Abstract
During meiosis, the formation of viable haploid gametes from diploid precursors requires that each homologous chromosome pair be properly segregated to produce an exact haploid set of chromosomes. Genetic recombination, which provides a physical connection between homologous chromosomes, is essential in most species for proper homologue segregation. Nevertheless, recombination is repressed specifically in and around the centromeres of chromosomes, apparently because rare centromeric (or pericentromeric) recombination events, when they do occur, can disrupt proper segregation and lead to genetic disabilities, including birth defects. The basis by which centromeric meiotic recombination is repressed has been largely unknown. We report here that, in fission yeast, RNAi functions and Clr4-Rik1 (histone H3 lysine 9 methyltransferase) are required for repression of centromeric recombination. Surprisingly, one mutant derepressed for recombination in the heterochromatic mating-type region during meiosis and several mutants derepressed for centromeric gene expression during mitotic growth are not derepressed for centromeric recombination during meiosis. These results reveal a complex relation between types of repression by heterochromatin. Our results also reveal a previously undemonstrated role for RNAi and heterochromatin in the repression of meiotic centromeric recombination and, potentially, in the prevention of birth defects by maintenance of proper chromosome segregation during meiosis.
Keywords: chromosome segregation, meiosis, Schizosaccharomyces pombe, DSB formation, genetic separation of heterochromatin functions
Meiosis entails a unique type of chromosome segregation that reduces the number of chromosomes per cell by half (1). The precursor cells have two copies of each chromosome, one copy inherited from each parent. The products of meiosis have only one copy of each chromosome, in most cases a mosaic of the two parental chromosomes produced by recombination between them. To achieve this outcome, each chromosome is replicated before the first meiotic nuclear division, and the resulting sister chromatids remain attached to each other. During the first division, each replicated chromosome aligns and pairs with its homologue; the homologues then segregate from each other into the daughter nuclei.
In most species, proper segregation of homologues requires genetic recombination, the breakage and reunion of DNA molecules of the homologues. The exchange of parts of chromosomes generates a genetic crossover, which provides a physical connection (chiasma) between homologues. Microtubules in the spindle attach to the centromeres of each homologue. As microtubule-based forces move the homologous centromeres apart, tension is generated only if there is a chiasma connecting the homologues. This tension signals that homologues are segregating properly, and only when all chromosome pairs are under tension does chromosome segregation and nuclear division proceed to completion (1).
Although crossing over is critical for proper segregation, crossovers too close to the centromere interfere with segregation. In the several species analyzed, crossing over occurs less frequently, per unit physical distance, in and near the centromere than in the arms of the chromosomes (2, 3). Notably, centromeric crossing over in humans is correlated with birth defects resulting from chromosome missegregation (2). (Here and subsequently, “centromeric” is meant to include “pericentromeric.”) Thus, repression of recombination specifically in the centromere is crucial for the proper segregation of meiotic chromosomes, but the mechanism by which centromeric recombination is repressed during meiosis has been largely unknown.
Centromeric heterochromatin in many species represses within its domain the abundance of transcripts and the expression of genes inserted into the heterochromatic region (4). In the fission yeast Schizosaccharomyces pombe, the formation of centromeric heterochromatin is facilitated by RNAi functions, which direct the histone methyltransferase Clr4 and its partner protein Rik1 to the centromere (5, 6) (Fig. 1). Histone H3 methylated by Clr4 at lysine 9 is bound by several chromodomain proteins, including Chp1 and two homologues of heterochromatin protein HP1: Swi6 and Chp2 (4, 7). All these functions contribute to reduction of native centromeric transcripts, abundance of RNA polymerase II at native centromeres, and expression of genes inserted into the centromeric region (4, 5, 8–10). Here, we tested the possibility that heterochromatin is also involved in repression of centromeric meiotic recombination.
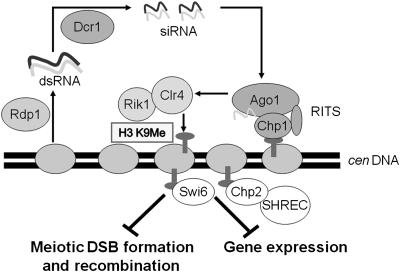
Fig. 1.
Repression of centromeric recombination and transcription by RNAi and heterochromatin components. In heterochromatic regions, such as the centromere and pericentric regions of S. pombe, transcripts are synthesized at low level by RNA polymerase II and converted to dsRNA by the RNA-dependent RNA polymerase Rdp1. dsRNA is converted into siRNA (approximately 22 bp) by the Dcr1 nuclease. The RNAi-induced transcriptional silencing complex (RITS), containing the Ago1 and Chp1 proteins, uses the siRNAs to direct the Clr4–Rik1 complex to methylate histone H3 lysine 9 (H3 K9) in and near the centromeres. Methylated H3 K9 is bound by chromodomain proteins Swi6 and Chp2, which directs the Clr3 histone deacetylase complex SHREC to further modify the histones in heterochromatic regions. The results presented here show that formation of centromeric DSBs, and consequently recombination in meiosis, are repressed by RNAi and Clr4–Rik1. Swi6, Chp2, and Clr3 are each required for repression of high-level mitotic transcription, but not for repression of meiotic DSB formation and recombination, although combined loss of Swi6 and Chp2 allows significant centromeric recombination.
Results and Discussion
Centromeric Meiotic Recombination Is Negligible in WT but as Frequent as in Chromosomal Arms in RNAi and Histone H3 Lys9 Methyltransferase Mutants.
Using markers closely flanking centromere 3 (cen3) of S. pombe (Fig. 2), we assayed centromeric recombination in WT and in mutants defective in RNAi and formation of heterochromatin. In WT cells, these markers recombined infrequently (one recombinant among 2141 meiotic spores; Table 1). This frequency is approximately 200 times less than the genome-wide average (approximately 0.16 cM/kb) for chromosomal arm markers separated by the physical distance between these centromere-flanking markers (approximately 125 kb) (3, 11). In a mutant dcr1Δ lacking the Dicer nuclease essential for generating the small RNAs that guide RNAi to its targets, centromeric recombinants were significantly more abundant than in WT (3.6% recombinants in dcr1Δ vs. <0.1% in dcr1+). To determine whether the nuclease activity, rather than some other function, of Dicer is required for the recombination block, we constructed and tested a double point mutant dcr1-5 altered in the nuclease motifs essential for nuclease activity of the related human Dicer protein and Escherichia coli RNase III (12). Like the dcr1Δ deletion mutant, dcr1-5 had abundant centromeric recombination (4.0%). Similar or higher frequencies were observed in other RNAi mutants—8.9% in rdp1Δ lacking RNA-dependent RNA polymerase and 3.1% in ago1Δ lacking the argonaute endoribonuclease. Collectively, these results show that the full complement of RNAi activities is required to repress meiotic centromeric recombination.
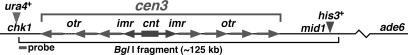
Fig. 2.
Chromosome structure surrounding centromere 3 (cen3) of S. pombe. The 4.9-kb central element (cnt) is flanked by 5.4-kb inverted innermost repeats (imr), which in turn are flanked by multiple 6.7-kb outermost repeats (otr) (35). The number of outermost repeats to the left of the central element is uncertain, but we estimate there are 6 ± 1. For genetic analysis the ura4+ gene was substituted for most of chk1, and his3+ was inserted between mid1 and the next centromere-distal gene cwf20; chk1 and mid1 are protein-coding genes separated from centromere 3 by, respectively, one and three protein-coding genes (35). Recombination between chk1::ura4+ and mid1-322::his3+ is used as a measure of centromere 3 recombination (Table 1). The ade6 gene is 166 kb from the mid1-322::his3+ insertion; recombination between these markers is used as a measure of cen3–ade6 recombination. Two ade6 markers, ade6-M26 and ade6-52, separated by 0.66 kb, are used to measure ade6 intragenic recombination. The small rectangle (bottom left) indicates the position of the radioactive probe for the Southern blot hybridizations of the indicated BglI DNA fragment (Fig. 3 and Table 1). The figure is drawn approximately to scale, except for the position ade6.
Table 1.
RNAi and some heterochromatin mutations derepress meiotic recombination in a centromere region but not in a chromosome arm region
| Mutant gene | Function of gene product | cen3 DSBs | Recombinant frequency in indicated interval | ||
| cen3, % | cen3–ade6, % | ade6 intragenic, Ade+ per 103 viable spores | |||
| WT | – | – | <0.1 | 15 | 7.7 |
| dcr1Δ* | Dicer nuclease | + | 3.6 | 12 | 5.2 |
| dcr1-5* | (Nuclease-deficient missense mutant) | ND | 4.0 | 14 | 6.7 |
| rdp1Δ* | RNA-dependent RNA polymerase | + | 8.9 | 13 | 5.1 |
| ago1Δ* | Endoribonuclease | + | 3.1 | 14 | 3.8 |
| clr4Δ* | Histone H3 lysine 9 methyl transferase | + | 9.1 | 8 | 6.0 |
| rik1Δ* | Clr4 partner protein | + | 5.7 | 8 | 9.6 |
| swi6Δ | Heterochromatin protein 1 | – | 0.3 | 11 | 4.1 |
| chp2Δ | Swi6 paralogue | ND | 0.3 | 15 | 7.4 |
| pli1Δ | SUMO E3 ligase | – | <0.1 | 4 | 0.6 |
| cid14Δ | PolyA polymerase | ND | <0.4 | 14 | 6.8 |
| clr3Δ | Histone deacetylase | – | 0.3 | 15 | 1.1 |
| swi6Δ chp2Δ* | (Double mutant) | ND | 2.2 | 13 | 1.9 |
| chp1Δ* | Chromodomain protein | + | 2.8 | 13 | 4.3 |
| clr3Δ dcr1Δ* | (Double mutant) | ND | 4.6 | 9 | 2.5 |
| clr4Δ rec8Δ | Sister chromatid cohesin | ND | <1.0 | <1.3 | <0.1 |
| clr4Δ rec10Δ | Linear element protein | ND | <1.4 | <1.1 | <0.1 |
Recombination data are the frequency of recombinants between markers flanking centromere 3 (cen3), between cen3 and ade6, or within ade6 (see diagram in Fig. 2). “<” indicates the upper 95% confidence limit based on the Poisson distribution when no recombinants were observed. For DSBs, “+” indicates readily visible meiosis-specific DSBs in centromere 3, and “–“ indicates no visible meiosis-specific DSBs (Fig. 3 and Fig. S1); the low level of DSBs at individual sites precludes accurate quantification. ND, not determined. Data are summarized from Table S3.
*Mutants strongly derepressed relative to WT; those without asterisks are not significantly derepressed.
In S. pombe, RNAi enables Clr4 to methylate centromeric histone H3 at lysine 9 (H3 K9) (5). We tested a role for Clr4 in repression of centromeric recombination and found that, like RNAi, it is required for repression: the clr4Δ mutant had 9.1% recombinants (Table 1), or approximately 0.1 cM/kb. Rik1, a Clr4 partner protein (13), is also required for repression: the rik1Δ mutant had 5.7% recombinants. Thus, this aspect of heterochromatin also appears essential for repression of meiotic centromeric recombination. Drosophila mutants lacking a homologue of Clr4 have increased numbers of foci of phosphorylated γH2Av histone, indicative of DSBs, in oocytes (14). In that study (14), recombination was not reported, and an increased number of MeiW68 (Spo11)–dependent DSBs, those specific for meiotic recombination as discussed later, was not established. Other mutations affecting Drosophila heterochromatin, however, do increase pericentromeric recombination (15). These results are consistent with ours and suggest that the mechanism of repression of centromeric recombination may be evolutionarily conserved.
Repression Occurs in the Absence of Heterochromatin Protein Swi6.
Methylation of H3 K9 by Clr4 enables Swi6 to bind to centromeric heterochromatin (7, 16). Relative to WT, swi6Δ mutants have more abundant transcripts and RNA polymerase II in unaltered (i.e., native) centromeric dg and dh repeats (5, 9), increased expression of genes inserted into the centromeric region (5, 8), and increased meiotic recombination in the heterochromatic native mating-type region (17). We therefore expected that Swi6 would also be required for repression of recombination at centromeres. Surprisingly, the swi6Δ mutant had only a low level of recombination (0.3%), not significantly different from that of WT (Table 1; P > 0.1 by Fisher exact test). This result suggests that the types of repression by heterochromatin can be genetically separated.
To test this suggestion further, we analyzed additional mutants derepressed for centromeric gene expression, either native or with inserted genes, and found that, like the swi6Δ mutant, they largely retained repression of centromeric recombination. The mutants tested were chp2Δ, which lacks a Swi6 paralogue (9, 10); pli1Δ, which lacks a SUMO E3 ligase (18); cid14Δ, which lacks a polyA polymerase (19); and clr3Δ, which lacks a histone deacetylase (20). These mutants are defective in a range of activities necessary for effective centromeric gene silencing, including transcriptional gene silencing [requiring Clr3 (20)] and posttranscriptional gene silencing [requiring Cid14 (19)]. Among these single mutants, none had significantly more centromeric recombination than WT (Table 1 and Table S1; P > 0.05 by Fisher exact test). The lack of derepression of centromeric recombination is particularly surprising in the case of the clr3Δ mutant. In this mutant, the abundance of RNA polymerase II at the native centromeric dg and dh repeats or at an inserted centromeric ura4+ gene is nearly as high as that in clr4Δ (9, 20). In the swi6Δ chp2Δ double mutant, we observed centromeric recombinants (2.2%) but significantly fewer than in clr4Δ (P < 0.0001 by Fisher exact test). Clearly, repression of centromeric transcription and meiotic recombination occur by genetically separable mechanisms, although Swi6 and Chp2 appear to have partially overlapping roles in both processes (Fig. 1).
We found that, unlike Swi6, the chromodomain protein Chp1 is required for repression of centromeric recombination (Table 1). This observation provides an interesting clue regarding the mechanism of repression by revealing a correlation between the extent to which a chromodomain protein represses recombination and the extent to which it facilitates H3 K9 methylation at a given location. The S. pombe chromodomain proteins Chp1, Swi6, and Chp2 cooperate with Clr4 to facilitate H3 K9 methylation through positive feedback loops, differentially at centromeres (where Chp1 has a greater effect than Swi6) and in the mating-type region (where Swi6 has a greater effect than Chp1). Sadaie et al. (21) found a comparatively stronger effect of chp1Δ at several centromeric locations, where swi6Δ has a negligible effect, than at a single location within the mating-type region. In a more comprehensive study, Hall et al. (22) found that swi6Δ decreases H3 K9me abundance approximately 10-fold across a large segment of the normally heterochromatic part of the mating-type region. We propose that the decrease in H3 K9me observed in the mating-type region but not at centromeres in swi6Δ accounts for swi6Δ allowing meiotic recombination in the mating-type region but not at centromeres.
It is notable that centromeric repeats are not completely devoid of H3 K9me in RNAi mutants; the residual H3 K9me in these mutants depends on Clr3 (23). We found, however, that the level of recombination in a mutant lacking RNAi (dcr1Δ) was not significantly further increased by deletion of clr3 (Table 1; P > 0.3 by contingency χ2 test), indicating that the low level of H3 K9me remaining in RNAi mutants does not limit meiotic recombination.
RNAi and Heterochromatin Mutants Are Not Derepressed for Chromosomal Arm Recombination.
The increased levels of centromeric recombination in the mutants discussed earlier might be the result of increased recombination throughout the genome. To test this possibility, we measured crossing over in the interval immediately to the right of cen3, extending to ade6 (Fig. 2). Crossing over in this interval in the derepressed mutants was not higher than that in WT (15%; Table 1); modest reductions were seen in some mutants (clr4Δ, rik1Δ, pli1Δ, and clr3Δ dcr1Δ). In addition, intragenic recombination in ade6, nearly exclusively gene conversion, was not significantly altered in the mutants tested, except for decreases in pli1Δ and clr3Δ (Table 1). Thus, the mutants tested here appear to derepress meiotic recombination specifically in the centromere.
Derepressed Mutants Have Meiosis-Specific DNA Double-Strand Breaks, Unlike WT.
Meiotic recombination in S. pombe, as in the distantly related budding yeast Saccharomyces cerevisiae, requires Rec12 (orthologue of Spo11 in S. cerevisiae), which forms DNA double-strand breaks (DSBs) (24). We measured DSBs in cen3 by Southern blot hybridization of DNA extracted after induction of meiosis; the ≈125-kb BglI fragment, which spans cen3, was analyzed for DSBs (Figs. 2 and 3). In WT, DSBs were below the level of detection. In contrast, DSBs were readily detectable in the mutants that manifested centromeric recombination (Fig. 3, Table 1, and Fig. S1). We note, however, that the DSB patterns differed among these mutants. As expected, the swi6Δ, pli1Δ, and clr3Δ mutants, which lack detectable recombination at cen3 (Table 1), also lacked detectable meiosis-specific DSBs at cen3. Each of the mutants analyzed had approximately WT levels of DSBs in the arm intervals analyzed. We infer that RNAi and heterochromatin block the formation of meiotic DSBs and, as a consequence, block recombination specifically in the centromere.
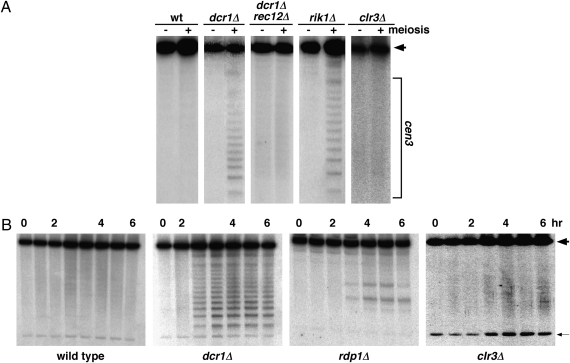
Fig. 3.
Meiosis-specific DSBs arise in centromere 3 (cen3) specifically in RNAi and heterochromatin mutants. (A) Cells were induced for meiosis and harvested 1 h (–) and 5 or 6 h (+) later. DNA was extracted, digested with BglI, and analyzed for DSBs by Southern blot hybridization (11). The unbroken BglI fragment (wide arrowhead) spanning cen3 is approximately 125 kb long (Fig. 2). Meiosis-specific DNA fragments (bracket) result from Rec12-dependent DNA breakage (i.e., DSBs) at specific sites in centromere 3. DSBs arise in dcr1Δ (lacking Dicer nuclease) and rik1Δ (lacking a partner of the Clr4 histone H3 Lys9 methyltransferase) mutants but not in WT, a clr3Δ mutant (lacking a histone deacetylase), or a swi6Δ mutant (lacking heterochromatin protein HP1; Fig. S1). (B) Complete time course of meiosis-specific DSB formation in representative mutants. DNA was analyzed as in A at hourly intervals; for strains GP3718 (WT) and GP4978 (dcr1Δ), additional samples at 3.5 h were analyzed, and for strain GP4978, the 1 h sample was not analyzed. The faint smears at 1, 3, 4, and 6 h for strain GP6891 (clr3Δ) likely represent cross-hybridization, as indicated by the relatively strong cross-hybridization with the 10.9-kb rDNA repeat (thin arrow) in this experiment. Additional data are shown in Table 1 and Fig. S1.
How might DSB formation in the centromere be repressed? DSBs are formed by the S. pombe Rec12 protein (24), but the action of Rec12 depends on multiple additional proteins. From genetic and cytological analyses, we have proposed (25) a pathway for DSB formation that begins with the loading of meiosis-specific sister chromatid cohesin subunits Rec8 and Rec11 at about the time of meiotic DNA replication. Loading of Rec8 and Rec11 enables the loading of Rec10, Rec25, and Rec27, which together form linear elements, structures distantly related to the synaptonemal complex of other species. Linear elements in turn enable the loading of Rec7 (26), which like Rec12 is essential for all detectable DSB formation throughout the genome (24). Additional proteins (e.g., Rec6, Rec14, and Rec15) are similarly essential and may, with Rec7, form a complex with Rec12 (25). Repression of centromeric recombination might act by blocking the loading of any or all of these proteins specifically in the centromere. Rec8 and Rec12, however, are present at the centromere during WT meiosis (27, 28). Therefore, we suppose that loading or activation of some other protein is the point at which RNAi and heterochromatin prevent recombination specifically in the centromere.
We tested a requirement for Rec8 and Rec10 in centromeric recombination in the highly derepressed mutant clr4Δ. In each of the double mutants, clr4Δ rec8Δ and clr4Δ rec10Δ, recombination was abolished—the frequency of centromeric recombinants was not significantly different from that in WT (Table 1; P > 0.2 by Fisher exact test). Furthermore, formation of centromeric DSBs requires Rec12, at least in the dcr1Δ mutant background tested (Fig. 3). Centromeric recombination therefore appears, at this level of analysis, to proceed by the same mechanism as arm recombination. We expect that DSBs, once formed, are repaired by the same mechanism as DSBs in chromosomal arms, which requires more than a dozen proteins (25). The primary target of repression of centromeric meiotic DSB formation and recombination is thus likely to be, directly or indirectly, the activation of Rec12 for DSB formation.
Conclusions
Our results reveal an unexpected, complex relation of three functions of heterochromatin—repression of transcription in and near the centromere during mitotic growth, repression of centromeric meiotic recombination, and repression of meiotic recombination at the mating-type locus. We infer that there are at least two genetically separable functional components of centromeric heterochromatin: one (exemplified by RNAi and Clr4-Rik1) blocks both centromeric mitotic transcription and meiotic recombination, whereas the other (exemplified by Clr3) blocks only gene expression (Fig. 1). Clearly, heterochromatin cannot be simply considered an impervious barrier to macromolecules. Rather, heterochromatin must act as a differential filter allowing some but not other proteins access to (or action on) the DNA, depending on which of its constituents are present.
The mechanism by which centromeric recombination interferes with chromosome segregation is not established. Presumably, a crossover too near the centromere places a spatial constraint on the centromere and the complex structure (kinetochore) built on it; spindle microtubules attach to the kinetochore and direct chromosome segregation. Repression of centromeric recombination is thus important to facilitate proper chromosome segregation in meiosis. Failure of this repression, as in the mutants studied here, can result in occasional meiotic missegregation in S. pombe (29). Repression of centromeric recombination in humans (2), perhaps by a closely related mechanism, appears to be requisite for preventing miscarriages and severe birth defects, such as Down syndrome (30).
Materials and Methods
Strains and Genetic Methods.
Genotypes of the S. pombe strains used and references for alleles are in Table S2. Genealogies are available upon request.
Crosses between heterothallic strains were conducted and random spores analyzed as described (11). Because some mutations, such as clr4Δ, derepress expression of the mat2 and mat3 loci and allow haploid meiosis and self-mating (31), in each cross, except some rik1Δ crosses, we analyzed segregation of the heterozygous lys4-95 marker on Chr. 2. If ade6 (on Chr. 3) and lys4 did not segregate randomly, the total number of interstrain spores analyzed was taken as twice the sum of colonies with any nonparental marker among each ade6 class (light red, ade6-52; dark red, ade6-M26) tested. In some rik1Δ crosses mid1-6 (a temperature-sensitive mutation also called dmf1-6) was used in place of mid1-322::his3+; mid1-6 was scored by replica-plating onto yeast extract agar medium supplemented with adenine (100 μg/mL) and phloxin B (20 μg/mL) and incubation at 37 °C. Diploids, determined as I2-staining spore colonies, were omitted from all analyses. Statistical significance was determined with the indicated tests on StatPages (http://statpages.org/).
The dcr1-5 allele, designed to eliminate Dcr1 nuclease activity on the basis of homology with human Dicer and E. coli RNase III (12), encodes alanine in place of two aspartate residues thought to bind divalent ions required for nuclease activity. dcr1-5 was constructed by site-directed mutagenesis using a QuikChange kit (Stratagene). A 1,244-bp fragment of the dcr1 gene (codons 872–1,287) was amplified by a PCR primed with oligonucleotides OL1595 and OL1596 (Table S1), digested with BamHI, and cloned into the BamHI site of plasmid pBluescript II KS (+) (Stratagene) to create plasmid pCE2. Mutations were introduced into this segment of dcr1 in two steps using oligonucleotides OL1591 and OL1592 to change GAC to GCA (D908A) and OL1593 and OL1594 to change GAT to GCA (D1127A), thereby creating pCE5. The 1254 bp BamHI fragment of pCE5 was used to transform strain GP6049 (dcr1-1::ura4+) to 5-fluoro-orotic acid resistance to create strain VT3110. Homologous replacement was confirmed by nucleotide sequence analysis. The dcr1-1::ura4+ allele was made by substituting bp 2,719 through 3,756 of the dcr1 coding sequence with the 1.8-kb HindIII ura4+ fragment using standard methods (32) and oligonucleotides OL1599 and OL1600.
The dcr1::hph1 allele was constructed by transforming strain TV293 (dcr1::kanMX6) with the hph1 gene amplified from pCR2.1-hph1, as described (33); primers were MD1 and MD2s. Hygromycin-resistant, G418-sensitive transformants were isolated, and the marker substitution was confirmed by PCR.
The mid1-322::his3+ allele, in which his3+ is inserted 322 bp to the cen3-distal side of the mid1 (dmf1) ORF (5′ of the ORF), was generated as follows. A 663-bp fragment of DNA spanning the 626 bp mid1–cwf20 intergenic region was amplified by a PCR primed with oligonucleotides OL1618 and OL1621, digested with HindIII, and inserted into the HindIII site of pBR322 to create plasmid pCE7. A BglII recognition sequence was created in pCE7 to generate the sequence 5′tcgaAGATCTaagc 3′ using a QuikChange kit (Stratagene) and oligonucleotides OL1619 and OL1620 to create pCE8. (The BglII recognition sequence is in uppercase; the mutated bp are in italics.) The 2,013-bp BglII fragment of pAF1, containing his3+ (34), was inserted into the BglII site of pCE8 to create pCE6. Strain GP5872 (his3-D1) was transformed to His+ with the (PCR amplified) 2.7-kb HindIII fragment of pCE6; a transformant, GP6138, was confirmed to have the expected insertion by PCR and nucleotide sequence analysis.
DNA Analysis.
DSBs were analyzed as described (11). In brief, synchronous meiosis of pat1-114 strains was induced by raising the temperature of a nitrogen-starved (i.e., G1-arrested) culture to 34 °C and restoring nitrogen. At the indicated times after induction, cells were harvested by centrifugation, washed, embedded in agarose plugs, and treated with lytic enzymes. DNA in the plugs was partially purified by incubation with proteinase K and digested with BglI restriction enzyme. The DNA fragments were separated by pulsed-field gel electrophoresis and analyzed by Southern blot hybridization, using radioactive probes complementary to the left and right ends of the BglI fragment spanning cen3. These probes spanned, respectively, bp 1,061,879 through 1,063,134 and bp 1,152,795 through 1,153,765 of the S. pombe Chr. 3 sequence (http://www.genedb.org/genedb/ContigMap?organism=pombe&name=chromosome3.contig).
Supplementary Material
Acknowledgments
We are grateful to S. Grewal, P. Russell, N. Walworth, Y. Murakami, and R. Egel for strains; V. Tseng for technical support and S. Amundsen, G. Cromie, S. Henikoff, N. Milman, and F. Steiner for helpful comments. This work was supported by National Institutes of Health Grant GM032194 (to G.R.S.) and by the Lundbeck Foundation and Danish Research Council (G.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914160107/-/DCSupplemental.
References
- 1.Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 2.Lynn A, Ashley T, Hassold T. Variation in human meiotic recombination. Annu Rev Genomics Hum Genet. 2004;5:317–349. doi: 10.1146/annurev.genom.4.070802.110217. [DOI] [PubMed] [Google Scholar]
- 3.Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 5.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 6.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 8.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 9.Motamedi MR, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thon G, Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155:551–568. doi: 10.1093/genetics/155.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Hong EJ, Villén J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 14.Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 17.Klar AJS, Bonaduce MJ. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot” of fission yeast. Genetics. 1991;129:1033–1042. doi: 10.1093/genetics/129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B. Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J. 2004;23:3844–3853. doi: 10.1038/sj.emboj.7600394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bühler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama T, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 23.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 25.Cromie GA, Smith GR. Meiotic recombination in Schizosaccharomyces pombe: A paradigm for genetic and molecular analysis. In: Egel R, Lankenau D-H, editors. Recombination and Meiosis: Models, Means, and Evolution (Genome Dynamics and Stability) Berlin: Springer-Verlag; 2008. pp. 195–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenz A, Estreicher A, Kohli J, Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006;115:330–340. doi: 10.1007/s00412-006-0053-9. [DOI] [PubMed] [Google Scholar]
- 27.Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- 28.Ludin K, et al. Sites of strong Rec12/Spo11 binding in the fission yeast genome are associated with meiotic recombination and with centromeres. Chromosoma. 2008;117:431–444. doi: 10.1007/s00412-008-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall IM, Noma K-I, Grewal SIS. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb NE, et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- 31.Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bähler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 33.Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 34.Ohi R, Feoktistova A, Gould KL. Construction of vectors and a genomic library for use with his3-deficient strains of Schizosaccharomyces pombe. Gene. 1996;174:315–318. doi: 10.1016/0378-1119(96)00085-6. [DOI] [PubMed] [Google Scholar]
- 35.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.