Abstract
Hepatitis B virus (HBV)-associated acute liver failure (ALF) is a dramatic clinical syndrome due to a sudden loss of hepatic cells leading to multiorgan failure. The mechanisms whereby HBV induces ALF are unknown. Here, we show that liver tissue collected at the time of liver transplantation in two patients with HBV-associated ALF is characterized by an overwhelming B cell response apparently centered in the liver with massive accumulation of plasma cells secreting IgG and IgM, accompanied by complement deposition. We demonstrate that the molecular target of these antibodies is the hepatitis B core antigen (HBcAg); that these anti-bodies display a restricted variable heavy chain (VH) repertoire and lack somatic mutations; and that these two unrelated individuals with ALF use an identical predominant VH gene with unmutated variable domain (IGHV1-3) for both IgG and IgM anti-HBc antibodies, indicating that HBcAg is the target of a germline human VH gene. These data suggest that humoral immunity may exert a primary role in the pathogenesis of HBV-associated ALF.
Keywords: fulminant hepatitis B, pathogenesis, gene expression profiling, B cell immunity, phage display Fab library
Acute liver failure (ALF), also known as fulminant hepatitis, is a dramatic clinical syndrome characterized by the sudden loss of hepatic cells leading to multiorgan failure in a person without preexisting liver disease (1). It has been estimated that approximately 2,000 cases of ALF per year occur in the United States. Viral infections and drugs are the principal causes of ALF (1). Hepatitis B virus (HBV) is a major cause of ALF worldwide: ≈1% of acute hepatitis B patients develop fulminant hepatitis (2, 3). Before the advent of orthotopic liver transplantation (OLT) the overall mortality of HBV-associated ALF exceeded 80% (1). Although liver damage in the classic form of acute hepatitis B is believed to be mediated by the cellular immune response (4), the pathogenesis of HBV-associated ALF is poorly understood. However, demonstrated differences in both viral and antibody kinetics distinguish ALF from classic acute hepatitis B (5). Fulminant hepatitis B is characterized by an unusually brisk antibody response to viral antigens and a more rapid viral clearance than is observed in traditional cases of acute hepatitis B (6–8). Whether this unusually rapid viral clearance and accelerated antibody seroconversion play a role in the pathogenesis of HBV-associated ALF has been for a long time a matter of debate. Variants of HBV containing precore or core promoter mutations have been associated with a fulminant course of acute hepatitis B (9–11), but these mutations are also commonly detected in inactive hepatitis B surface antigen (HBsAg)-positive carriers as well as in patients with chronic hepatitis B (12).
The advent of gene array technologies has provided a powerful tool to study the pathogenesis of complex human diseases. However, to our knowledge, studies of gene expression profiling of ALF in humans have not been reported. Access to well-preserved explanted liver tissue specimens from two well-defined HBV-associated ALF patients who underwent OLT provided us with the unique opportunity to define the gene expression profile of HBV-associated ALF and to correlate the molecular analysis with the clinical, virologic, and histopathologic features. Here, we show that HBV-associated ALF is characterized by an overwhelming B cell response centered in the liver with massive intrahepatic production of IgG and IgM by infiltrating plasma cells against the hepatitis B core antigen (HBcAg) of HBV, implicating a major role of B cell immunity in the pathogenesis of HBV-associated ALF.
Results
Course of HBV-Associated ALF.
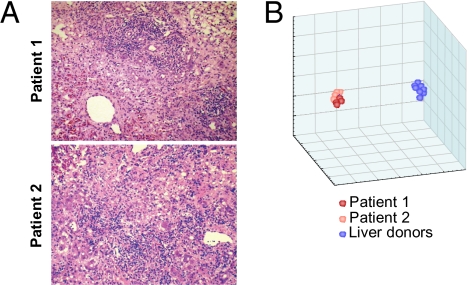
Two previously healthy, young adults suddenly developed typical HBV-associated ALF that was documented by clinical, biochemical (Table S1), serologic and virologic (Table 1), and histopathologic criteria (Fig. 1A). The HBV serologic markers were compatible with an accelerated antibody response, low levels of HBV replication, and an early clearance of HBV antigens (Table 1), as documented in previous studies (5–8). At presentation, HBsAg was positive in patient 1 and borderline in patient 2 concomitant with the appearance of antibody to HBsAg (anti-HBs), whereas hepatitis B e antigen (HBeAg), a replication marker, was negative in both. The two patients were positive for IgM and IgG antibodies to HBcAg, and to HBeAg (anti-HBe), although the latter was borderline positive in patient 1. Both patients developed grade IV coma and underwent OLT within 3 days of admission and 8 days of the onset of symptoms. At the time of OLT, HBsAg became negative in patient 2 (who showed a slight increase in anti-HBs titer from 5.9 to 8 mIU) and detectable in patient 1, albeit at low levels (1.74 μg/mL). In both patients, IgM anti-HBc titers were extremely high (>1:500,000) in parallel with an increase in total IgM serum levels, especially in patient 1 (Table 1). Serum HBV DNA decreased from 185,000 copies/mL at admission to 7,500 at the time of OLT in patient 2, whereas it was 218,000 copies/mL at the single time point tested in patient 1. Serum levels of complement factors C3 and C4 and complement hemolytic activity (CH50) were dramatically reduced in both cases, with lower values in patient 1 (Table S1). At the time of OLT, four pieces of the native liver were obtained from each patient. Microscopic examination documented massive hepatic necrosis in patient 1 and submassive hepatic necrosis in patient 2 (Fig. 1A). Detailed pathology review is provided in SI Materials and Methods.
Table 1.
Serologic markers of HBV infection in two patients with HBV-associated ALF
| Marker | On admission | Day 3 | Day 4 |
| Patient 1 | |||
| HBsAg | Positive | Positive | Positive |
| Concentration (μg/mL) | 1.74 | ||
| Anti-HBs (mIU/mL) | 0 | 0 | 0 |
| Anti-HBc | Positive | Positive | Positive |
| Titer | 1:128 | ||
| IgM anti-HBc | Positive | Positive | Positive |
| Titer | 1:524,288 | ||
| HBeAg | Negative | Negative | Negative |
| Anti-HBe | Borderline positive | Positive | Positive |
| HBV DNA (copies/mL) | 218,000 | ||
| Patient 2 | |||
| HBsAg | Borderline positive | Negative | Negative |
| Concentration (μg/mL) | 0 | ||
| Anti-HBs (mIU/mL) | 5.9 | 8.0 | |
| Anti-HBc | Positive | Positive | Positive |
| Titer | 1:256 | ||
| IgM anti-HBc | Positive | Positive | Positive |
| Titer | 1:524,288 | ||
| HBeAg | Negative | Negative | Negative |
| Anti-HBe | Positive | Positive | Positive |
| HBV DNA (copies/mL) | 185,000 | 7,500 | |
Fig. 1.
(A) Histopathologic features of HBV-associated ALF. Liver specimens were obtained from the native livers of patients 1 and 2 at the time of OLT. In patient 1, the parenchyma shows massive hepatic necrosis with the presence of vascular spaces. There are no viable hepatocytes; lobular lymphoid cell infiltrates and Kupffer cells are prominent. The collapse of the lobules has resulted in the proximity of the portal tracts, which show extensive lymphoid infiltrates and bile duct regeneration. In patient 2, the parenchyma shows submassive liver necrosis with prominent lobular lymphoid cell infiltrates and Kupffer cell hyperplasia. Portal areas show prominent lymphoid cell infiltrates. (hematoxylin and eosin; magnification: ×75.) (B) Gene expression profiling of HBV-associated ALF. Multidimensional-scaling plot showing the 3D projection of eight liver specimens from two patients with HBV-associated ALF (four specimens per patient) and eight from individual liver donors with the use of all 15,652 transcripts that passed the filtering criteria. In the scatterplot, each point represents a liver specimen, and the distance between points is proportional to the overall dissimilarity of gene expression profiles. The eight samples from patients with ALF are shown in red (dark red dots, patient 1; light red dots, patient 2). The eight samples from the control liver donors are shown in blue. This plot illustrates how the gene expression profiles differentiate between the two classes.
Gene Expression Profiling of HBV-Associated ALF.
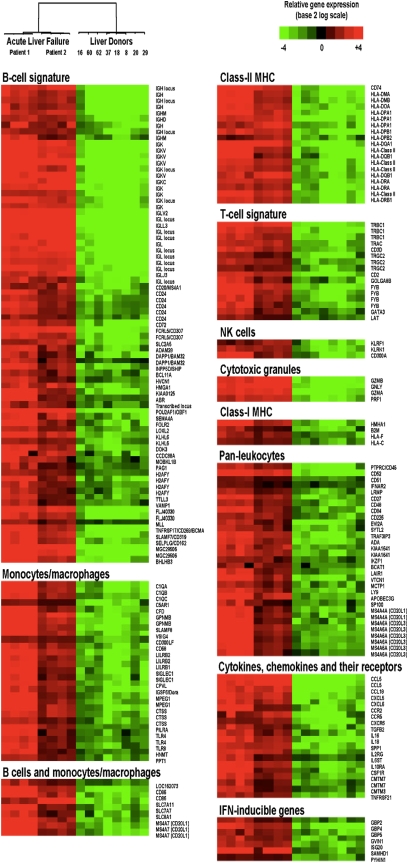
As a step toward understanding the pathogenesis of ALF, we performed gene expression profiling on eight liver specimens obtained from the two patients with ALF; as a control group, we studied individual liver specimens from eight HBV-negative liver donors (Table S2). Microarray was performed using Affymetrix Human U133 Plus 2 arrays, which contain 54,675 probe sets representing 29,486 unique human genes. Multidimensional scaling of all 16 liver specimens disclosed two distinct patterns of gene expression between ALF and liver donors, as shown by two clearly separated clusters (Fig. 1B), providing evidence of a common gene expression profile in ALF. Using a multivariate permutation test with a false discovery rate of <1%, a total of 1,368 transcripts were differentially expressed in ALF; 709 were up-regulated and 659 down-regulated. Supervised hierarchical clustering heat maps of the 16 individual liver specimens illustrated that both up- and down-regulated transcripts were clearly segregated between ALF and liver donors (Fig. S1 A and B). Genes involved in the immune response were the most represented ones among the up-regulated transcripts and showed the highest fold changes (Table S3). The most striking finding in ALF was an overriding B cell gene signature with a total of 77 B cell–related transcripts (Fig. 2 and SI Results), 31 of which were among the 50 most up-regulated transcripts (Table S4). Among the immune response genes, the Ig family was the most represented, with a similar distribution of κ and λ light chains, indicating the presence of a polyclonal B cell population. The B cell gene signature was also defined by cell-surface receptors, molecules involved in signal transduction, transcriptional regulators important for B cell development (including POU2AF1/OCA-B that was more than 40-fold up-regulated), genes selectively expressed or up-regulated in plasmablasts or plasma cells, and other molecules (SI Results for more details). Several other genes expressed within the B cell lineage but whose expression is shared with monocytes or macrophages, T cells, or other hematopoietic cells were also up-regulated (Fig. 2). Another prominent gene expression signature in ALF was that of the monocyte or macrophage lineage (Fig. 2). The presence of both B cells and macrophages in ALF livers was reflected by a strong intrahepatic expression of class II major histocompatibility complex genes (Fig. 2).
Fig. 2.
Supervised hierarchical cluster analysis showing the differential expression of immune response-related transcripts in HBV-associated ALF and control liver donors. Each row represents data for a particular human transcript, and each column the expression of transcripts in a single liver specimen. The color in each cell reflects the level of expression of the corresponding transcript in the corresponding sample, relative to its mean level of expression in the entire set of 16 samples. Mean-centered ratios of transcript expression are depicted by a log2-transformed scale. According to the color scale, up-regulated transcripts are shown in shades of red and down-regulated transcripts in shades of green.
In contrast with the prominent B cell gene signature, a limited number of genes associated with a T cell signature were up-regulated in ALF (Fig. 2), with only two transcripts among the 50 most up-regulated genes (Table S4). Also up-regulated were four transcripts encoding proteins contained in cytotoxic granules (Fig. 2), which are expressed by both cytotoxic T cells and natural killer cells. In accordance with the limited T cell gene signature, no prominent signs of IFN-γ response were seen in ALF, with only seven IFN-inducible transcripts recognized (Fig. 2). Strikingly, several cell-surface receptors that act as negative regulators of T cell activation were up-regulated (VSIG4, VTCN1/B7-H4, SLA, LAIR, LILRB1/CD85J, LILRB2/CD85D, and LAX1). Another major negative regulator, CTLA-4, showed substantial up-regulation (>7-fold changes; SI Results), although it did not achieve the stringent level of statistical significance that we had set (SI Materials and Methods). Another category of up-regulated genes was that of cytokines, chemokines, and their receptors (Fig. 2), including the chemokine receptors CCR2 and CXCR6 that are expressed by plasmablasts and plasma cells and have been implicated in the migration of these cells to inflamed tissues. We also found up-regulated keratin 7 and 19, which is consistent with the bile duct regeneration effort observed histologically in the liver. Additional functional classes are listed in the Table S3. The profile of down-regulated transcripts in ALF was indicative of massive hepatocellular damage with consequent shut-off of hepatic synthesis and the metabolic pathways that are regulated by the liver (Table S3).
Although there was a remarkable concordance in the up- and down-regulated transcripts between the two cases, consistent with the degree of histopathologic severity, there was a trend toward higher fold changes in patient 1 (massive hepatic necrosis) as compared with patient 2 (submassive hepatic necrosis), especially for the down-regulated transcripts (Fig. S2A), denoting a dramatic switch-off of gene expression in this extremely severe condition. The analysis of ALF gene expression profiles, using liver donors as controls, was further supported by a comparative analysis including a second group of normal liver specimens obtained from 11 patients who underwent liver resection for angioma (Fig. S2B). The analysis of differentially expressed genes detected by a multivariate permutation F test with a 0.0001 maximum proportion of false discoveries revealed a strong correlation between the two groups of normal livers relative to ALF.
Immunohistochemistry.
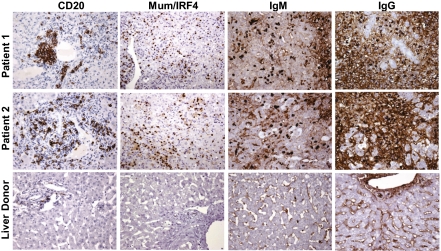
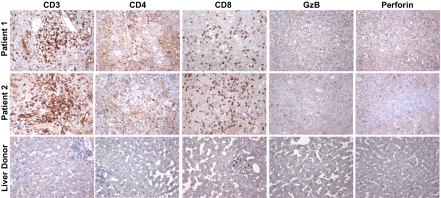
To validate the microrarray data using a different experimental approach, we performed an extensive immunohistochemical study on formalin-fixed, paraffin-embedded tissue sections. This analysis confirmed the prominent B cell signature observed by gene expression profiling, showing CD20-positive mature B cells present as clusters of different size in the portal areas and as single cells within the lobule (Fig. 3). Strikingly, there was an extensive infiltration of the lobules (and to a lesser extent of the portal areas) by plasmablasts and plasma cells expressing Mum1/IRF4 (Fig. 3) and CD138 (Fig. S3). Plasma cells were strongly positive for cytoplasmic IgM and IgG (Fig. 3), with a similar distribution of light chains (κ and λ) indicative of a polyclonal pattern (Fig. S4). The number of cells showing Mum/IRF4 positivity was very high and comparable to the number of cells expressing cytoplasmic IgM and IgG (Fig. 3). IgG-positive cells showed a greater range of differentiation with numerous plasmacytoid cells showing more open chromatin and prominent nucleoli (Fig. S5 A and B). Despite the limited expression of T cell genes, the liver was diffusely infiltrated by CD3-positive T cells, distributed both as aggregates in the portal areas and as single cells within the lobules (Fig. 4). The vast majority of the T cells were CD8 positive, whereas CD4-positive T cells were few and dispersed. The proportion of cells expressing granzyme B and perforin, two markers of cytotoxic function, was limited and markedly lower than the total CD8-positive population (Fig. 4). Expression of the natural killer cell marker CD56 was not observed. We also detected a massive liver infiltration by cells of the mononuclear phagocytic lineage expressing CD163 (Fig. S4), which correlated with the extent of liver necrosis. There was also a strong staining for keratin 7, reflecting extensive bile duct regeneration (Fig. S4). Staining for HBV antigens (HBsAg and HBcAg) and hepatitis D virus (HDV) antigen (HDAg) was negative.
Fig. 3.
Immunohistochemical staining of B cell lineage markers in liver tissue from two patients with HBV-associated ALF and a representative control liver donor. Liver specimens obtained at the time of OLT from the native livers of patient 1 with massive hepatic necrosis and patient 2 with submassive hepatic necrosis, and from a control liver donor, were stained with monoclonal antibodies against CD20, MUM1/IRF4, IgM, and IgG with the use of immunoperoxidase. Sections from patient 1 and patient 2 show the presence of B cell clusters (CD20) around the portal areas; strong nuclear staining for MUM1/IRF4 is seen in a large number of plasmacytoid cells and plasma cells diffusely distributed in the lobules; IgM-secreting mature plasma cells are seen predominantly in the lobules; IgG-secreting plasma cells show a greater range of differentiation with plasmacytoid cells containing more open chromatin and often a prominent nucleolus (Fig. S5 A and B). In addition, a diffuse staining for IgG, and to a lesser extent IgM, is seen in the intercellular spaces and in patient 2 on the surface of residual hepatocytes. Sections from the control liver show few CD20-positive cells within the portal space and limited Ig deposits in the sinusoids. (Magnification: ×75.)
Fig. 4.
Immunohistochemical staining of T cell lineage markers in liver tissue from two patients with HBV-associated ALF and a representative control liver donor. Liver specimens obtained at the time of OLT from the native livers of patient 1 with massive hepatic necrosis and patient 2 with submassive hepatic necrosis, and from a control liver donor, were stained with monoclonal antibodies against CD3, CD4, CD8, granzyme B (GzB), and perforin with the use of immunoperoxidase. Liver sections from patient 1 and patient 2 show the presence of T cell clusters (CD3), with CD8-positive cells diffusely distributed within the lobules and few dispersed CD4-positive cells. Low-level diffuse CD4 staining reflects the extensive tissue infiltration by macrophages. Only a small proportion of the CD8-positive cells express granzyme B, and an even smaller proportion express perforin. Sections from the control liver show only rare cells expressing T cell markers, granzyme B, or perforin. (Magnification: ×65.)
Immunohistochemical staining of liver tissues from the eight control liver donors was essentially negative. Only few lymphocytes, predominantly CD3- and CD8-positive T cells (Fig. 4) and very rare B cells (Fig. 3), were detected in the portal areas, and CD163-positive Kupffer cells within the sinusoids (Fig. S4).
Complement Deposition.
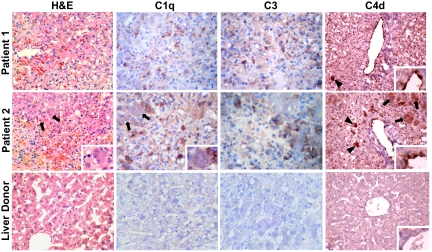
Because we had observed a strong intrahepatic expression of immunoglobulins, we investigated the deposition of complement factors in the liver tissue of patients with ALF. In both patients we documented a significant deposition of C1q, C3, and C4d in the liver parenchyma (Fig. 5). In patient 2, who had submassive hepatic necrosis, there was a strong signal for all complement markers associated with residual hepatocytes, as well as C1q staining focally along the intercellular spaces, suggestive of plasma–membrane binding. In both patients there was a strong signal for all complement markers associated with Kupffer cells showing a granular pattern, suggesting the presence of immune complexes internalized by these cells. Deposition of C4d was also detected on the surface of endothelial cells in the central veins (Fig. 5). All of the control liver tissues were negative for C1q, C3, and C4d (Fig. 5).
Fig. 5.
Degree of liver necrosis in HBV-associated ALF and immunohistochemical staining of complement in liver tissue from two patients with HBV-associated ALF and a representative control liver donor. Liver sections were stained with hematoxylin and eosin. In patient 1, the parenchyma shows massive hepatic necrosis with no viable hepatocytes; in patient 2, the parenchyma shows submassive liver necrosis with residual hepatocytes (arrows and Inset). The control liver donor shows normal liver histology. Liver sections obtained from patient 1 and patient 2, and from a normal liver donor, were also stained with antibodies against C1q, C3, and C4d with the use of immunoperoxidase. In patient 1, C4d staining is seen on the endothelial surface of the central vein (Inset), as well as focally in a macrophage-like cell (arrowhead). Deposition of C1q and C3 is seen mainly within macrophage-like cells, focally with a granular pattern suggestive of immune complexes. No residual hepatocytes are visible. In patient 2, C4d staining is seen on the endothelial surface of the central vein (Inset), as well as in macrophage-like cells (arrowheads) and in residual hepatocytes (arrows). Deposition of C1q and C3 is seen within macrophage-like cells, focally with a granular pattern suggestive of immune complexes. There is also C1q staining along intercellular spaces (Inset), suggesting plasma membrane deposition, and focal C3 staining of residual hepatocytes. Liver tissue from the control liver donor shows no significant staining for any of these complement proteins. (Magnification: ×90; Insets, ×270.)
Molecular Target of the IgG and IgM Antibodies Produced Within the Liver.
Gene expression profiling and immunohistochemistry provided evidence for an extensive production of immunoglobulins by infiltrating plasma cells in the liver of patients with HBV-associated ALF. To identify the target of these antibodies massively produced in the liver of patients with ALF, we generated phage display Fab libraries of both IgG1 and IgM from the liver tissue of each of the two patients. The phages were panned against several HBV proteins, namely HBsAg ad and ay, pre-S1 and pre-S2, HBcAg, and HBeAg. After two rounds of panning, a single viral antigen, HBcAg, was identified as the major target of the antibodies produced in the liver of both patients with ALF. Phage ELISA performed with 96 individual clones randomly selected for each library demonstrated a marked enrichment of HBcAg-specific clones for both the IgG1 (74% and 92% in patient 1 and 2, respectively) and the IgM (96% and 98%, respectively) libraries, whereas only a limited number of clones specific for HBsAg (14.5%) was detected, and solely in the IgG1 library of patient 2 who was also the only patient whose HBsAg converted to negative before OLT (Table 1).
Nucleotide sequence analysis of antibody heavy and light chains from a total of 276 clones obtained from the IgG1 and IgM libraries of the two patients demonstrated that the selected anti-HBc Fab clones used only two families of VH genes, with 184 clones (67%) using VH1 and 92 clones (33%) using VH3 (Table S5). A restricted repertoire was also found among light chains, with 215 clones (77.9%) using VK1, 15 clones (5.4%) using VK2, 43 clones (15.6%) using VK3, and 3 clones (1.1%) using VL1. Moreover, despite the isotype switch, most Ig genes lacked somatic mutations. Strikingly, 127 of the 276 anti-HBc clones (46%) contained an identical dominant heavy-chain gene with the variable domain in germline configuration (IGHV1-3) (Table 1). This predominant heavy chain was detected in both the IgG1 and IgM libraries of both patient 1 (34 and 57 clones, respectively) and patient 2 (8 and 28 clones, respectively) and paired with light chains from essentially a single family, VK1 (98.4%), indicating structural constraints (Fig. S6).
Discussion
The dramatic clinical course of ALF has posed major limitations for pathogenesis studies. Here we report a molecular definition of HBV-associated ALF, providing evidence for a primary role of humoral immunity in the pathogenesis of this disease. By studying liver tissue obtained at the time of OLT, we found that HBV-associated ALF is characterized by an overwhelming B cell response apparently centered in the liver, with massive intrahepatic production of IgM and IgG by plasma cells infiltrating the hepatic lobules. Extralymphatic Ig production has been documented in persistent viral infections (13, 14) and autoimmune diseases (15), but no evidence has been reported, to our knowledge, in acute viral infection (14). Our study demonstrates the ectopic production of specific antibodies during an acute viral infection, implicating a major role of B cell immunity in the pathogenesis of ALF. In contrast to the prominent B cell gene signature, only a limited number of transcripts related to the T cell lineage were detected in the liver of patients with ALF, with the notable absence of IFN-γ and its inducible chemokines, CXCL-9, -10, and -11, which are invariably up-regulated in the course of acute hepatitis B (16), a disease in which the liver damage is T cell mediated (4). Despite the up-regulation of few genes associated with cytotoxic granules, immunohistochemistry did not show a prominent expression of granzyme B and perforin in ALF. Thus, our data suggest that in ALF, in contrast to classic acute hepatitis B, the liver damage is not T cell mediated. The generation of an effective T cell response is regulated by a complex network of positive and negative signals. Interestingly, we documented the intrahepatic expression of several negative regulators of T cell activation, such as VSIG4 (17), VTCN1 (also known as B7-H4) (18), SLA (19), LAIR1 (20), LILRB1 and -2 (21, 22), LAX1 (23), and CTLA4 (24), providing a possible mechanism for the lack of T cell dominance in ALF.
Using phage display technology, we discovered that the massive intrahepatic antibody response in both patients with ALF was essentially directed against a single viral antigen, the HBcAg. This finding is consistent with the inherently enhanced immunogenicity of HBcAg, which unlike HBsAg and HBeAg was shown to function as a T cell–independent antigen (25). The lack of somatic mutations in the intrahepatic anti-HBc antibodies indeed supports a T cell–independent B cell response. A lack of somatic mutations is typically associated with B cell stimulation by T cell–independent antigens, which are recognized by germline antibodies. Thus, our data suggest that HBcAg was able to directly activate B cells to produce specific antibodies in an extralymphatic milieu in the absence of T cell–mediated help. But the most striking observation was the detection of a single predominant heavy chain with the variable domain in germline configuration among both IgG1 and IgM anti-HBc of two unrelated individuals with ALF. This result identifies HBcAg as the target of a germline human VH segment, providing an explanation for the T cell–independent nature of this antigen. This finding is in line with emerging evidence that the germline antibody repertoire is not shaped by purely stochastic phenomena but rather by a positive evolutionary selection (26), presumably driven by conserved antigenic structures of specific pathogens (27).
The significant deposition of complement components C1q, C3, and C4d documented in the liver tissue of both cases of ALF further supports a key role of humoral immunity in the pathogenesis of HBV-associated ALF. Thus, in addition to defective synthesis (28), complement consumption may contribute to the dramatic reduction of serum complement levels seen in ALF. In particular, C4d is considered a specific marker of humoral immune response in renal allografts (29), and more recently it has been detected in acute liver allograft humoral rejection (30). Our data are consistent with the hypothesis that anti-HBc antibodies massively produced in the liver may combine with HBcAg, leading to the extensive formation of antigen–antibody complexes on the surface of hepatocytes and Kupffer cells, resulting in the activation of the classical complement pathway and massive liver necrosis. Several studies performed in patients with chronic HBV infection have shown that HBcAg, albeit commonly detected in the nucleus, is also expressed on the liver cell membrane (31–34).
In summary, the results presented in this study provide insights into the pathogenesis of HBV-associated ALF, illustrating the potential of gene expression profiling for elucidating the molecular basis of complex human diseases. Our data strongly suggest that HBV-associated ALF is mediated by a T cell–independent intrahepatic B-cell response against the core antigen of HBV that is associated with complement-mediated hepatocellular damage. The immunologic memory of these intrahepatic B cells has been cloned, and the presumably pathogenic antibodies are being characterized. These results may have implications for the development of novel therapeutic strategies for the treatment of HBV-associated ALF.
Materials and Methods
Study Subjects and Controls.
We studied two young adults with HBV-associated ALF (SI Materials and Methods). As a control group, we studied eight liver donors; a second control group comprised normal liver specimens obtained from 11 subjects during resection for liver angioma (SI Materials and Methods and Table S2).
Methods.
Details of serologic and virologic assays, gene expression profiling, and immunohistochemistry, as well as details on the construction of Fab-display phage libraries, selection of specific Fab clones, and sequence analysis of anti-HBc positive Fab clones, are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank P. Lusso, F. Marincola, and R. Dalla-Favera for their valuable comments; H. J. Alter for critically reviewing the manuscript; S. Dedola for taking care of the patients; V. Congias, M. Cuccuru, R. Strazzera, S. Farci, D. Cao, and R. Scioscia for their technical help; and K. Prims for editorial assistance. W. H. Gerlich generously supplied prototype monoclonal antibodies against pre-S1 and pre-S2. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
Data deposition: The complete-microarray data set is available at the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE14668).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003854107/-/DCSupplemental.
References
- 1.Lee WM. Acute liver failure. N Engl J Med. 1993;329:1862–1872. doi: 10.1056/NEJM199312163292508. [DOI] [PubMed] [Google Scholar]
- 2.Escorsell A, Mas A, de la Mata M, Spanish Group for the Study of Acute Liver Failure Acute liver failure in Spain: Analysis of 267 cases. Liver Transpl. 2007;13:1389–1395. doi: 10.1002/lt.21119. [DOI] [PubMed] [Google Scholar]
- 3.Koskinas J, et al. Aetiology and outcome of acute hepatic failure in Greece: Experience of two academic hospital centres. Liver Int. 2008;28:821–827. doi: 10.1111/j.1478-3231.2008.01782.x. [DOI] [PubMed] [Google Scholar]
- 4.Thimme R, et al. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fagan EA, Williams R. Fulminant viral hepatitis. Br Med Bull. 1990;46:462–480. doi: 10.1093/oxfordjournals.bmb.a072410. [DOI] [PubMed] [Google Scholar]
- 6.Trepo CG, et al. Hepatitis B antigen (HBSAg) and/or antibodies (anti-HBS and anti-HBC) in fulminant hepatitis: Pathogenic and prognostic significance. Gut. 1976;17:10–13. doi: 10.1136/gut.17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabor E, Gerety RJ, Hoofnagle JH, Barker LF. Immune response in fulminant viral hepatitis, type B. Gastroenterology. 1976;71:635–640. [PubMed] [Google Scholar]
- 8.Gimson AE, Tedder RS, White YS, Eddleston AL, Williams R. Serological markers in fulminant hepatitis B. Gut. 1983;24:615–617. doi: 10.1136/gut.24.7.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang TJ, Hasegawa K, Rimon N, Wands JR, Ben-Porath E. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N Engl J Med. 1991;324:1705–1709. doi: 10.1056/NEJM199106133242405. [DOI] [PubMed] [Google Scholar]
- 10.Sato S, et al. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med. 1995;122:241–248. doi: 10.7326/0003-4819-122-4-199502150-00001. [DOI] [PubMed] [Google Scholar]
- 11.Wai CT, et al. US Acute Liver Failure Study Group. Clinical outcome and virological characteristics of hepatitis B-related acute liver failure in the United States. J Viral Hepat. 2005;12:192–198. doi: 10.1111/j.1365-2893.2005.00581.x. [DOI] [PubMed] [Google Scholar]
- 12.Brunetto MR, Rodriguez UA, Bonino F. Hepatitis B virus mutants. Intervirology. 1999;42:69–80. doi: 10.1159/000024968. [DOI] [PubMed] [Google Scholar]
- 13.Moskophidis D, Löhler J, Lehmann-Grube F. Antiviral antibody-producing cells in parenchymatous organs during persistent virus infection. J Exp Med. 1987;165:705–719. doi: 10.1084/jem.165.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moskophidis D, Frei K, Löhler J, Fontana A, Zinkernagel RM. Production of random classes of immunoglobulins in brain tissue during persistent viral infection paralleled by secretion of interleukin-6 (IL-6) but not IL-4, IL-5, and gamma interferon. J Virol. 1991;65:1364–1369. doi: 10.1128/jvi.65.3.1364-1369.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin F, Chan AC. B cell immunobiology in disease: Evolving concepts from the clinic. Annu Rev Immunol. 2006;24:467–496. doi: 10.1146/annurev.immunol.24.021605.090517. [DOI] [PubMed] [Google Scholar]
- 16.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogt L, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sica GL, et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 2003;18:849–861. doi: 10.1016/s1074-7613(03)00152-3. [DOI] [PubMed] [Google Scholar]
- 19.Sosinowski T, Pandey A, Dixit VM, Weiss A. Src-like adaptor protein (SLAP) is a negative regulator of T cell receptor signaling. J Exp Med. 2000;191:463–474. doi: 10.1084/jem.191.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyaard L. The inhibitory collagen receptor LAIR-1 (CD305) J Leukoc Biol. 2008;83:799–803. doi: 10.1189/jlb.0907609. [DOI] [PubMed] [Google Scholar]
- 21.Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: Witness of the past, actors of the future. Nat Rev Immunol. 2004;4:190–198. doi: 10.1038/nri1306. [DOI] [PubMed] [Google Scholar]
- 22.Liang S, Zhang W, Horuzsko A. Human ILT2 receptor associates with murine MHC class I molecules in vivo and impairs T cell function. Eur J Immunol. 2006;36:2457–2471. doi: 10.1002/eji.200636031. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro MJ, Nguyen CT, Aghajanian H, Zhang W, Shapiro VS. Negative regulation of TCR signaling by linker for activation of X cells via phosphotyrosine-dependent and -independent mechanisms. J Immunol. 2008;181:7055–7061. doi: 10.4049/jimmunol.181.10.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teft WA, Kirchof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. 2006;24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535. [DOI] [PubMed] [Google Scholar]
- 25.Milich DR, McLachlan A. The nucleocapsid of hepatitis B virus is both a T-cell-independent and a T-cell-dependent antigen. Science. 1986;234:1398–1401. doi: 10.1126/science.3491425. [DOI] [PubMed] [Google Scholar]
- 26.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson CA, et al. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008;27:2592–2602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyke RJ, Rajkovic IA, Eddleston AL, Williams R. Defective opsonisation and complement deficiency in serum from patients with fulminant hepatic failure. Gut. 1980;21:643–649. doi: 10.1136/gut.21.8.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racusen LC, Haas M. Antibody-mediated rejection in renal allografts: Lessons from pathology. Clin J Am Soc Nephrol. 2006;1:415–420. doi: 10.2215/CJN.01881105. [DOI] [PubMed] [Google Scholar]
- 30.Lorho R, et al. C4d: A marker for hepatic transplant rejection. Transplant Proc. 2006;38:2333–2334. doi: 10.1016/j.transproceed.2006.06.120. [DOI] [PubMed] [Google Scholar]
- 31.Gudat F, Bianchi L. Evidence for phasic sequences in nuclear HBcAg formation and cell membrane-directed flow of core particles in chronic hepatitis B. Gastroenterology. 1977;73:1194–1197. [PubMed] [Google Scholar]
- 32.Trevisan A, et al. Core antigen-specific immunoglobulin G bound to the liver cell membrane in chronic hepatitis B. Gastroenterology. 1982;82:218–222. [PubMed] [Google Scholar]
- 33.Chu CM, Liaw YF. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology. 1987;92:220–225. doi: 10.1016/0016-5085(87)90863-8. [DOI] [PubMed] [Google Scholar]
- 34.Saito T, Kamimura T, Ishibashi M, Shinzawa H, Takahashi T. Electron microscopic study of hepatitis B virus-associated antigens on the infected liver cell membrane in relation to analysis of immune target antigens in chronic hepatitis B. Gastroenterol Jpn. 1992;27:734–744. doi: 10.1007/BF02806526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.