Abstract
Cytochrome P450-mediated detoxification is one of the most important mechanisms involved in insecticide resistance. However, the molecular basis of this mechanism and the physiological functions of P450s associated with insecticide resistance remain largely unknown. Here, we exploited the functional genomics and reverse genetic approaches to identify and characterize a P450 gene responsible for the majority of deltamethrin resistance observed in the QTC279 strain of Tribolium castaneum. We used recently completed whole-genome sequence of T. castaneum to prepare custom microarrays and identified a P450 gene, CYP6BQ9, which showed more than a 200-fold higher expression in the deltamethrin-resistant QTC279 strain when compared with its expression in the deltamethrin-susceptible Lab-S strain. Functional studies using both double-strand RNA (dsRNA)-mediated knockdown in the expression of CYP6BQ9 and transgenic expression of CYP6BQ9 in Drosophila melanogaster showed that CYP6BQ9 confers deltamethrin resistance. Furthermore, CYP6BQ9 enzyme expressed in baculovirus metabolizes deltamethrin to 4-hydroxy deltamethrin. Strikingly, we also found that unlike many P450 genes involved in insecticide resistance that were reported previously, CYP6BQ9 is predominantly expressed in the brain, a part of the central nervous system (CNS) containing voltage-gated sodium channels targeted by deltamethrin. Taken together, the current studies on the brain-specific insect P450 involved in deltamethrin resistance shed new light on the understanding of the molecular basis and evolution of insecticide resistance.
Keywords: functional genomics, insecticide resistance, RNA interference, transgenic expression, insecticide metabolism
Understanding the molecular basis of resistance mechanisms, including discovering the function of resistance-related genes, is critical to designing novel resistance management strategies and developing new insecticidal compounds. With its whole-genome sequencing completed, Tribolium castaneum has become an ideal model organism for research on insecticide resistance (1, 2). T. castaneum belongs to the most species-rich eukaryotic order (Coleoptera), containing about 25% of animal species. It is a notorious worldwide pest of stored grains and farinaceous materials (3, 4), has developed resistance to all five classes of insecticides and fumigants used against it, and ranks among the top 20 arthropods in the number of cases of resistance development reported (http://www.pesticideresistance.org/). Moreover, the functional genomics method, RNA interference (RNAi), works systemically in any tissue and developmental stage of T. castaneum (5, 6).
Pyrethroid insecticides are one of the most widely used classes of insecticides for the control of insect pests that feed on crops and transmit diseases (7). However, pyrethroid resistance has broadly developed in many insect pests (http://www.pesticideresistance.org/). The most significant mechanisms involved in pyrethroid resistance are increased metabolic detoxification by cytochrome P450s (8–10) and decreased target-site sensitivity of sodium channels (11). P450s constitute the largest gene superfamily and are found in a variety of organs and tissues of many organisms (8). Insect P450s are known to play an important role in detoxifying insecticides (8, 12) and plant toxins (13), resulting in the development of resistance to insecticides and facilitating the adaptation of insects to their plant hosts. An important character of insect P450s that are involved in resistance is the constitutively transcriptional overexpression in insecticide-resistant strains, causing enhanced metabolic detoxification of insecticides (8, 14). The insect voltage-gated sodium channel is the primary target of pyrethroids and dichlorodiphenyltrichloroethane (DDT) (15). Mutations in sodium channels confer pyrethroid resistance by reducing the binding of pyrethroids to the sodium channel. The most common sodium channel mutation (kdr, L to F/H/S) has been identified in many insects (16). In addition, nine other amino acid mutations were also shown to cause kdr and kdr-type resistance (11).
A T. castaneum strain, QTC279, exhibits high levels of resistance to deltamethrin, a pyrethroid insecticide. This resistance was almost completely suppressed by piperonyl butoxide (PBO), an inhibitor of cytochrome P450s, suggesting that P450-mediated detoxification is the major mechanism involved in deltamethrin resistance in the QTC279 strain (3). The current studies were conducted to test this hypothesis and identify P450s involved in deltamethrin resistance of the QTC279 strain.
Results
Identification of Genes Differentially Expressed in Resistant and Susceptible Strains.
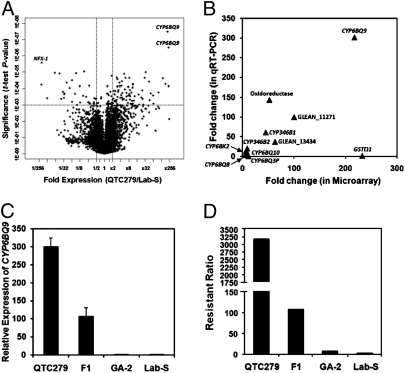
A T. castaneum custom microarray containing 15,008 probe sets (60-mer oligonucleotides) was hybridized with probes prepared using total RNA samples isolated from the QTC279 (deltamethrin resistant) and Lab-S (deltamethrin susceptible) adult beetles. The spot intensity data for these probe sets were statistically analyzed using GeneSpring software (Agilent Technologies). Fold differences in expression and the significance of difference (P value) for 10,879 probe sets detected by at least one of the six probes hybridized are shown as a volcano plot (Fig. 1A). Among the genes tested, 108 probe sets showed significant differences in expression between QTC279 and Lab-S (P ≤ 0.001 and ≥ 2-fold differential expression). Of these 108 probe sets, 75 showed an increase and 33 showed a decrease in the QTC279 strain when compared with their expression in the Lab-S strain. To correct for occurrence of false positives, we introduced two multiple-testing corrections to adjust P values. Benjamini and Hochberg multiple-testing corrections with a false discovery rate of 0.001 revealed 64 probe sets detected genes that are differentially expressed between QTC279 and Lab-S. In QTC279 strain, the signal intensities of 47 probe sets showed an increase and 17 probe sets showed a decrease when compared with their expression in the Lab-S strain. Among these 47 probe sets, eight hybridized to genes that code for P450 enzymes and one hybridized to a gene coding for GST. However, when we applied a more stringent Bonferroni multiple-testing correction, only three probe sets were identified with significantly different expression between QTC279 and Lab-S. One of the three probe sets recognized GLEAN_02938, which codes for a transcription factor similar to a NFX-1-type zinc finger-containing protein (NFX-1). The NFX-1 is conserved in both invertebrates and vertebrates, but the function of this protein is not well known. The signal intensities for this gene were >200-fold lower in the QTC279 strain compared with the Lab-S strain. The other two probe sets recognized GLEAN_07318 and GLEAN_07319, and showed >200-fold higher signal intensities in the QTC279 strain compared with the Lab-S strain. T. castaneum whole-genome sequencing and gene prediction data (http://beetlebase.org/) showed that both GLEAN_07318- and GLEAN_07319-predicted ORFs are located adjacent to each other in chromosome LG4. Gene cloning, sequence alignment (Fig. S1), and Northern hybridization (Fig. S2) showed that these two ORFs predicted by the whole-genome sequencing project are part of the same gene, CYP6BQ9, named by the P450 nomenclature committee.
Fig. 1.
Differential expression of T. castaneum genes. (A) The V plot of differentially expressed genes identified by microarray analysis. The P values of t test were plotted against fold suppression or overexpression. The horizontal bar in the plot shows the nominal significant level 0.001 for the t test under the assumption that each gene had a unique variance. The vertical bars separate the genes that are a minimum of 2.0-fold difference in the QTC279 strain when compared with their levels in the Lab-S strain. Three independent samples each containing 10 adult QTC279 or Lab-S beetles were analyzed. The genes identified by the Bonferroni multiple-testing correction are shown. (B) Correlation between microarray data and qRT-PCR data of genes selected from the list shown in Table S2. Both microarray and qRT-PCR data showed similar fold changes for most genes with only two exceptions, GSTΩ and CYP6BQ3P. The genes were labeled with gene names for those with identified gene products, and the rest of them were labeled with Glean numbers. (C) Relative expression of CYP6BQ9 in QTC279, F1 (offspring of QTC279 male and Lab-S female), GA-2, and Lab-S strain. The data shown are the mean + SEM (n = 4). (D) The resistant ratio of deltamethrin in QTC279, F1 (offspring of QTC279 male and Lab-S female), GA-2, and Lab-S strains. The resistance ratios were calculated by comparing LC50 values obtained with QTC279, F1 (offspring of QTC279 male and Lab-S female), and GA-2 with the LC50 obtained with Lab-S strain.
To validate differential expression of genes identified by microarray analyses, quantitative real-time PCR (qRT-PCR) was employed with primers (Table S1) designed based on sequences of genes identified in microarray analyses. Twelve genes (including seven genes coding for P450 enzymes, one gene each coding for GST, oxidoreductase, a conserved hypothetical protein, a transcription factor, and a protein with unknown function) identified by Benjamini and Hochberg false discovery rate multiple-testing correction applied to microarray data were selected for qRT-PCR analysis. These genes were selected based on their differential expression between the QTC279 and Lab-S strains as well as their potential role in insecticide detoxification. The qRT-PCR analysis confirmed data obtained by microarray analysis for 10 of 12 genes (Fig. 1B and Table S2). Six of the confirmed genes code for P450 enzymes. The microarray data showed overexpression of GSTΩ1 and CYP6BQ3P in the QTC279 strain, but qRT-PCR analysis did not show such an increase in the QTC279 strain for these two genes. Based on the consistent data derived from both microarray and qRT-PCR analyses, CYP6BQ9 showed the most remarkable difference between the QTC279 and Lab-S strains. Therefore, we hypothesized that the P450 gene, CYP6BQ9, may play the most important role in the deltamethrin resistance of the QTC279 strain.
To test our hypothesis, we quantified the expression level of CYP6BQ9 (Fig. 1C) and deltamethrin resistance (Fig. 1D) in QTC279, F1 (offspring of QTC279 male and Lab-S female), and two susceptible T. castaneum strains, GA-2 and Lab-S. As shown in Fig. 1 C and D, a positive correlation between relative expression levels of CYP6BQ9 and deltamethrin resistance was observed among the tested strains of beetles. These data suggest that the P450 gene, CYP6BQ9, plays an important role in deltamethrin resistance.
RNAi Aided Suppression of Deltamethrin Resistance.
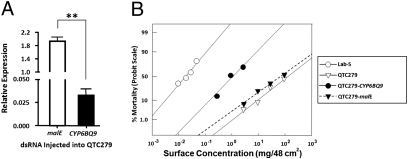
To further investigate the function of CYP6BQ9, we employed RNAi technology to knock down the expression of CYP6BQ9 in the QTC279 beetles. Our RNAi experiment showed that the CYP6BQ9 mRNA levels decreased by >97% at 5 days after injection of CYP6BQ9 dsRNA (Fig. 2A), indicating that this gene was successfully silenced by RNAi.
Fig. 2.
Knockdown in the expression of CYP6BQ9 in T. castaneum QTC279 strain reduced its resistance to deltamethrin. (A) The mRNA levels of CYP6BQ9 were quantified by qRT-PCR at 5 days after dsRNA injection. The relative mRNA levels were shown as a ratio in comparison with the levels of rp49 mRNA. The data shown are mean + SEM (n = 3). Statistical significance of the gene expression between two samples was calculated using Student's t test. **P < 0.01. (B) Dose-response curves (log dose versus mortality on a probit scale) for T. castaneum adults exposed to deltamethrin. Bioassays were performed 5 days after dsRNA injection. Lab-S (○◯), a susceptible strain; QTC279 (▽), a deltamethrin resistant strain; QTC279-CYP6BQ9 RNAi (●), QTC279 strain injected with CYP6BQ9 dsRNA; and QTC279-malE RNAi (▼), QTC279 strain injected with malE dsRNA as a control were exposed to various doses of deltamethrin, and the mortality was recorded and graphed.
To determine whether knocking down the expression of CYP6BQ9 causes suppression of deltamethrin resistance in the QTC279 strain, we performed bioassays to compare deltamethrin resistance levels among uninjected QTC279 beetles and Lab-S beetles and CYP6BQ9 or malE dsRNA injected QTC279 beetles. As shown in Fig. 2B, 0.1 mg/48 cm2 or higher concentrations of deltamethrin caused 100% mortality in the Lab-S strain (n = 186; slope = 1.20 ± 0.17; LC50 = 0.02 mg/48 cm2 [95% CI = 0.01–0.02]). In contrast, 100% mortality was not achieved even after treating the resistant QTC279 strain with 1,000 mg/48 cm2 deltamethrin (n = 68; slope = 0.50 ± 0.15; LC50 = 80.13 mg/48 cm2 [95% CI = 38.53–541.85]). The resistance ratio between the QTC279 and Lab-S strains is 4,007. A similar dose response to deltamethrin was observed in the QTC279 beetles injected with malE dsRNA (n = 84; slope = 0.44 ± 0.12; LC50 = 46.02 mg/48 cm2 [95% CI = 23.62–167.91]; resistance ratio = 2,301). However, the QTC279 beetles injected with CYP6BQ9 dsRNA showed a much higher susceptibility to deltamethrin (n = 78; slope = 0.77 ± 0.19; LC50 = 0.79 mg/48 cm2 [95% CI = 0.37–1.35]). As a result, the resistance ratio between the QTC279 strain injected with CYP6BQ9 dsRNA and the Lab-S strain decreased dramatically (to <40). These data suggest that CYP6BQ9 is required for the high level of deltamethrin resistance observed in the QTC279 strain.
Brain-Specific Expression.
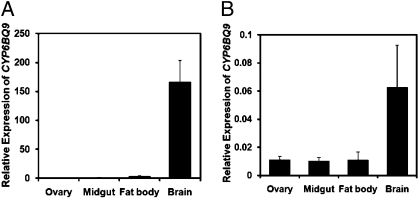
Insect P450s are known to be expressed in various tissues in response to diverse physiological and environmental stimuli (8). Therefore, the expression of CYP6BQ9 was quantified in the ovary, midgut, fat body (thorax and abdomen), and brain (free from fat body and other tissues) isolated from the QTC279 and Lab-S beetles. As shown in Fig. 3 A and B, CYP6BQ9 mRNAs were detected predominantly in the brain of both QTC279 and Lab-S strains. Because midgut and/or fat body tissues were suggested as the primary detoxification organs where most insect P450s are expected to be expressed (8), we used transgenic Drosophila melanogaster to test whether the predominant CYP6BQ9 expression in the CNS observed in our studies is sufficient to cause deltamethrin resistance.
Fig. 3.
Tissue-specific expression of CYP6BQ9. Relative expression levels of CYP6BQ9 in the ovary, midgut, fat body, and brain of the QTC279 (A) and Lab-S (B) strains. Tissues were dissected and total RNAs were isolated to quantify CYP6BQ9 mRNA levels by qRT-PCR as described in Materials and Methods. Relative expression levels were normalized by the expression of rp49. The data shown are mean + SEM (n = 9).
Transgenic Expression of CYP6BQ9 in D. melanogaster.
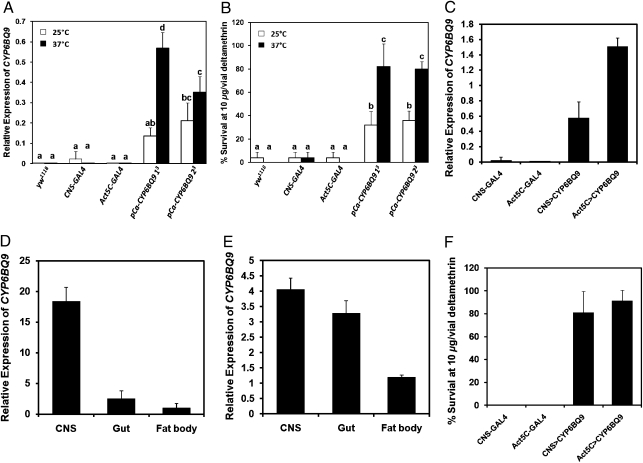
To identify if the expression of CYP6BQ9 is sufficient to cause deltamethrin resistance, CYP6BQ9 was introduced into the vector pCaSpeR-hs (under the control of the heat shock protein 70 promoter). After embryonic microinjections of D. melanogaster w1118 with pCaSpeR-hs-CYP6BQ9 (pCa-CYP6BQ9) plasmids, 20 lines of flies, in which the expression of CYP6BQ9 could be enhanced after heat shock, were identified. Two independent transformed lines with homozygous CYP6BQ9 expression in the chromosome III were chosen to perform contact bioassays and expression studies. We quantified mRNA produced from the CYP6BQ9 transgene using qRT-PCR in two pCa-CYP6BQ9 transformed lines. CYP6BQ9 mRNA was detected in both transformed lines, and the expression was enhanced by heat shock in both lines (Fig. 4A). In addition, the CYP6BQ9 mRNA was not detected in three control D. melanogaster strains (yw1118, CNS-GAL4, and Act5C-GAL4), which were not transformed with pCa-CYP6BQ9 construct (Fig. 4A). Bioassays showed that both transgenic lines of flies transformed with pCa-CYP6BQ9 constructs were tolerant to deltamethrin at a diagnostic dose of 10 μg deltamethrin per vial with higher survival rates (around 40% at 25 °C and 80% at 37 °C) than the control flies (less than 10%; Fig. 4B). These findings suggest that the expression of CYP6BQ9 alone is sufficient to cause deltamethrin resistance.
Fig. 4.
The expression of T. castaneum CYP6BQ9 and its effect on deltamethrin resistance in D. melanogaster. (A) The mRNA levels of CYP6BQ9 were quantified by qRT-PCR in three control nontransgenic strains (yw1118, CNS-GAL4, and Act5C-GAL4) and two different transgenic fly lines (pCa-CYP6BQ9 13 and pCa-CYP6BQ9 23; both of them showed CYP6BQ9 insertion in the third chromosome). The expression levels of CYP6BQ9 in both transformed lines were enhanced by heat shock. The data shown are the mean + SEM (n = 4). (B) The percent survival of three control nontransgenic strains and two different transgenic fly lines exposed to 10 μg deltamethrin/vial at 25 °C and 37 °C. The data shown are mean + SEM (n = 5). The data (A and B) were analyzed by ANOVA, followed by Bonferroni multiple mean separation techniques at P < 0.05 (SAS v9.0 software; SAS Institute Inc.). There is no significant difference in the level of expression among samples marked with the same letter. (C) The relative expression of CYP6BQ9 in two control strains (CNS-GAL4 and Act5C-GAL4) and two transgenic lines (CNS > CYP6BQ9 and Act5C > CYP6BQ9) are shown. The data shown are mean + SEM (n = 4). (D and E) Tissue-specific expression of CYP6BQ9 in CNS > CYP6BQ9 and Act5C > CYP6BQ9 strains, respectively. Relative expression levels were normalized using rp49 expression as a control. The data shown are mean + SEM (n = 4). (F) The percent survival of two control strains and two transgenic lines exposed to 10 μg deltamethrin/vial. The data shown are mean + SEM (n = 8).
Transgenic D. melanogaster were also produced to express CYP6BQ9 in the CNS alone or in the whole body using the GAL4/UAS system. We compared the expression of CYP6BQ9 in two control strains and two strains expressing UAS-CYP6BQ9 under the control of a CNS-GAL4 driver (CNS > CYP6BQ9) or an Act5C-GAL4 driver (Act5C > CYP6BQ9). As shown in Fig. 4C, higher levels of CYP6BQ9 mRNA were detected in the whole body of Act5C > CYP6BQ9 transgenic flies when compared with its expression in the whole-body samples of CNS > CYP6BQ9 flies. CYP6BQ9 was expressed in all three tissues (midgut, fat body, and CNS) tested in Act5C > CYP6BQ9 transgenic flies, whereas CYP6BQ9 was expressed predominantly in the CNS but with low levels in both midgut and fat body of CNS > CYP6BQ9 transgenic flies (Fig. 4 D and E). In contact bioassays, 80% of both lines of transgenic flies (Act5C > CYP6BQ9 and CNS > CYP6BQ9) survived at a diagnostic dose of 10 μg deltamethrin per vial (Fig. 4F). However, none of the control flies survived this dose of deltamethrin (Fig. 4F). These data suggest that the expression of CYP6BQ9 in the CNS is sufficient for causing deltamethrin resistance.
Pyrethroid Metabolism.
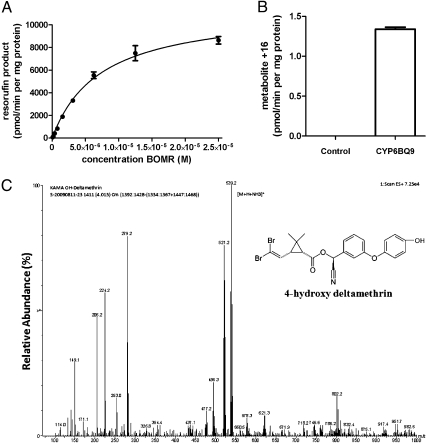
Experiments were conducted to determine whether CYP6BQ9 could metabolize deltamethrin. The CYP6BQ9 protein was expressed in Sf9 insect cells using a baculovirus expression system. To ensure an intact electron supply, the P450 was coexpressed with a cytochrome P450 reductase (CPR) from D. melanogaster (GenBank accession no. Q27597). The recombinant CYP6BQ9 and CPR proteins efficiently debenzylated the artificial model substrate, benzyloxyresorufin (resorufin benzyl ether, BOMR), to its red-fluorescent product resorufin with a Km value of 4.8 ± 0.9 μM (Fig. 5A; activity given in pmol/min per mg total protein) and a Vmax of 11.5 ± 0.5 mg/nmol per min protein. In control experiments, no turnover was caused by proteins isolated from cells infected with the baculovirus containing vector with no insert of CYP6BQ9.
Fig. 5.
The metabolic activity of recombinant CYP6BQ9 enzyme. (A) Rate of dealkylation reaction of recombinant CYP6BQ9 with the fluorescent substrate BOMR (Vivid; Invitrogen). The Km value was determined as 4.8 ± 0.9 μM. (B) In vitro activity assays with recombinant CYP6BQ9. Microsomes from Sf9 cells expressing QTC279 CYP6BQ9 and D. melanogaster CPR were incubated with deltamethrin for 3 h followed by mass spectrometric analyses. The empty baculovirus vector containing cells were used as a control. The peak area of the major metabolite (+16) was quantified. The data shown are mean + SEM (n = 3). (C) Mass spectrum of 4-hydroxy deltamethrin, the only metabolite detected in in vitro experiments incubating deltamethrin and CYP6BQ9 reconstituted protein complex.
More importantly, deltamethrin itself could be metabolized by the recombinant CYP6BQ9/NADPH CPR microsomes in in vitro assays. Samples from such metabolism experiments were subjected to tandem mass spectrometry (HPLC-MS/MS). As shown in Fig. 5B, only the CYP6BQ9 protein displayed in vitro activity, and the control that used protein from cells infected with the baculovirus vector without insert showed no activity. Subsequently, the deltamethrin metabolite was identified as 4-hydroxy deltamethrin (m/z 539.2) via HPLC-MS/MS measurement (Fig. 5C). These experiments provided direct evidence for CYP6BQ9 metabolizing deltamethrin, to which the QTC279 strain resists, thus further confirming the function of CYP6BQ9 in deltamethrin resistance of the QTC279 strain.
Discussion
In this study, we employed the recently completed genome sequence of T. castaneum and functional genomics methods to uncover the molecular mechanisms of deltamethrin resistance in T. castaneum QTC279 strain. Based on the data presented here, we conclude that P450-mediated detoxification is the major mechanism of resistance against the synthetic pyrethroid, deltamethrin, in the QTC279 strain. We identified a P450 gene, CYP6BQ9, which is responsible for the majority of pyrethroid resistance observed in this strain. The most surprising finding was the brain-specific expression of this deltamethrin-detoxifying P450 gene, CYP6BQ9. Deltamethrin, like most other pyrethroids, is a lipophilic low-molecular-weight neurotoxin. It is capable of penetrating the blood-brain barrier and binding to voltage-gated sodium channels (15). The high expression level of CYP6BQ9 in the brain could greatly enhance the ability of brain cells to metabolize the insecticide and thus significantly deplete the levels of insecticide at the target site necessary for insect control. CYP6D1, a house fly P450 responsible for pyrethroid resistance in the LPR strain, was shown to metabolize pyrethroids in the thoracic ganglia (17). The authors suggested that the expression of P450 in the nervous system might provide the last line of defense to protect the target site. In vertebrates, many P450 genes have been demonstrated to be present in the brain (partially or predominantly), catalyzing the metabolism of neurosteriods and drugs (18, 19). Drosophila P450 gene CYP4G15 is primarily expressed in the brain of third instar, but there is no evidence of its role in insecticide resistance (8, 20, 21). Our studies make a causal connection between brain-specific expression of an insect P450 gene and its function in the detoxification of an insecticide. Because most of the insecticides are neurotoxins, the central nervous system—especially the brain—could be considered as a target tissue to identify more insect P450s that are involved in insecticide resistance.
Cytochrome P450s are an abundant family of metabolic enzymes found in all living organisms (8). Previous studies suggested that multiple P450s commonly share the role associated with insecticide resistance through coup regulation in resistant strains. For example, CYP6G1 and CYP12D1 are overexpressed in DDT-resistant strains of D. melanogaster (22). In a permethrin-resistant housefly strain ALHF, CYP4D4v2, CYP4G2, CYP6A38, and CYP6A36 are up-regulated through induction (23) or constitutive expression (14). The overexpression of two duplicated P450s, CYP6P4 and CYP6P9, is associated with pyrethroid resistance in A. funestus strains (10). Our microarray and qRT-PCR data identified several genes that are up-regulated in the QTC279 strain. Although our data suggest that CYP6BQ9 may play a major role in deltamethrin resistance in the QTC279 strain, the contribution from other genes to deltamethrin resistance cannot be ruled out. Because dsRNA-aided knockdown in the expression of CYP6BQ9 did not completely block deltamethrin resistance in the QTC279 strain, we hypothesize that one or more of the other overexpressed genes in the QTC279 strain may play minor roles in deltamethrin resistance. Experiments to test this hypothesis are currently underway. CYP6BQ9 is a member of a 12-P450 gene cluster, and two other members of this cluster are also overexpressed in the QTC279 strain. Work is in progress to determine the role of these two P450s in deltamethrin resistance of the QTC279 strain.
Transgenic overexpression of P450s in D. melanogaster was successfully exploited to evaluate the function of fruit fly P450s in insecticide resistance. The GAL4/UAS system has been used to express CYP6G1 in all tissues (24) or in specific tissues (25), and the expression of this gene in transgenic fruit flies conferred resistance to insecticides tested. The CYP12A4 expressed specifically in the midgut and Malpighian tubules of D. melanogaster conferred resistance to the insecticide lufenuron (26). In another study, eight D. melanogaster P450s were expressed in the midgut, Malpighian tubules, and fat body, and some of them conferred resistance to multiple insecticides (27). In the present study, a T. castaneum P450 expressed ubiquitously or in the CNS of D. melanogaster conferred resistance to deltamethrin. Though the P450 sequences are highly divergent across the taxa, the CPR and cytochrome b5 sequences remain more evolutionary conserved (28). Taken together, these studies suggest that the power of genetics and genomics in fruit flies could be exploited for studying the function of P450s from pest insects.
Our studies showed that the brain-specific overexpression of one P450 gene is responsible for the majority of deltamethrin resistance observed in the QTC279 strain, but the mechanism of the overexpression of CYP6BQ9 in the QTC279 strain is not known. Microarray and qRT-PCR analyses showed that the expression of a transcription factor, NFX-1, was reduced in the QTC279 strain when compared with its expression in the Lab-S strain (Table S2). However, RNAi-aided knockdown in the expression of NFX-1 in the Lab-S strain (Fig. S3A) neither affected deltamethrin susceptibility (Fig. S3B) nor caused an increase in the expression of CYP6BQ9 (Fig. S3C). Combined with the lack of positive correlation between expression of NFX-1 and deltamethrin resistance (Fig. S3D) and tissue-specific expression of NFX-1 (Fig. S3E), we concluded that NFX-1 may not play an important role in the regulation of CYP6BQ9 expression in T. castaneum. We hypothesize that either the differences in cis-regulatory elements present in the promoters of CYP6BQ9 from the QTC279 and Lab-S strains and/or the expression of transcription factors that bind to these elements may be responsible for enhanced expression of CYP6BQ9 in the QTC279 strain. Work is in progress to elucidate the mechanism of CYP6BQ9 expression in the QTC279 strain of T. castaneum.
Materials and Methods
Red Flour Beetle Strains.
Three red flour beetle strains were used in this study. QTC279, originally collected from a wheat storage facility in Malu, Queensland, Australia, in 1984, was selected with pyrethroids for 10 generations until it was homozygous for the major pyrethroid resistance factor (3). Lab-S is a laboratory strain that is susceptible to most of the insecticides tested. GA-2 was used in the whole-genome sequencing project. Beetles were reared in whole wheat flour with yeast (10% by weight) and maintained in the dark at 32 °C and 55 ± 2% relative humidity.
RNA Extraction, Analyses, and RNAi.
Standard procedures as described in SI Materials and Methods were used to prepare RNA and probes for microarray analysis. Three biological replicates were included for each sample analyzed. The 60-mer oligonucleotides designed based on 15,008 genes selected from the 16,000 genes predicted by Tribolium genome annotations and 736 control probe sets were printed onto glass slides at Agilent Technologies. Probes for microarray analysis were labeled using Low RNA Input Linear Amplification Kit with one color (Agilent) following manufacturer's instructions. The hybridization of labeled probes to arrays was performed using the Agilent Gene Expression Hybridization Kit at 65 °C for 17 h. The raw data files (.txt) were imported into GeneSpring (GX 7.3), and the data were normalized and analyzed. Further details on microarray methods can be found in SI Materials and Methods. qRT-PCR and Northern blot analysis were performed following standard procedures as described in SI Materials and Methods. For RNAi experiments, dsRNAs were synthesized using the MEGAscript RNAi Kit (Ambion, Inc.) and 300- to 400-bp-long PCR-amplified DNA from unique regions of genes using gene-specific primers containing T7 promoter at the end. One- to 2-week-old adults were injected with dsRNA (0.8–1 μg). Further details on RNAi methods can be found in SI Materials and Methods.
Tribolium Bioassays.
Beetles were exposed to filter paper surface treated with serial dilutions of technical grade deltamethrin (Bayer Environmental Science) prepared in ethanol. Beetle mortality was scored after 24-h exposure in the dark at 32 °C and 55 ± 2% relative humidity. Mean and standard errors for each time point were obtained from at least three independent bioassays. All data were analyzed by using MINITAB statistical software (MINITAB 14; Minitab, Inc.).
Construction of Transgenic Fly Strains and Drosophila Contact Bioassays.
CYP6BQ9 cloned from the QTC279 strain was inserted into pCaSpeR-hs and pUAST vectors to prepare pCa-CYP6BQ9 and UAS-CYP6BQ9 constructs, and the constructs were transformed into the germline of D. melanogaster yw1118 strain using standard P-element-mediated transformation techniques (29). The transgenic flies were assayed for deltamethrin susceptibility as described in SI Materials and Methods.
BOMR Metabolism.
BOMR metabolism by CYP6BQ9 microsomes was carried out in a 384-well plate as described in the Vivid CYP450 screening kit (Invitrogen) manual with the prepared microsomes of CYP6BQ9/NADPH CPR (0.1 mg/mL protein content) and 0.1–50 μM benzyloxyresorufin substrate and an assay volume of 50 μL. Fluorescence was detected upon incubation for 60 min at 22 °C on a Tecan Infinite M1000 fluorescence reader (λex = 535 nm, λem = 590 nm, 10 nm bandwidth), revealing an apparent Km of 4.8 ± 0.9 μM.
Deltamethrin Metabolism.
Deltamethrin metabolism was assayed by incubation of the recombinant CYP6BQ9/NADPH CPR microsomes (0.2 mg/mL total protein content) in 0.1 M potassium phosphate buffer with NADPH-regenerating system (Promega; 1.3 mM NADP+, 3.3 mM glucose-6-phosphate, 3.3 mM MgCl2, 0.4 U/mL glucose-6-phosphate dehydrogenase) and deltamethrin (10 μM) at 27 °C for 3 h. The total assay volume was 200 μL. The assay was quenched by the addition of acetonitrile (to 80% final concentration) and analyzed by tandem mass spectrometry.
Quantitative HPLC-MS/MS Measurements.
The samples were measured on an Applied Biosystems API 4000 QTrap MS/MS system running in positive electrospray MRM mode with a capillary voltage of 5 kV and Turbo V gas temperature of 500 °C. The HPLC system was a Waters Acquity UPLC consisting of binary solvent manager, column manager, and sample manager. The samples were run on a Waters Atlantis HSS T3 1.8-μm column (50 × 2.1 mm) running in reversed-phase gradient mode with methanol/2 mM NH4OAc/1% acetic acid as eluent. For the quantification of deltamethrin and 4-OH deltamethrin, the MRM transition 523.0 > 280.9 and 539.0 > 280.9 was monitored, respectively. The peak integrals were calibrated externally against a standard calibration curve with a correlation coefficient of r > 0.99. The limit of quantification (S/n > 10) was 0.1 ng/mL for deltamethrin.
Supplementary Material
Acknowledgments
We thank three anonymous reviewers for their helpful comments, Dr. R. W. Beeman (US Department of Agriculture) for supply of beetles, Dr. Yoonseong Park (Kansas State University) for help with microarray design, Dr. Nelson (University of Tennessee) for help with P450 nomenclature, Dr. Byron Reid (Bayer Environmental Science) for the gift of deltamethrin, Prof. Paul O´Neill (University of Liverpool) for providing 4-OH deltamethrin, Dr. Nigel Cooper and Ms. Xiaohong Li (University of Louisville) for help with microarray experiment, Dr. Nannan Liu (Auburn University) for comments on an earlier version of the manuscript, and Dr. Al Fournier (University of Arizona) for help on language editing. This work was supported by the US Department of Agriculture National Research Initiative Cooperative State Research, Education, and Extension Service Grant 2007-04636 (to S.R.P.). The University of Louisville microarray facility is supported by National Center for Research Resources Institutional Development Award (IDeA) NBRE-P20 RR016481 and COBRE-P20RR018733.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GU727868 and GU727869).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000059107/-/DCSupplemental.
References
- 1.Brown SJ, Denell RE, Beeman RW. Beetling around the genome. Genet Res. 2003;82:155–161. doi: 10.1017/s0016672303006451. [DOI] [PubMed] [Google Scholar]
- 2.Richards S, et al. Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 3.Collins PJ. A new resistance to pyrethroids in Tribolium castaneum (Herbst) Pestic Sci. 1990;28:101–115. [Google Scholar]
- 4.Stuart JJ, Ray S, Harrington BJ, Neal JJ, Beeman RW. Genetic mapping of a major locus controlling pyrethroid resistance in Tribolium castaneum (Coleoptera: Tenebrionidae) J Econ Entomol. 1998;91:1232–1238. doi: 10.1093/jee/91.6.1232. [DOI] [PubMed] [Google Scholar]
- 5.Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr Biol. 2002;12:R85–R86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- 6.Tomoyasu Y, et al. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008;9:R10. doi: 10.1186/gb-2008-9-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khambay BPS, Jewess PJ. Pyrethroids. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 6. Oxford: Elsevier; 2004. pp. 1–29. [Google Scholar]
- 8.Feyereisen R. Insect cytochrome P450. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 4. Oxford: Elsevier; 2005. pp. 1–77. [Google Scholar]
- 9.Müller P, et al. Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. PLoS Genet. 2008;4:e1000286. doi: 10.1371/journal.pgen.1000286. 10.1371/journal.pgen.100286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wondji CS, et al. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 2009;19:452–459. doi: 10.1101/gr.087916.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong K. Insect sodium channels and insecticide resistance. Invert Neurosci. 2007;7:17–30. doi: 10.1007/s10158-006-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karunker I, et al. Structural model and functional characterization of the Bemisia tabaci CYP6CM1vQ, a cytochrome P450 associated with high levels of imidacloprid resistance. Insect Biochem Mol Biol. 2009;39:697–706. doi: 10.1016/j.ibmb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 14.Zhu F, Feng J-N, Zhang L, Liu N. Characterization of two novel cytochrome P450 genes in insecticide-resistant house-flies. Insect Mol Biol. 2008;17:27–37. doi: 10.1111/j.1365-2583.2008.00777.x. [DOI] [PubMed] [Google Scholar]
- 15.Vijverberg HPM, van der Zalm JM, van der Bercken J. Similar mode of action of pyrethroids and DDT on sodium channel gating in myelinated nerves. Nature. 1982;295:601–603. doi: 10.1038/295601a0. [DOI] [PubMed] [Google Scholar]
- 16.Soderlund D. Sodium channels. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 5. Oxford: Elsevier; 2005. pp. 1–24. [Google Scholar]
- 17.Korytko PJ, Scott JG. CYP6D1 protects thoracic ganglia of houseflies from the neurotoxic insecticide cypermethrin. Arch Insect Biochem Physiol. 1998;37:57–63. doi: 10.1002/(SICI)1520-6327(1998)37:1<57::AID-ARCH7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 18.Miksys S, Tyndale RF. Brain drug-metabolizing cytochrome P450 enzymes are active in vivo, demonstrated by mechanism-based enzyme inhibition. Neuropsychopharmacology. 2009;34:634–640. doi: 10.1038/npp.2008.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strobel HW, Thompson CM, Antonovic L. Cytochromes P450 in brain: Function and significance. Curr Drug Metab. 2001;2:199–214. doi: 10.2174/1389200013338577. [DOI] [PubMed] [Google Scholar]
- 20.Maïbèche-Coisne M, Monti-Dedieu L, Aragon S, Dauphin-Villemant C. A new cytochrome P450 from Drosophila melanogaster, CYP4G15, expressed in the nervous system. Biochem Biophys Res Commun. 2000;273:1132–1137. doi: 10.1006/bbrc.2000.3058. [DOI] [PubMed] [Google Scholar]
- 21.Chung H, et al. Characterization of Drosophila melanogaster cytochrome P450 genes. Proc Natl Acad Sci USA. 2009;106:5731–5736. doi: 10.1073/pnas.0812141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Festucci-Buselli RA, et al. Expression of Cyp6g1 and Cyp12d1 in DDT resistant and susceptible strains of Drosophila melanogaster. Insect Mol Biol. 2005;14:69–77. doi: 10.1111/j.1365-2583.2005.00532.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu F, Li T, Zhang L, Liu N. Co-up-regulation of three P450 genes in response to permethrin exposure in permethrin resistant house flies, Musca domestica. BMC Physiol. 2008;8:18. doi: 10.1186/1472-6793-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daborn PJ, et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- 25.Chung H, et al. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics. 2007;175:1071–1077. doi: 10.1534/genetics.106.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogwitz MR, et al. Cyp12a4 confers lufenuron resistance in a natural population of Drosophila melanogaster. Proc Natl Acad Sci USA. 2005;102:12807–12812. doi: 10.1073/pnas.0503709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daborn PJ, et al. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol. 2007;37:512–519. doi: 10.1016/j.ibmb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Korytko PJ, MacLntyre RJ, Scott JG. Expression and activity of a house-fly cytochrome P450, CYP6D1, in Drosophila melanogaster. Insect Mol Biol. 2000;9:441–449. doi: 10.1046/j.1365-2583.2000.00207.x. [DOI] [PubMed] [Google Scholar]
- 29.Spradling A. In: Drosophila: A Practical Approach. Roberts D, editor. Oxford: IRL; 1986. pp. 175–197. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.