations were established using the Bio-Rad Quick Start Bradford Protein Assay Kit (Bio-Rad Laboratories, Hercules, Calif., USA) with the SmartSpec 3000 (Bio-Rad Laboratories). Normalized cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to Immobilon-P membranes (Millipore), and incubated in primary antibody for 2–4 h at room temperature. We used KLF13 (RFLAT-1, C-13) and FGFR3 (C-15) antibodies from Santa Cruz Biotechnology (Santa Cruz, Calif., USA) at a 1:1,000 dilution. To verify equal protein loading, membranes were sectioned and probed for β-tubulin (H-235) (Santa Cruz Biotechnology) at a dilution of 1:1,000. Proteins were visualized using the Western Lightning™ Chemiluminescence Reagent Plus kit (Perkin-Elmer Life Sciences, Boston, Mass., USA). Densitometric analyses were done using Un-Scan-It Gel™ software (Silk Scientific, Orem, Utah, USA).
Clonogenic Survival Assays
To determine if reducing the levels of KLF13 and FGFR3 would increase the sensitivity of OSCC cells to ionizing radiation, we carried out clonogenic survival assays [Parikh et al., 2007]. Cells were seeded into 60-mm dishes and transfected with KLF13 siRNA, FGFR3 siRNA, or the negative control siRNA. At 48 h post-transfection, 1,000 cells were seeded into 60-mm dishes and allowed to attach overnight. The following day, the cells were irradiated with gamma radiation, using the Gammacell 1000 Elite irradiator (MDS Nordion, Ottawa, Ont., Canada) at 0, 1, 2.5, 5, and 10 Gy. The medium was changed 7 days after irradiation. At 12 days post-irradiation, dishes were stained with Giemsa stain (Sigma, St. Louis, Mo., USA) and colonies were counted. We defined a colony as a cluster of 50 or more cells that presumably originated from a single cell.
QRT-PCR
To determine the expression levels of KLF13 and FGFR3 in tumors, we carried out QRT-PCR. Reverse transcription reactions were performed as previously described [Huang et al., 2002] with 2 total RNA inputs (400 ng and 100 ng) for each sample using MMLV Reverse Transcriptase (Epicentre, Madison, Wisc., USA) and random hexamers. For Quantitative PCR (QPCR), we used TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, Calif., USA) for KLF13, FGFR3, and the 18s RNA control, using TaqMan 2× Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems) on the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems). All QPCR samples were done in triplicate and each QPCR plate included no reverse transcriptase and no template controls. The data were analyzed using the comparative CT method.
Statistical Analysis
Data are presented as either the mean ± standard error or the mean ± standard deviation, as noted. Assessment of statistical significance in all of our experiments was done using 2-sided t tests with p values <0.05 being considered statistically significant.
Results
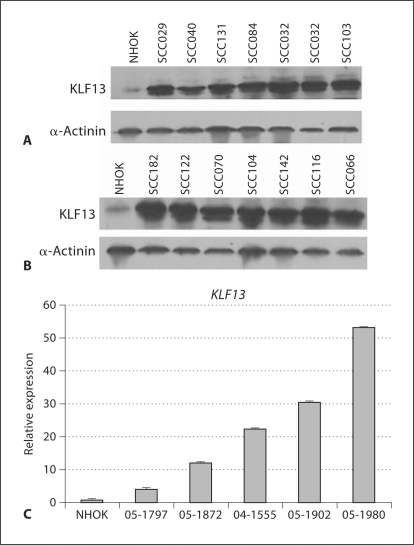
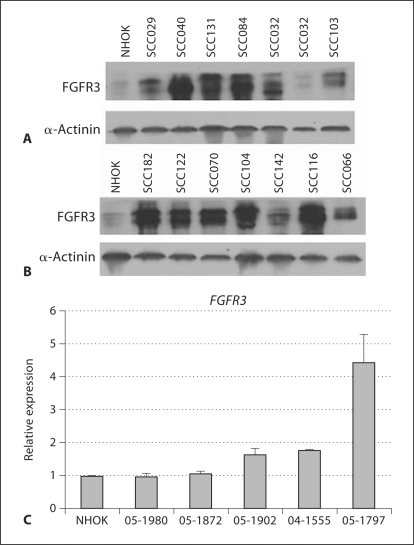
We found that KLF13 protein levels were substantially higher in all OSCC cell lines than in NHOK controls (fig. 1A, B). We also found that KLF13 transcript levels were considerably higher in all OSCC tumors than in NHOK controls (fig. 1C). Our data also indicate the levels of FGFR3 protein were considerably higher in all but one of our cell lines than in the controls (fig. 2A, B), and that FGFR3 mRNA levels were higher in 3 of the 5 tumors than in the controls (fig. 2C). Given the elevated levels of KLF13 and FGFR3 in our OSCC cell lines and tumors, we sought to determine whether they play a role in cell proliferation and radiosensitivity in oral cancer cells.
Fig. 1.
Overexpression of KLF13 in UPCI:SCC oral cancer cell lines, tumors and NHOK controls. A, B Comparison of KLF13 protein levels in OSCC cell lines to NHOK controls. C Comparison of KLF13 transcript levels between OSCC primary tumors and NHOK controls, with standard error of the mean shown.
Fig. 2.
Overexpression of FGFR3 in UPCI:SCC oral cancer cell lines, tumors and NHOK controls. A, B Comparison of FGFR3 protein levels in OSCC cell lines to NHOK controls. C Comparison of FGFR3 transcript levels between OSCC primary tumors and NHOK controls, with standard error of the mean shown.
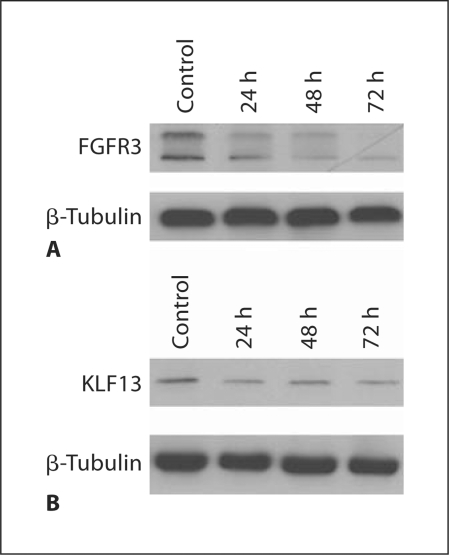
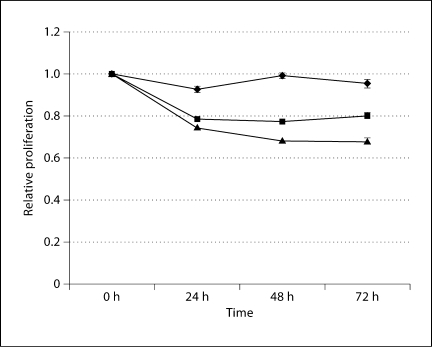
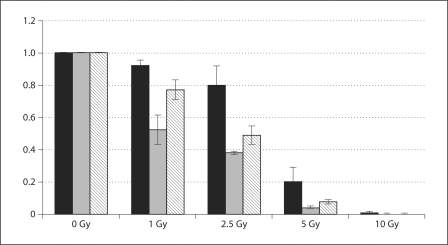
To determine if the overexpression of KLF13 and FGFR3 contributes to malignancy, we reduced cellular levels of KLF13 and FGFR3 (fig. 3) and assessed changes in cell proliferation and sensitivity to ionizing radiation. We found that reducing levels of KLF13 led to a ∼20% decrease in proliferation, and reducing levels of FGFR3 resulted in a ∼35% decrease in proliferation (fig. 4). Therefore, our data suggest that KLF13 and FGFR3 contribute to increased cell proliferation in oral cancer cells. Reducing levels of KLF13 in conjunction with ionizing radiation led to a 25% reduction in colony formation with 1 Gy, a 50% reduction in colony formation with 2.5 Gy, and a 93% reduction in colony formation with 5 Gy, all of which were substantially lower than the negative control (fig. 5). Reducing levels of FGFR3 in conjunction with ionizing radiation led to a 50% reduction in colony formation with 1 Gy, a 60% reduction in colony formation with 2.5 Gy, and a 96% reduction in colony formation with 5 Gy, all of which were considerably lower than the negative control (fig. 5). Thus, the overexpression of KLF13 and FGFR3 in OSCC appears to contribute to resistance to ionizing radiation.
Fig. 3.
Knockdown of FGFR3 (A) and KLF13 (B) levels in UPCI:SCC029 cells transfected with FGFR3 siRNA and KLF13 siRNA, respectively. Cells were collected at 24 h, 48 h, and 72 h post-transfection and compared to cells transfected with the negative control.
Fig. 4.
Results of the MTT assays. Cells transfected with KLF13 siRNA are represented by squares, cells transfected with FGFR3 siRNA are represented by triangles, and cells transfected with the negative control are represented by diamonds, with standard error of the mean indicated. All experimental groups were normalized to the mock-transfection control at all time points.
Fig. 5.
Results of clonogenic survival assays showing the relative survival of UPCI:SCC029 cells transfected with the negative control (black bars), FGFR3 siRNA (gray bars), or KLF13 siRNA (hatched bars), and treated with ionizing radiation, with standard deviation shown.
Discussion
Both KLF13 and FGFR3 have important biological functions and are altered in a number of malignancies. Here we show that KLF13 and FGFR3 are overexpressed in most of the OSCC cell lines and tumors we examined. This suggests that KLF13 and FGFR3 could be used as biomarkers for early detection of this debilitating disease. Others have suggested that FGFR3 be used as a biomarker for early detection of bladder cancer [Knowles, 2007]. Our data indicate that artificially reducing KLF13 and FGFR3 levels negatively impacted cell proliferation and resistance to ionizing radiation, indicating that the overexpression of KLF13 and FGFR3 contributes to malignancy in OSCC. This could be due in part to KLF13 being required for the expression of the oncogenes CCL5, CYP1A1, and CCND1, and that FGFR3 is known to stimulate the PI3K, MAPK, p38, STAT1, STAT3, and STAT5 pathways [Eswarakumar et al., 2005; Toll and Real, 2008; Matsushita et al., 2009], which have known roles in cancer.
KLF13 and FGFR3 probably do not hold any great promise as individual therapeutic targets in OSCC; however, in combination with other therapeutic agents they may be beneficial. For instance, reducing cellular levels of KLF13 and FGFR3, in conjunction with ionizing radiation, led to a substantial decrease in cellular survival. Others have suggested that FGFR3 be used as a therapeutic target to treat bladder cancer [Tomlinson et al., 2007a; Martinez-Torrecuadrada et al., 2008]. This is of interest because FGFR3, among other proteins, has been implicated in contributing to resistance to ionizing radiation in OSCC cells [Ishigami et al., 2007]. This point is supported by the data presented here. In summary, the overexpression of KLF13 and FGFR3 appears to contribute to malignancy in the oral cavity, and may be utilized as biomarkers for early detection and new targets for therapy.
Acknowledgements
This research was supported in part by NIH grant RO1DE14729 to S.M.G., NIH grant P50CA097190 to J.R. Grandis, and an NIH NRSA Kirschstein Fellowship Award to B.J.H., F32CA119725. We would like to thank Dr. Gerard Nau for the use of his laboratory equipment for these studies.
References
- Agundez JA. Cytochrome P450 gene polymorphism and cancer. Curr Drug Metab. 2004;5:211–224. doi: 10.2174/1389200043335621. [DOI] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Borczuk AC, Papanikolaou N, Toonkel RL, Sole M, Gorenstein LA, et al. Lung adenocarcinoma invasion in TGFBRII-deficient cells is mediated by CCL5/RANTES. Oncogene. 2008;27:557–564. doi: 10.1038/sj.onc.1210662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM, Johns DC, Geiman DE, Marban E, Dang DT, et al. Krüppel-like factor 4 (gut-enriched Krüppel-like factor) inhibits cell proliferation by blocking G(1)/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, Brents LA, Fly SA, Bais C, Robbiani DF, et al. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–736. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- Chuang JY, Yang WH, Chen HT, Huang CY, Tan TW, et al. CCL5/CCR5 axis promotes the motility of human oral cancer cells. J Cell Physiol. 2009;220:418–426. doi: 10.1002/jcp.21783. [DOI] [PubMed] [Google Scholar]
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Krüppel-like factor (Krüppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Fact Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Henson BJ, Bhattacharjee S, O'Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Gene Chromosomes Cancer. 2009;48:569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and head and neck cancer risk (review) Intl J Oncol. 2008;32:945–973. [PubMed] [Google Scholar]
- Huang X, Gollin SM, Raja S, Godfrey TE. High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc Natl Acad Sci USA. 2002;99:11369–11374. doi: 10.1073/pnas.172285799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigami T, Uzawa K, Higo M, Nomura H, Saito K, et al. Genes and molecular pathways related to radioresistance of oral squamous cell carcinoma cells. Int J Cancer. 2007;120:2262–2270. doi: 10.1002/ijc.22561. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao YP, Xu JQ, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1-and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Karoui M, Hofmann-Radvanyi H, Zimmermann U, Couvelard A, Degott C, et al. No evidence of somatic FGFR3 mutation in various types of carcinoma. Oncogene. 2001;20:5059–5061. doi: 10.1038/sj.onc.1204651. [DOI] [PubMed] [Google Scholar]
- Knowles MA. Role of FGFR3 in urothelial cell carcinoma: biomarker and potential therapeutic target. World J Urol. 2007;25:581–593. doi: 10.1007/s00345-007-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MA. Molecular pathogenesis of bladder cancer. Int J Clin Oncol. 2008;13:287–297. doi: 10.1007/s10147-008-0812-0. [DOI] [PubMed] [Google Scholar]
- Lavallee G, Andelfinger G, Nadeau M, Lefebvre C, Nemer G, et al. The Krüppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006;25:5201–5213. doi: 10.1038/sj.emboj.7601379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Millikan RC, Bell DA, Cui L, Tse CKJ, et al. Cigarette smoking, cytochrome P4501A1 polymorphisms, and breast cancer among African-American and white women. Breast Cancer Res. 2004;6:R460–R473. doi: 10.1186/bcr814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CL, Reshmi SC, Ried T, Gottberg W, Wilson JW, et al. Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature. Oral Oncol. 2008;44:369–382. doi: 10.1016/j.oraloncology.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KM, Ellis PD, Metcalfe JC, Kemp PR. Selective modulation of the SM22 alpha promoter by the binding of BTEB3 (basal transcription element-binding protein 3) to TGGG repeats. Biochem J. 2003;375:457–463. doi: 10.1042/BJ20030870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Torrecuadrada JL, Cheung LH, Lopez-Serra P, Barderas R, Canamero M, et al. Antitumor activity of fibroblast growth factor receptor 3-specific immunotoxins in a xenograft mouse model of bladder carcinoma is mediated by apoptosis. Mol Cancer Ther. 2008;7:862–873. doi: 10.1158/1535-7163.MCT-07-0394. [DOI] [PubMed] [Google Scholar]
- Matsushita T, Wilcox WR, Chan YY, Kawanami A, Bukulmez H, et al. FGFR3 promotes synchondrosis closure and fusion of ossification centers through the MAPK pathway. Hum Mol Genet. 2009;18:227–240. doi: 10.1093/hmg/ddn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Eklund EA, Peddinghaus ML, Cao ZJ, Fernandes N, et al. Krüppel-like factor 4 regulates laminin alpha 3a expression in mammary epithelial cells. J Biol Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- Mori N, Krensky AM, Ohshima K, Tomita M, Matsuda T, et al. Elevated expression of CCL5/RANTES in adult T-cell leukemia cells: Possible transactivation of the CCL5 gene by human T-cell leukemia virus type I tax. Int J Cancer. 2004;111:548–557. doi: 10.1002/ijc.20266. [DOI] [PubMed] [Google Scholar]
- Negus RPM, Stamp GWH, Hadley J, Balkwill FR. Quantitative assessment of the leukocyte infiltrate in ovarian cancer and its relationship to the expression of C-C chemokines. Am J Pathol. 1997;150:1723–1734. [PMC free article] [PubMed] [Google Scholar]
- Nemer M, Horb ME. The KLF family of transcriptional regulators in cardiomyocyte proliferation and differentiation. Cell Cycle. 2007;6:117–121. doi: 10.4161/cc.6.2.3718. [DOI] [PubMed] [Google Scholar]
- Outram SV, Gordon AR, Hager-Theodorides AL, Metcalfe J, Crompton T, Kemp P. KLF13 influences multiple stages of both B and T cell development. Cell Cycle. 2008;7:2047–2055. doi: 10.4161/cc.7.13.6234. [DOI] [PubMed] [Google Scholar]
- Parikh RA, White JS, Huang X, Schoppy DW, Baysal BE, et al. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Gene Chromosomes Cancer. 2007;46:761–775. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- Robinson SC, Scott KA, Wilson JL, Thompson RG, Proudfoot AEI, Balkwill FR. A chemokine receptor antagonist inhibits experimental breast tumor growth. Cancer Res. 2003;63:8360–8365. [PubMed] [Google Scholar]
- Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10:93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- Toll A, Real FX. Somatic oncogenic mutations, benign skin lesions and cancer progression: Where to look next? Cell Cycle. 2008;7:2674–2681. doi: 10.4161/cc.7.17.6523. [DOI] [PubMed] [Google Scholar]
- Tomlinson D, Hurst CD, Knowles MA. Knockdown by shRNA identifes S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007a;26:5889–5899. doi: 10.1038/sj.onc.1210399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson DC, Baldo O, Hamden P, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol. 2007b;213:91–98. doi: 10.1002/path.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaday GG, Peehl DM, Kadam PA, Lawrence DM. Expression of CCL5 (RANTES) and CCR5 in prostate cancer. Prostate. 2006;66:124–134. doi: 10.1002/pros.20306. [DOI] [PubMed] [Google Scholar]
- White JS, Weissfeld JL, Ragin CCR, Rossie KM, Martin CL, et al. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007;43:701–712. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]