Abstract
We report the crystal structure of release factor 2 bound to ribosome with an aminoacyl tRNA substrate analog at the ribosomal P site, at 3.1 Å resolution. The structure shows that upon stop-codon recognition, the universally conserved GGQ motif packs tightly into the peptidyl transferase center. Nucleotide A2602 of 23S rRNA, implicated in peptide release, packs with the GGQ motif in release factor 2. The ribose of A76 of the peptidyl-tRNA adopts the C2′-endo conformation, and the 2′ hydroxyl of A76 is within hydrogen-bond distance of the 2′ hydroxyl of A2451. The structure suggests how a catalytic water can be coordinated in the peptidyl transferase center and, together with previous biochemical and computational data, suggests a model for how the ester bond between the peptidyl tRNA and the nascent peptide is hydrolyzed.
Keywords: ribosome structure, translational termination, X-ray crystallography
The ribosome translates the genetic information present in mRNA into proteins (1). A crucial step of this process is the termination of protein synthesis, which involves the cleavage and release of the nascent peptide chain from the P-site tRNA when the end of the coding sequence is reached. Translational termination by the ribosome is a precise and complex step that occurs when a stop codon of an mRNA is encountered in the A site of the small ribosomal subunit. The stop codons are recognized by proteins called class I release factors (RFs). In bacteria, two class I RFs recognize the three stop codons with overlapping specificity: RF1 recognizes UAG and RF2 recognizes UGA, whereas both factors recognize UAA (2). Upon stop-codon recognition, the class I RF promotes the hydrolysis of the ester bond between the nascent polypeptide and the peptidyl tRNA at the ribosomal P site, leading to the release of the nascent polypeptide and the termination of protein synthesis (3). Eukaryotes and archaea possess a single “omnipotent” class I RF, eRF1 or aRF1, respectively, which recognizes all three stop codons (4, 5). eRF1 and aRF1 are highly homologous with each other and were proposed to have evolved independently from their bacterial counterpart (4).
A major advance in our understanding in the specificity of stop-codon recognition and peptide release at the molecular level was achieved when three high-resolution crystal structures of the 70S ribosome complexed with class I RFs and their cognate stop codons were solved at atomic resolution (6–8). An analysis of the interactions of RF1 and RF2 with the decoding center in these structures provides explanations for the specificity of stop-codon recognition by these factors. However, because all three structures represent the product state with a deacylated tRNA in the P site, the mechanism of peptide release is less clear.
A universally conserved GGQ motif (9) that was shown to be required for catalytic activity (10, 11) was indeed found in the peptidyl transferase center (PTC) in earlier structures by cryoEM (12, 13) or crystallography at ∼6 Å (14). Mutations introduced at the first two conserved glycines of the GGQ motif completely abolished the peptide release activity in both bacteria and eukaryotes, whereas mutating the conserved glutamine to alanine, arginine, and glutamates leads to only about 50–80% loss of the peptide release activities in bacteria (11) and eukaryotes (9, 15). More recently, results from in vitro pre-steady-state kinetic studies suggest that the side chain of the glutamine is important for specifically selecting water as a nucleophile for the hydrolysis of the peptidyl tRNA (16). Apart from protein RFs, the ribosome also conceivably plays an important role in translational termination. Experimental evidence suggesting that both the peptidyl transferase and release reactions take place in the large ribosomal subunit and are closely related dates back to 1969 (17). The fact that the presence of a cognate tRNA in the ribosomal A site in 30% acetone can trigger peptide release in the absence of a class I RF provides important evidence that the PTC is directly involved in peptide release (18). Mutations at A2451, U2506, U2585, and A2602 in the 23S rRNA affect peptide release to varying degrees, with the most deleterious effect observed in the A2602 mutants (19, 20). Similar to the peptidyl transferase reaction, the 2′ hydroxyl (2′ OH) of A76 from the peptidyl tRNA was shown to be critical for peptide release (21). The importance of A2602 and A76 of peptidyl tRNA was also demonstrated in molecular dynamics calculations (22).
The high-resolution structures (6–8) show that the two glycines in the GGQ adopt conformations that are not possible for other amino acids, explaining why they are conserved and their mutation to alanines results in a reduction of peptide release activity. However, the role of the glutamine in catalysis remains uncertain. Two of the three studies suggested that the main-chain amide group of the conserved glutamine was involved in catalysis (6, 8), whereas the third also postulated a role for the glutamine side chain (7). Additional high-resolution structures that represented the substrate and transition state complexes would therefore be useful to shed light on the mechanism of catalysis. Among this the key questions are as follows: (i) Does the glutamine side chain contribute to the catalysis of peptide release? (ii) Do any nucleotides in the 23S rRNA also possibly contribute to catalysis? (iii) What is the structural basis for the catalytic role of the 2′ OH group in A76 of the peptidyl tRNA? (iv) How could a catalytic water be positioned by important structural features of the ribosome, peptidyl tRNA, and RF for optimal nucleophilic attack on the ester bond between tRNA and nascent peptide?
Here we report the crystal structure at 3.1 Å resolution of the 70S ribosome in complex with RF2, a cognate UAA stop codon in the A site and a Phe-NH-tRNAPhe at the P site (Table S1). In the Phe-NH-tRNAPhe, the ester bond between the amino acid phenylalanine and the 3′ carbon of A76 of tRNAPhe is replaced by an amide bond that is not hydrolyzed under our experimental conditions (Fig. S1). The crystal structure therefore mimics the substrate complex prior to catalysis. The interactions of ribosome, RF2, and peptidyl tRNA in the PTC prior to catalysis can be directly visualized, as can some possible water molecules that could be of importance. These interactions, together with the previously available biochemical and computational data, lead to a proposal for the mechanistic model for peptide release.
Results and Discussion
As expected, the overall structure of the release complex we report here is similar to that of the three previous release complexes (6–8) (Fig. S2). The PTC of the ribosome is in an activated or “induced” state in our structure (19, 23). Superposition of the 23S rRNA from this structure on that from the complex of the 50S subunit with a transition state analog of the peptidyl transferase reaction [1VQN (23)] reveals that these two are very similar, with the notable exception of A2602. The binding of RF2 to the ribosome, similar to the binding of a cognate tRNA to the ribosome A site, induces changes in the conformation of U2584, U2585, and U2506 that expose the ester bond between the last amino acid in the nascent peptide and P-site tRNA to the nucleophilic attack of a water molecule.
The Conformation of the GGQ Motif.
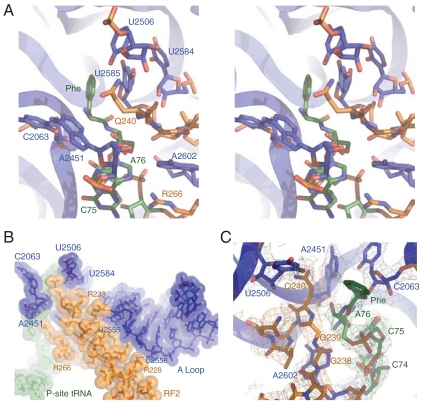
The catalytic GGQ motif makes extensive interactions in the PTC (Fig. 1). This catalytic motif is positioned in the PTC by a network of hydrogen-bond interactions between the highly conserved residues in class I RFs and the 23S rRNA. The positively charged guanidino groups of the highly conserved arginines R233 and R228 are found forming hydrogen-bond interactions with the 2′ OH of the nucleotide U2555 and the phosphate backbones of the nucleotide C2558 in the A loop (nucleotide 2547-2561, Escherichia coli numbering) of the 23S rRNA, respectively (Fig. 1B). Q268 in RF2 forms hydrogen bond interaction with the 2′ OH of the nucleotide U2492 (Fig. S3). The extended stretch GPGGQ (residue 236 to residue 240) in RF2 is positioned in between 23S rRNA nucleotide U2584 and U2585 on one side and A2602 on the other (Fig. 1A). As what was observed in the previous structures of the release complex (6–8), the backbone of the two conserved glycines, G238 and G239, adopts a conformation that is not possible for any other amino acid, explaining why their mutation reduces catalytic activity significantly (16, 24). The conformation of these glycines also helps the proper placement of the conserved Q240 in the core of the PTC, where it forms a densely packed catalytic pocket with the A76 of the peptidyl tRNA and A2451, C2063, U2585, and U2506 of the 23S rRNA (Fig. 2A). The side-chain amine of the Q240 in the refined structure is 3.66 Å in distance with the 2′ OH of U2506. The main-chain amide of the Q240 is close in proximity to the 2′ OH of the A76 with a distance of 3.27 Å. The distances between the carbonyl oxygen and the amide nitrogen of the Q240 to the attacking center, the carbonyl carbon in the Phe-NH-tRNAPhe, are 6.78 and 4.94 Å, respectively.
Fig. 1.
GGQ motif in the PTC. (A) Stereo view of the PTC showing the GGQ motif of RF2 (Orange), important bases of 23S rRNA (Blue), and the CCA-NH-Phe of P-site tRNA (Green). (B) Overview of the interactions between RF2 (Orange) and the A loop of the 23S rRNA (Blue) that help position the GGQ motif in the PTC. P-site tRNA is colored in green, and important nucleotides at the PTC are colored in blue. (C) σA weighted 3mFobs-2DFcal map from experiments: GGQ motif of RF2 (Orange), bases in 23S rRNA (Gray), and CCA-NH-Phe of P-site tRNA (Green) displayed with a map calculated at 3.1 Å.
Fig. 2.
The core of the PTC in translation termination. (A) Catalytic pocket created by Q240 in the PTC from the structure. Upon stop-codon recognition, the Q240 of the RF2 extends into the core of the PTC and creates a catalytic pocket to accommodate one water molecule for the peptide release. Important nucleotides in the PTC are shown in blue, RF2 is shown in orange, and the A76 of the P-site tRNA is shown in green. (B) Conformation of important residues in the PTC. A 3mFobs-2DFcal electron density (Raspberry) in the PTC from the experiment showing a possible coordinated water molecule. 3mFobs-2DFcal electron density for A76-NH-Phe of the P-site substrate (Green), A2451 and A2602 in the 23S rRNA (Gray), and GGQ motif of the RF2 (Orange). The sugar pucker of the A76 adopts the C2′-endo conformation. The 2′ OH of the A76 is within hydrogen-bond distance of the 2′ OH of the A2451.
Possible Role of the Glutamine of the GGQ Motif.
This structure sheds light on the role that the universally conserved Q240 in the GGQ motif plays in peptide release at the molecular level. Q240 creates a tightly packed catalytic pocket to accommodate a water molecule (Fig. 2A). Such a packing role of the Q240 was initially proposed from biochemical studies (16). By using hydroxylamine as a competitor nucleophile to water, Shaw and Green showed that the serine, alanine, and glycine substitutions for the conserved glutamine resulted in a substantial increase of the rate of aminolysis, whereas the tryptophan, lysine, and leucine substitutions did not, even though all these mutations gave rise to a modest decrease in the observed rate of hydrolysis. Therefore, the optimal packing by the glutamine is important for specifically selecting water as a nucleophile in peptide release. Consistent with these results, recent molecular dynamic simulations showed that the side chain of the glutamine made an entropic contribution to the hydrolytic reaction (25).
The packing of this pocket by class I RFs in the ribosome should be further facilitated by the natural posttranslational modification of the glutamine. In bacteria, this glutamine residue is methylated at the N5 position. The methylation was reported to have a noticeable effect on the RF2- but not RF1-dependent release activity in vitro (26). The same modification is also found in eukaryotes and is required for optimal cell growth (27). The glutamine residue of the GGQ motif in RF2 under this study is not fully methylated. Methylation could help to position the amine away from the catalytic center and thus orient the carbonyl oxygen of the Q240 toward the catalytic water for optimal attack on the ester bond. Such a catalytic role of the glutamine methylation was demonstrated in recent molecular dynamics simulation studies (22, 25).
The observation that a Q240E mutant has reasonable peptide release activity supports a role for the carbonyl oxygen of Q240 in coordinating the catalytic water (16). However, a water molecule, in addition to its capacity to form a strong hydrogen bond with its neighboring atoms, can rapidly rotate, which leads to the rapid reorientation of the hydrogen bonds that it can form alternatively with other nearby functional groups when the catalytic glutamine is absent. Apart from the functional groups on the conserved glutamine, there are other functional groups in the catalytic center that can potentially coordinate a water molecule for the nucleophilic attack of the ester bond. These functional groups include the 2′ OH of the A76, the N3 and 2′ OH of the A2451, the O2 and the 2′ OH of the C2063, and also possibly the backbone amide NH of the terminal polypeptide. The existence of these functional groups is likely to be one of the reasons that mutations in Q240 result only in a reduction but not loss of the peptide release activity in vitro. In addition, other functional groups introduced in some mutational studies have the potential to coordinate a water molecule. A molecular dynamics simulation suggests the involvement of a second water in the case of a Q240A mutation (22). Taken together, it is very likely that the side chain of the conserved glutamine directly contributes to the catalysis of peptide release. However, other studies in both bacteria (11) and eukaryotes (15) show a significant drop in activity in the Q240E mutant, suggesting that apart from creating a confined space to accommodate the catalytic water molecule at the PTC, as seen in this structure, the precise roles of the different functional groups in the conserved glutamine still remain elusive.
Mutational studies show that any other base substitution for A2451, U2506, and C2063 results in an insignificant rate reduction in the ester peptidyl-tRNA hydrolysis (19, 20). It is very possible that the mutant ribosomes are still able to properly orient the glutamine side chain for coordinating a water molecule, because other functional groups in the substituted nucleotides are able to make similar interactions with them. The potential positions in the heterocyclic base, O2 vs. N3, or the 2′ OH in the ribose, are “equivalent” in all mutations as far as their capacity to serve as hydrogen-bond donors or acceptors are concerned. Therefore, the mutant ribosome at these positions can still maintain a reasonable level of peptide release activity. Finally, because of the confined space in the catalytic core, it is possible that a water molecule, even when only partially coordinated, could be effective in hydrolyzing the ester bond between peptidyl tRNA and the nascent peptide.
A Possible Water Molecule in the Core of the Catalytic Center.
Because the complex under our study represents a substrate bound to the ribosome prior to catalysis, it is reasonable to assume that it has acquired the conformation that is necessary for nucleophilic attack by a water molecule. This water molecule, if visible, should be located in the catalytic pocket created by Q240. In our initial refinement, the unbiased Fo - Fc map shows an obvious positive density between A76-NH-Phe and the Q240 at a contour level of 2.5σ, which suggests the presence of a water molecule. After a water molecule was placed into this density, further refinement showed the density for this molecule. This water appears to be coordinated by the 2′ OH of the A76, N3 of the A2451, and NH2 of the Phe attached to the A76 (Fig. 2B). A recent computational study suggested a catalytic water at a similar position that can be coordinated by the 2′ OH of the A76, the N3 of the A2451, and the carbonyl oxygen of the methylated Q240 for peptide release (25).
Previous studies on small organic molecules show that the preferred angle for a nucleophilic attack on a carbonyl carbon is about 107° from the targeted carbonyl plane (28). The electron density described above enables building of one water molecule in a position nearly suitable for such an attack. It would be attractive if this water could be positioned by donating its two hydrogen atoms to the carbonyl oxygen of the Q240 and 2′ OH of A76 and possibly accepting one hydrogen atom from the main-chain amide of the Q240 (7). As such, the available lone pair of the structured water could attack the ester carbonyl carbon from the preferred angle. In the current model, the assigned water is too far to hydrogen bond with carbonyl oxygen of Q240, at a distance of 4.17 Å. Though it is possible that the water molecule we observe in the densely packed catalytic core is the one that is crucial for peptide release, it is likely that, though trapped in the confined space of the catalytic pocket, it is not positioned optimally and would have to shift into proper position prior to catalysis.
Compared to the proposed ideal position for catalysis (28), the assigned water molecule is displaced by ∼2.9 Å away from the glutamine. Although substrate analogs similar to what we have used in this study have been used in several studies on the PTC (23, 29, 30), its modified chemical nature can account for the displacement of the free water molecule relative to its optimal position for catalysis. First, the NH2 in the Phe in our current Phe-NH-tRNAPhe substrate analog would normally be a main-chain amide in the nascent polypeptide in translational termination. Thus the substrate analog introduces a free NH2 group compared to a more restrained amide group in the nascent polypeptide during termination. The introduced amine is not protonated under our experimental conditions and is a relatively better hydrogen-bond donor than the amide hydrogen; therefore, it is more likely to be involved in coordinating a water molecule. Furthermore, the identity of the last two amino acids of the nascent peptide was shown to affect translational termination efficiency at a specific stop codon (31), suggesting that the structure of the C terminus of the nascent peptide is important for termination. Second, the ester oxygen is more electronegative than the amide nitrogen. In addition, the peptide bond (HN-CO) that resulted from introducing an amide linkage in the Phe-NH-tRNAPhe has partial double bond character. These differences will result in a carbonyl carbon atom connected to an amide bond to be less suitable for a nucleophilic attack by a water molecule compared with the one with an ester linkage. Third, the amide bond in the Phe-NH-tRNAPhe could also introduce a potential hydrogen-bond donor, thus changing the local electrostatic environment around the targeted carbonyl group. Finally, as mentioned previously, in its natural form, the position of the carbonyl oxygen of the Q240 should be different because of the methylation of its N5 position. The methylation could facilitate in positioning the carboxyl group of the Q240 closer to optimally coordinate the catalytic water. All these differences with the real substrate complex may well account for the less-than-optimal location of the observed water molecule in the structure.
The Role of the 2′ Hydroxyl of the Terminal A76 of tRNA.
The 2′ hydroxyl of A76 of the peptidyl tRNA was shown to be critical to the efficiency of peptide release. Both F and H substitutions to the 2′ OH resulted in a drastic decrease to the rate of RF1-mediated peptide release (21). The sugar pucker of the A76 of the peptidyl tRNA adopts the C2′-endo conformation in our structure, as has been found previously for aminoacyl tRNA analogues in the P site (32). In the C2′-endo conformation, the bond between C3 and N3 is approximately perpendicular to the sugar plane. This conformation helps to position the ester carbonyl carbon optimally for catalysis. The 2′ OH of A76 is also within the hydrogen-bond distance of the 2′ OH of A2451 (Fig. 2B).
On the basis of the structure, the functional groups that are most likely to coordinate a catalytic water are the 2′ OH of A76, the N3 of A2451, the side-chain carbonyl oxygen, and the backbone amide hydrogen of the Q240. Among them, the 2′ OH of A76, which can act as both a hydrogen-bond donor and acceptor, is probably the most effective. This functional group was suggested to facilitate the peptidyl transferase reaction through a proton shuttle mechanism (29, 33–35). A similar proton shuttle mechanism for the peptide release was suggested from biochemical studies (21) and on the basis of molecular dynamic calculations (22, 25).
Possible Role of the Nucleotide A2602.
Among the four essential nucleotides located in close proximity (the so-called “inner shell”) of the PTC (19), A2602 undergoes the most pronounced conformational change upon the binding of RF2. Structural studies on the 50S ribosomal subunit using a transition state analog suggest that A2602 and U2584 coordinate a water molecule that is likely to stabilize the negatively charged oxyanion of the transition state intermediate during peptide bond formation (29). Such an interaction is broken in our current structure. Instead, the conformation of the A2602 changes to interact with the conserved elements in RF2. The base moiety of the A2602 packs between the smooth backbone of the GGQ in RF2 (Fig. 1). Although functionally important, A2602 is unlikely to directly activate or coordinate a catalytic water molecule for peptide release, because the distance of A2602 and the amide carbonyl carbon is too far for a direct interaction. Rather, A2602 probably helps to stabilize the GGQ loop in a catalytically active conformation.
Whether A2602 could also help to guide the potential catalytic water molecule into the catalytic pocket that is then activated for the ester bond hydrolysis is a question for future investigation. A possible water molecule was modeled into the density adjacent to A2602 and A2451 so that it has the potential to be coordinated by the NH2 of the A2602 and the 2′ OH of the A2451 in the 23S rRNA. However, conclusive proof of the existence of additional water molecules requires a 70S/RF2 structure at higher resolution.
A Possible Mechanistic Model for Peptide Release.
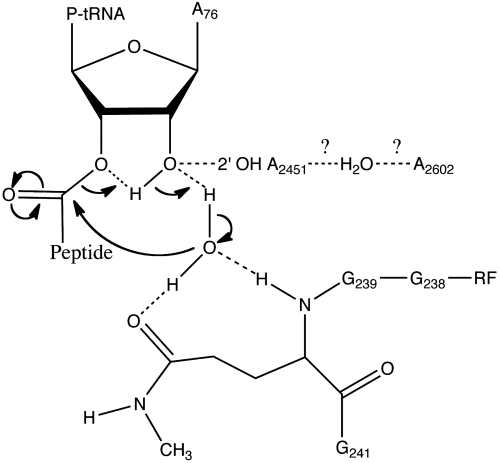
Previous biochemical and computational data suggest that a proton shuttle mechanism could be involved in translational termination (21, 22, 25). This study provides direct structural support for this idea. On the basis of the available data, a possible mechanism by which the ester bond between nascent polypeptide and peptidyl tRNA is hydrolyzed in translational termination is suggested (Fig. 3). During this process, a class I RF binds to the ribosomal A site where it recognizes the stop codon in mRNA. Binding of the protein release factor to the ribosome induces changes at the PTC to allow the entry of a water molecule into the catalytic pocket, a tightly packed space created by the conserved GGQ motif in class I RFs. Numerous interactions between highly conserved residues in the RFs, 23S rRNA, and the CCA end of the peptidyl tRNA help induce a structure specific to the RF-bound ribosome. The 23S rRNA nucleotide A2602, in particular, packs with the GGQ motif in the RF. It might also coordinate a water molecule with the nucleotide A2451, though such a role of A2602 in termination remains to be determined. The catalytic glutamine creates a tight pocket for the specific selection of a catalytic water molecule. The positioning of the catalytic water is likely to be achieved by the hydrogen-bond interactions with the 2′ OH of the A76 and the highly conserved glutamine in the RF. The chemical reaction likely proceeds through a concerted proton shuttle mechanism involving the 2′ OH of peptidyl-tRNA A76 (Fig. 3). In the catalytic pocket of the release complex structure, no monovalent and divalent ions were observed, suggesting that metal ions are unlikely to directly coordinate a water molecule in peptide release. The main-chain amide of the Q240 forms a hydrogen bond with the 3′ OH of the deacylated peptidyl tRNA and thus is involved in the stabilization of the product, as seen in previous structures (6–8), although it is not clear whether this interaction has any role in catalysis. It is conceivable that peptide release shares certain mechanistic similarities with peptidyl transfer reaction (36), considering the similar chemistry as well as their common catalytic site in the ribosome. However, because of the involvement of a protein release factor and a completely different nucleophile (i.e., water) in peptide release, it is also conceivable that the two reactions will also have some differences.
Fig. 3.
A possible mechanistic model for the peptide release. Upon stop-codon recognition, class I RFs coordinate with the active site of the ribosome to create a tightly packed catalytic pocket. The highly conserved 23S rRNA nucleotide A2602 is likely involved in stabilizing the GGQ motif. The catalytic water molecule is coordinated by the 2′ OH of the A76 and the Q240 to launch a nucleophilic attack on the carbonyl carbon of the ester linkage. The hydrolytic chemical reaction likely proceeds through a concerted proton shuttle mechanism involving 2′ OH of the A76 that is the similar to the one employed in peptide bond formation (21, 22, 25).
Although the substrate analog we used in this study binds effectively to the ribosomal P site, we are aware of the possibility that the conformational and chemical differences caused by an amide substitution to a biological ester bond could prevent the catalytic water from being optimally positioned for the nucleophilic attack. In addition, terminal amino acids were shown affecting translational termination efficiency (31). Furthermore, the phenylalanine attached to the A76 of the peptidyl tRNA in our structure is not particularly well ordered, as was also the case in a previous structure (30). Given the resolution, water molecules were modeled in only one of the two molecules in the asymmetric unit where the density is better resolved, and we cannot fully exclude the possibility of the existence of an alternative conformation of the glutamine in the GGQ motif of the RF2 in the structure. Moreover, the precise conformation of the conserved glutamine is also likely to be subtly influenced by the N5 methylation that normally occurs in vivo. Clearly, higher resolution crystal structures of the ribosome with posttranslationally modified RF2 and transition state analogs as well as dipeptides or longer substrate analogs will help to further elucidate the mechanism of peptide release in translational termination.
Although eukaryotic and bacterial class I RFs share little sequence or structural identity, the GGQ motif involved in catalysis is conserved from E. coli to humans, as is the PTC of the ribosome. Mutations in the GGQ motif in E. coli and human class I RFs have similar effects for cell survival in vivo (10, 11) as well as on the rate of peptide release in vitro (15, 24). These results suggest strongly that, despite the differences in class I RFs among kingdoms, the mechanism for peptide release is likely to be conserved from bacteria to humans.
Conclusion
The catalytic mechanism of peptide release has been a fundamental question in molecular biology for decades. Using a substrate analog that prevents hydrolysis of the amino acid from the peptidyl tRNA, we have determined the ribosome structure complexed with release factor in the state just prior to catalysis. Together with available biochemical and computational data, the structure sheds light on a possible mechanism by which the ester bond between the last amino acid of the nascent polypeptide chain and P-site tRNA is hydrolyzed.
Materials and Methods
Thermus thermophilus ribosomes, T. thermophilus RF2, and E. coli tRNAPhe were purified as described previously (7, 32). Chemically modified Phe-NH-tRNAPhe containing an amide bond between A76 and the amino acid phenylalanine was obtained by incorporating the chemically synthesized 3′-amino-3′-deoxy adenosine nucleotide as the 3′ A76 of the purified tRNAPhe (30, 37). The mRNA used was chemically synthesized by Dharmacon and had the sequence 5' GGC AAG GAG GAA AAA UUC UAA UAC A 3′, which contained a UUC Phe codon in the P site (bold) and an UAA stop codon in the A site (underlined bold).
Complexes of RF2 with the ribosome containing Phe-NH-tRNAPhe and mRNA with a UAA codon in the A site were crystallized as described previously (7). X-ray diffraction data were collected at beamline X10SA at the Swiss Light Source, Villigen. A detailed description on the crystallization and structure determination is given in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank T. Martin Schmeing for a critical review of the manuscript; Chyan Leong Ng, Kathrin Lang, and Albert Weixlbaumer for helpful discussions; and Clemens Schulze-Briese, Meitian Wang, and Chitra Rajendran for advice and help with data collection at the Swiss Light Source. This work was supported by the Medical Research Council UK, the Welcome Trust, Agouron Institute, and the Louis-Jeantet Foundation. H.J. is the recipient of a Ruth L. Kirschstein postdoctoral fellowship from the National Institute of Health.
Footnotes
Conflict of interest statement: V.R. is on the Scientific Advisory Board of Rib-X Pharmaceuticals, a company dedicated to develop antibiotics that target the ribosome.
Data deposition: The structure has been deposited at the Protein Data Bank with the accession codes 2x9r, 2x9s, 2x9t, and 2x9u.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003995107/-/DCSupplemental.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci USA. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caskey CT, Tompkins R, Scolnick E, Caryk T, Nirenberg M. Sequential translation of trinucleotide codons for the initiation and termination of protein synthesis. Science. 1968;162:135–138. doi: 10.1126/science.162.3849.135. [DOI] [PubMed] [Google Scholar]
- 4.Dontsova M, et al. Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett. 2000;472:213–216. doi: 10.1016/s0014-5793(00)01466-6. [DOI] [PubMed] [Google Scholar]
- 5.Frolova L, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 6.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 7.Weixlbaumer A, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H, et al. The crystal structure of human eukaryotic release factor eRF1–Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 11.Mora L, et al. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol. 2003;47:267–275. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- 12.Rawat UB, et al. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 13.Klaholz BP, et al. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 14.Petry S, et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 15.Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: Invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw JJ, Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel Z, Zamir A, Elson D. The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry. 1969;8:5161–5168. doi: 10.1021/bi00840a070. [DOI] [PubMed] [Google Scholar]
- 18.Caskey CT, Beaudet AL, Scolnick EM, Rosman M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci USA. 1971;68:3163–3167. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 20.Polacek N, et al. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- 21.Brunelle JL, Shaw JJ, Youngman EM, Green R. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl-tRNA substrate. RNA. 2008;14:1526–1531. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trobro S, Aqvist J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell. 2007;27:758–766. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 23.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 24.Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 25.Trobro S, Aqvist J. Mechanism of the translation termination reaction on the ribosome. Biochemistry. 2009;48:11296–11303. doi: 10.1021/bi9017297. [DOI] [PubMed] [Google Scholar]
- 26.Dincbas-Renqvist V, et al. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heurgue-Hamard V, et al. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J Biol Chem. 2005;280:2439–2445. doi: 10.1074/jbc.M407252200. [DOI] [PubMed] [Google Scholar]
- 28.Burgi HB, Dunitz JD, Shefter E. Geometrical reaction coordinates. 2. Nucleophilic addition to a carbonyl group. J Am Chem Soc. 1973;95:5065–5067. [Google Scholar]
- 29.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjornsson A, Mottagui-Tabar S, Isaksson LA. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- 32.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 33.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 34.Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proc Natl Acad Sci USA. 2005;102:12395–12400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorner S, Panuschka C, Schmid W, Barta A. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res. 2003;31:6536–6542. doi: 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodnina MV, Beringer M, Wintermeyer W. Mechanism of peptide bond formation on the ribosome. Q Rev Biophys. 2006;39:203–225. doi: 10.1017/S003358350600429X. [DOI] [PubMed] [Google Scholar]
- 37.Fraser TH, Rich A. Synthesis and aminoacylation of 3′-amino-3′-deoxy transfer RNA and its activity in ribosomal protein synthesis. Proc Natl Acad Sci USA. 1973;70:2671–2675. doi: 10.1073/pnas.70.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.