Abstract
For self-assembly of the bacterial flagellum, most of the flagellar component proteins synthesized in the cytoplasm are exported by the flagellar type III export apparatus to the growing, distal end. Flagellar protein export is highly organized and well controlled in every step of the flagellar assembly process. Flagellar-specific chaperones not only facilitate the export of their cognate proteins, as well as prevent their premature aggregation in the cytoplasm, but also play a role in fine-tuning flagellar gene expression to be coupled with the flagellar assembly process. FliT is a flagellar-specific chaperone responsible for the export of the filament-capping protein FliD and for negative control of flagellar gene expression by binding to the FlhDC complex. Here we report the crystal structure of Salmonella FliT at 3.2-Å resolution. The structural and biochemical analyses clearly reveal that the C-terminal segment of FliT regulates its interactions with the FlhDC complex, FliI ATPase, and FliJ (subunits of the export apparatus), and that its conformational change is responsible for the switch in its binding partners during flagellar protein export.
Keywords: bacterial flagellum, crystal structure, flagellar gene expression, molecular chaperone, type III protein export
The bacterial flagellum is a locomotive organelle that propels the cell body in liquid environments. The flagellum is a huge protein complex consisting of at least three parts: the basal body, the hook, and the filament. The basal body, which works as a motor, is embedded within the cell envelope, whereas the hook and the filament, which function as a universal joint and a propeller, respectively, extend outward from the cells (1–3). Most of the flagellar proteins are translocated into the central channel and to the distal end of the growing flagellar structure for self-assembly by the flagellar type III protein export apparatus, which consists of six integral membrane proteins (FlhA, FlhB, FliO, FliP, FliQ, and FliR) and three soluble proteins (FliH, FliI, and FliJ) (4, 5). Proton motive force across the cytoplasmic membrane provides an energy source for flagellar protein export (6, 7). Interestingly, the export process is well regulated according to the stages of the flagellar assembly process (3).
The flagellar genes can be divided into three classes according to their transcriptional hierarchy (8). The class 1 flhDC operon is at the top of the hierarchy. FlhD and FlhC form the FlhD4C2 complex and activate the expression of the class 2 genes encoding components of the hook–basal body (HBB) and some regulators, including the flagellar-specific sigma factor σ28 (FliA) and the anti-σ28 factor FlgM. During HBB assembly, the activity of σ28 is suppressed by FlgM. On completion of HBB assembly, FlgM is secreted into the culture media, freeing σ28 and allowing it to transcribe class 3 genes responsible for filament formation, motor function, and chemotaxis (9, 10). This regulatory system is well conserved among gram-negative enteric bacteria (11). Other bacteria, such as Caulobacter spp. and Helicobacter pylori, have similar transcriptional hierarchy and feedback regulation in flagellar biogenesis, but use distinct regulatory pathways (12, 13).
The cytoplasmic proteins FlgN, FliS, and FliT act as flagellar-specific chaperones to facilitate the export of their cognate substrates on completion of HBB assembly (14–16). They bind not only to their cognate substrates to prevent premature aggregation in the cytoplasm (15, 16) but also to the soluble export component proteins FliI and FliJ (17, 18). These chaperones also play other distinct roles in the flagellar construction process. FlgN is responsible for the translational regulation of FlgM (19), FliS is responsible for the negative regulation of FlgM export (20), and FliT is responsible for the negative regulation of class 2 gene expression (21). Thus, the flagellar chaperons seem to fine-tune the flagellar assembly process. Many virulence-related type III secretion system chaperones are also known to play multiple roles, such as facilitating the secretion of effector proteins and transcriptional or translational regulation and fine-tuning the secretion process (22, 23); the molecular mechanism through which they regulate these multiple functions remains obscure, however.
FliT is a chaperone specific for the filament-capping protein FliD. FliT forms a stable complex with FliD through an interaction between the C-terminal half of FliT and a C-terminal region of FliD (14, 15). FliT also is known to interact with FliJ (18). Because FliT binds to the FlhD4C2 complex, thereby inhibiting the activation of the class 2 promoters, FliT is proposed to be an anti-FlhD4C2 factor (21). These observations suggest that the interactions between FliT and these FliT-binding proteins are highly regulated during flagellar assembly. To elucidate the regulatory mechanisms of the interactions of FliT with its binding partners, we crystallized (24) and determined the structure of FliT of Salmonella enterica serovar Typhimurium, which is composed of 122 amino acid residues with a molecular mass of 13.7 kDa.
Results
Overall Structure of FliT.
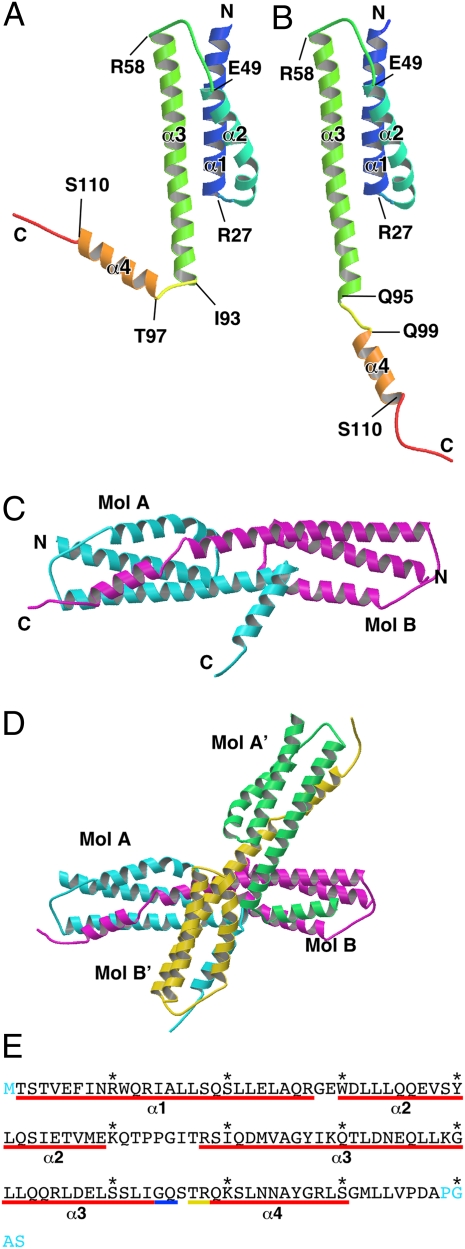
The crystal structure of FliT has been solved at 3.2-Å resolution. The asymmetric unit of the crystal contains two FliT molecules, Mol A and Mol B (Fig. 1 A and B), which form a dimer related by a pseudo-twofold local symmetry (Fig. 1C). The dimer and its adjacent dimer (Mol A′ and Mol B′) related by a crystallographic twofold symmetry produce a tetramer (ABA′B′) in the crystal (Fig. 1D). The atomic model was built from residues 2–116 for Mol A and residues 2–118 for Mol B. The N-terminal methionine residues and several C-terminal residues (117–122 for Mol A and 119–122 for Mol B) were invisible in the electron density map, likely due to the map's conformational flexibility. FliT consists of four α-helices (α1, α2, α3, and α4). The amino acid sequence and secondary structure of FliT are summarized in Fig. 1E. The core of the molecule is formed by antiparallel α-helical bundle structure composed of α1, α2, and α3 (Fig. 1 A and B). Helices α2 and α3 form a hydrophobic cleft with Phe-7 and Trp-11 of α1 (Fig. 2). The core structures of Mol A and B can be superimposed with an rms displacement of 0.84 Å for corresponding Cα atoms (residues 2–93). In contrast, the conformation of the remaining C-terminal segment including α4, which is an amphipathic α-helix, is quite distinct between the two molecules. Helix α4 of Mol A makes a sharp turn from α3 (Fig. 1A) and interacts with Mol B′ (Fig. 1D), whereas α4 of Mol B extends straight from α3 (Fig. 1B) and interacts with Mol A (Fig. 1D). The α4 helices of Mol B and Mol B′ cover the hydrophobic cleft of the core of their dimeric partners, Mol A and Mol A′, respectively (Fig. 2 A and B). The α4 helices of Mol A and Mol A′ interact with the hydrophobic clefts of Mol B′ and Mol B, respectively (Fig. 2 C and D). Thus, the domain swapping of the C-terminal segments produces the FliT tetramer. Despite the conformational flexibility of the C-terminal segment and the differences in length of α3 and α4 between Mol A and Mol B (Fig. 1 A and B), the interactions between the hydrophobic cleft and α4 of an adjacent molecule are almost the same for the Mol A–Mol B pair and the Mol B–Mol A′ pair (Fig. 2). These structural features imply that the flexible C-terminal segment might play some significant role in FliT function.
Fig. 1.
Structure of FliT. (A and B) Mol A (A) and Mol B (B) in a color-coded Cα ribbon drawing from the N terminus (blue) to the C terminus (red). (C and D) FliT dimer (C) and FliT tetramer (D) found in the crystal. Mol A is shown in cyan; Mol B, in magenta; Mol A′, in green; and Mol B′, in yellow. (E) Amino acid sequence of FliT with indications of various structural features. The regions of α-helix are indicated by underscores: red line, α-helix in both Mol A and Mol B; blue line, only in Mol B; yellow line, only in Mol A. The labels α1–α4 correspond to those in A and B. The terminal residues missing in the atomic model are shown in cyan.
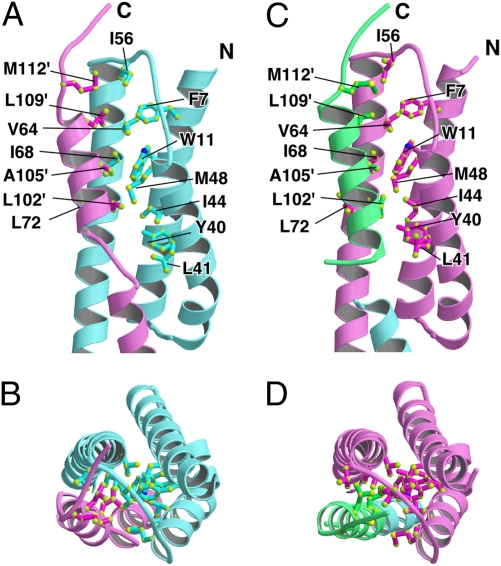
Fig. 2.
Close-up view of the hydrophobic cleft covered by the C-terminal segment of an adjacent molecule. (A and B) Side view (A) and end-on view (B) of the hydrophobic cleft of Mol A (cyan) covered by the C-terminal segment of Mol B (pink). (C and D) Side view (C) and end-on view (D) of the hydrophobic cleft of Mol B (pink) covered by the C-terminal segment of Mol A′ (green). Hydrophobic residues in the cleft and on the C-terminal segment are indicated by ball and stick model.
Oligomeric State of FliT in Solution.
Size-exclusion chromatography has shown that FliT forms a stable homodimer in solution (15). In contrast, our crystal structure exhibited a tetramer. Consequently, we reexamined the oligomeric state of FliT in solution by analytical ultracentrifugation and analytical size-exclusion chromatography. The apparent molecular mass of FliT determined by the sedimentation equilibrium analysis was 20.5 kDa, and it gradually increased with increasing protein concentration (Fig. S1A), indicating that FliT is present as a mixture of monomer and oligomers in solution. The sedimentation datasets were best fitted by assuming the presence of monomer and dimer with a self-association constant of 3,100 L/mol (Fig. S1B). Size-exclusion chromatography was performed under various solution conditions with a pH of 7–10 and NaCl concentrations of 150–500 mM, as well as that used for the sedimentation equilibrium measurement. FliT eluted as a single peak at the same position with tailing (Fig. S1C), demonstrating that the equilibrium is not significantly affected by pH and ionic strength under our conditions.
FliT and FliD Forms a Heterodimer in Solution.
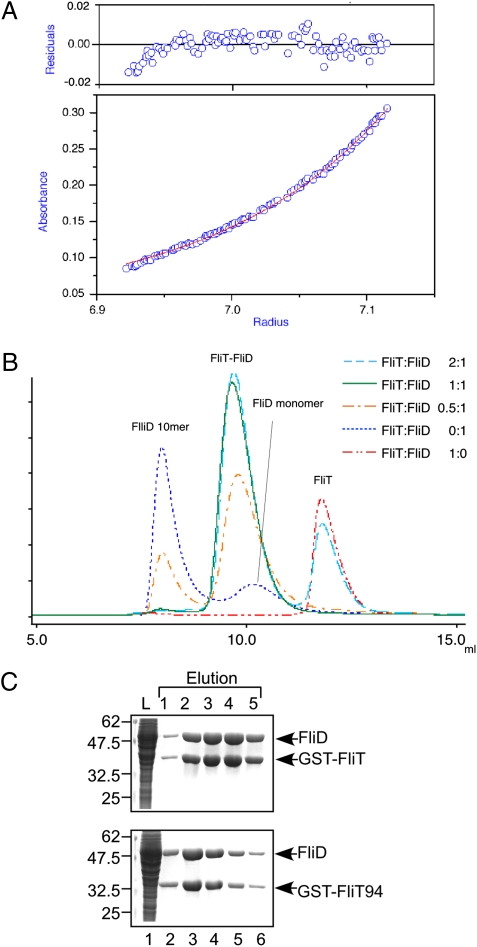
We also analyzed the oligomeric state of the FliT–FliD complex in solution using analytical ultracentrifugation. The apparent molecular mass determined by the sedimentation equilibrium measurement was 62.0 kDa (Fig. 3A), which closely corresponds to the 63.4-kDa molecular mass of the FliT–FliD heterodimer.
Fig. 3.
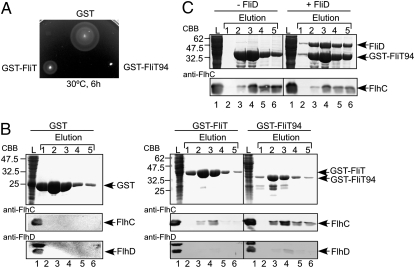
Heterodimer formation of FliT and FliD in solution. (A) Sedimentation equilibrium measurement of the FliT–FliD complex. (Lower) Sedimentation equilibrium profile (initial absorbance of 0.2 at 280 nm, measured at 12,000 rpm). Open circles are data points, and the continuous red line is a model fit curve for a single molecular species with a molecular mass of 62.0 kDa, which corresponds well to the molecular mass of a FliT–FliD heterodimer (63.4 kDa). (Upper) Plots of the residuals of each data point from the fitting curve. (B) Elution profiles of mixtures of FliT and FliD at various FliT:FliD molar ratios: 2:1 (cyan dashed line), 1:1 (green solid line), 0.5:1 (orange dotted-dashed line), 0:1 (blue dotted line), and 1:0 (red two-dotted–dashed line). (C) Pull-down assay by GST affinity chromatography. Soluble fractions (L) prepared from a ΔflhDC-cheW mutant expressing GST-FliT or GST-FliT94 were mixed with those from the ΔflhDC-cheW mutant producing FliD and loaded onto the GST affinity column. After extensive washing, proteins were eluted with a buffer containing 10 mM reduced glutathione. The eluted fractions were analyzed by CBB staining.
To confirm this, we performed analytical size-exclusion chromatography. FliT and FliD were mixed at molar ratios of 2:1, 1:1, or 0.5:1, and the mixture was loaded onto a size-exclusion column (Fig. 3B). Most of the FliD molecules formed a decamer at pH 7.8, as reported previously (25). The FliT–FliD mixture with the molar ratio of 1:1 had a single elution peak at a position slightly larger than the FliD monomer (49.7 kDa). The 2:1 FliT–FliD mixture eluted with two peaks, one at the peak position of the 1:1 mixture and the other at the position of monomeric FliT. The 0.5:1 FliT–FliD mixture also showed two peaks, one at the peak position of the 1:1 mixture and the other at the position of the FliD decamer. These results indicate that FliT and FliD form a heterodimer in solution.
Interaction Between FliT and FliD.
It has been shown that residues 42–122 of FliT are responsible for the binding to FliD (15). To test whether the C4 α-helix is required for the FliT–FliD interaction, we investigated the effect of C-terminal truncation of FliT on its interaction with FliD through a pull-down assay with GST affinity chromatography (Fig. 3C). FliD copurified with FliT(2-94) (hereinafter GST-FliT94) at the same level as with GST-FliT(2-122) (hereinafter GST-FliT), indicating that the C-terminal segment of FliT does not contribute to its binding to FliD.
To identify residues of FliT responsible for the interaction with FliD, we chose eight highly conserved, surface-exposed residues for site-directed mutation experiments: Ile-68 and Leu-72, which are located within the hydrophobic cleft formed by α2 and α3; Asn-74, Leu-81, and Gln-83, which are opposite to the hydrophobic cleft; and Glu-75, Lys-79, and Leu-82, which are under the cleft (Fig. S2A). We replaced each of these with alanine. The K79A mutation abolished the interaction between GST-FliT94 and FliD, the E75A mutation partially diminished these interactions, and the others demonstrated no effect (Fig. S2B), indicating that Lys-79 is critical for binding to FliD and Glu-75 is involved in this binding. Although these two residues are covered in the crystal by the C-terminal segment of FliT including α4, the flexible linker between α3 and α4 would allow FliD to bind to these two residues even in the presence of FliT.
Interactions of FliT with Flagellar Export Apparatus Proteins FliI and FliJ.
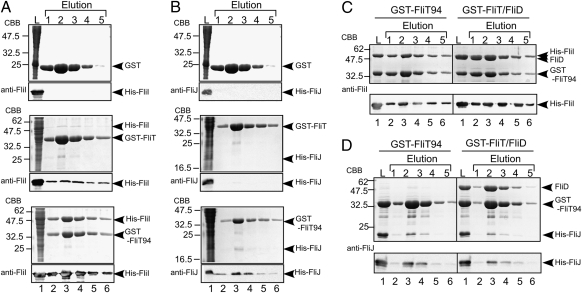
FlgN, a flagellum-specific chaperone for the hook–filament junction proteins FlgK and FlgL, has been reported to bind to two soluble components of the flagellar type III export apparatus, FliI and FliJ (17, 18). Because FliT is required for efficient export of FliD, FliT also is expected to bind to the soluble components of the export apparatus. In fact, an interaction between FliT and FliJ has been shown (18). Therefore, we analyzed the interactions of FliT with FliH, FliI, and FliJ through a pull-down assay with GST affinity chromatography (Fig. 4). His-FliI bound strongly to GST-FliT94, very weakly to GST-FliT, and not at all to GST (Fig. 4A), and His-FliJ bound weakly to GST-FliT94 but not to GST-FliT or GST (Fig. 4B). These results indicate that the C-terminal segment of FliT significantly reduces FliT's binding affinity to FliI and FliJ. Neither GST-FliT nor GST-FliT94 interacted with FliH. These results suggest that the C-terminal segment of FliT regulates its interactions with FliI and FliJ.
Fig. 4.
Interactions of FliT with the soluble component proteins of the flagellar protein export apparatus. (A and B) Interactions of GST-FliT and GST-FliT94 with FliI and FliJ by pull-down assay using GST affinity chromatography. Soluble fractions (L) prepared from a ΔflhDC-cheW mutant expressing GST, GST-FliT, or GST-FliT94 were mixed with those from the ΔflhDC-cheW mutant producing either His-FliI (A) or His-FliJ (B) and loaded onto the GST affinity column. The eluted fractions were analyzed by both CBB staining and immunoblotting with polyclonal anti-FliI (A) or anti-FliJ (B) antibodies. (C and D) Effect of FliD on the interactions of FliT94 with FliI or FliJ. Purified His-FliI (C) or His-FliJ (D) was mixed with purified GST-FliT94 or the GST-FliT94–GST-FliD complex and incubated overnight at 4 °C, after which GST affinity chromatography was performed.
We also examined whether FliD affects the interaction of GST-FliT94 with FliI or FliJ. Both FliI and FliJ coeluted with the GST-FliT94–GST-FliD complex from the column at the same levels as with GST-FliT94 alone (Fig. 4 C and D), indicating that FliD and the export component proteins FliI and FliJ bind to different surface areas on the FliT molecule.
Interaction of FliT with the FlhD4C2 Complex.
Overexpression of FliT is known to reduce the expression levels of the class 2 genes by its binding to the FlhD4C2 complex, leading to the suppression of flagellar formation (21, 26). To test whether GST-FliT94 retains the ability to interact with the FlhD4C2 complex, we first analyzed the motility of WT cells carrying a plasmid that overexpresses GST-FliT or GST-FliT94 on soft tryptone agar plates (Fig. 5A). In agreement with a previous report (26), the expression of GST-FliT considerably inhibited WT motility (Fig. 5A), due to a significant decrease in the flagellar number as established by dark-field optical microscopy. Interestingly, GST-FliT94 completely suppressed WT motility (Fig. 5A), indicating that GST-FliT94 has a much stronger inhibitory effect than GST-FliT.
Fig. 5.
Interaction between FliT and the FlhDC complex. (A) Swarming motility of SJW1103 (WT) transformed with pGEX-6p-1–based plasmids: GST, pGEX-6p-1–expressing GST; GST-FliT, N-terminally GST-tagged FliT(2-122); GST-FliT94, and N-terminally GST-tagged FliT(2-94). The plate was incubated at 30 °C for 6 h. (B) Pull-down assay of FlhC and FlhD by GST affinity chromatography. The soluble fractions from WT cells overproducing GST, GST-FliT, or GST-FliT94 were loaded onto a GST column. The eluted factions were analyzed by CBB staining for GST, GST-FliT, and GST-FliT94 (Top) and by immunoblotting for FlhC and FlhD with polyclonal anti-FlhC (Middle) and FlhD antibodies (Bottom), respectively. (C) Effect of FliD on the interaction between GST-FliT94 and the FlhDC complex.
We next analyzed the ability of GST-FliT and GST-FliT94 to bind to FlhD and FlhC by pull-down assay with GST affinity chromatography (Fig. 5B). FlhD and FlhC both coeluted with GST-FliT and GST-FliT94 from the column, but not with GST. The interactions of FlhD and FlhC with GST-FliT94 were much stronger than those with GST-FliT (Fig. 5B), which is consistent with the motility assay. Interestingly, the amount of eluted FlhD was much lower than that of eluted FlhC. Given the observation that FliT binds to FlhC alone as well as to the FlhDC complex, but not to FlhD alone (21), our data suggest that the interaction between FliT and the FlhDC complex weakens the FlhD–FlhC interaction. These results strongly suggest that the C-terminal region of FliT partially suppresses its interactions with the FlhDC complex.
We also investigated whether FliD affects the interaction of GST-FliT94 with FlhC. FlhC coeluted with the GST-FliT94–GST-FliD complex from the column at the same level as with GST-FliT94 alone (Fig. 5C), indicating that the binding site of the FlhDC complex on FliT differs from that on FliD.
Discussion
The core structure of FliT is an antiparallel α-helix bundle. This structural architecture is similar to that of FliS, which is a flagellar chaperone specific for FliC (16, 27), although the arrangement of α-helices is rather different (Fig. S3 A and D). FliS forms a heterodimer with its cognate substrate FliC (27). The C-terminal region of FliC interacts with all of the α-helices of FliS in an extended conformation containing three α-helices (27). In this study, we found that FliT forms a heterodimer with FliD in solution (Fig. 3) through an interaction between FliT and the C-terminal region of FliD (14), raising the possibility that the FliT–FliD complex might have a similar structure to that of the FliS–FliC complex. However, our present data demonstrating that only Lys-79 in α3, which is one of the highly conserved, surface-exposed residues (Fig. S2), is critical for the interaction with FliD suggests that the C-terminal region of FliD might interact with α3 of FliT in a different manner from the FliS–FliC interaction. This difference may reflect the functional difference between FliS and FliT. The atomic coordinates of two other putative flagellar chaperones, FliT from Bordetella bronchiseptica and FlgN from Pseudomonas aeruginosa, have been deposited into the PDB database by structural genomic group (PDB ID codes 3H3M and 2FUP). Their structural features including the arrangement of the helices are more or less similar to our structure. It should be noted that the C-terminal segment is missing in the structure model of Bordetella FliT. Interestingly, Bordetella FliT shows a parallel dimer, in contrast to the antiparallel arrangement of Salmonella FliT.
Despite the extensive similarities between the flagellar type III protein export system and the virulence type III protein secretion system (28), the structure of FliT is completely different from the structures of any of the virulence type III chaperones such as SicP, SycD, CesA, and PseE (Fig. S3) (29–35), suggesting that FliT's action on protein export may differ from that of these virulence-related chaperones.
The crystal structure of FliT is a tetramer composed of two asymmetric dimers (Fig. 1). The sedimentation equilibrium measurement clearly showed that FliT exists in the equilibrium between monomer and dimer (Fig. S1 A and B). The elution profile of size-exclusion chromatography showed a single peak with tailing (Fig. S1C), indicating a rapid monomer–dimer conversion of FliT. In addition, FliT eluted in fractions calibrated by molecular size markers to contain proteins significantly larger than the molecular mass of dimeric FliT (27.4 kDa) in our size-exclusion column assay (Fig. S1C). This anomalous mobility is presumably due to the elongated shape of the FliT molecule. Therefore, we conclude that the tetramer is an artifact of the crystal packing.
FliT94, which is missing α4 in the C-terminal region, showed significantly higher binding affinities for the FlhDC complex, FliI, and FliJ compared with FliT (Figs. 4 and 5), suggesting that the C-terminal α4 helix of FliT controls the binding affinity for its binding partners. Because FliD binding does not affect the interactions of FliT94 with the FlhDC complex, FliI, and FliJ, their binding sites on the FliT molecule are different from the surface area containing Lys-79 in α3, which is critical for the interaction with FliD. Because the C-terminal α4 helix binds to the hydrophobic cleft formed by α2 and α3, as observed in the crystal structure, and because the deletion of α4 does not affect the interaction between FliT and FliD, the interaction between α4 and the hydrophobic cleft formed by α2 and α3 appear to regulate the binding affinities of FliT for its binding partners. The conformational difference of the C-terminal region between Mol A and Mol B suggests that the C-terminal segment is flexible in solution; thus, the α4 helix could possibly cover the hydrophobic cleft of the same molecule, even though it requires unfolding of the C-terminal by 3 turns of the 10-turn α3 helix. Another possibility is that the C-terminal segment of another FliT molecule competes with FlhDC, FliI, and FliJ for binding to the hydrophobic cleft. In fact, FliT is in the monomer–dimer equilibrium under physiological conditions. Therefore, it also is plausible that FliT forms a homodimer in the cytoplasm through the interaction between α4 and the hydrophobic cleft to regulate the interactions with FlhDC, FliI, or FliJ.
Based on the available information, we propose a model in which dynamic interactions of FliT with its binding partners play important roles. The export apparatus switches its export substrate specificity from the rod-and-hook type to the filament type on completion of the HBB assembly (3). FliD is not exported during HBB assembly, implying that the interactions of FliT with FliI and FliJ must be suppressed by α4, covering their putative binding cleft until the HBB assembly is completed. After completion of HBB assembly, α4 is somehow released from the cleft to allow the FliT–FliD complex to bind to the FliH–FliI–FliJ complex through the specific interactions of FliT with FliI and FliJ, and the entire complex binds to the docking platform of the export gate formed by the cytoplasmic domains of FlhA and FlhB. After FliD is unfolded and translocated into the flagellar channel through the export gate by proton motive force across the cytoplasmic membrane (7, 8), released FliT forms a complex with the FlhDC complex through an interaction between FliT and FlhC, thereby suppressing class 2 gene expression. When α4 interacts with the hydrophobic cleft again, FliT dissociates from the FlhDC complex, allowing the free FlhDC complex to activate the transcription from the class 2 promoters. Thus, the interactions of FliT with its binding partners must be well organized and regulated in a timely manner during the flagellar assembly process.
Materials and Methods
Detailed descriptions of the materials and methods used in this study—including bacterial strains, plasmids, media, purification of proteins, crystal structure determination, swarming motility assay, preparation of whole cell proteins and immunoblotting, pull-down assays by GST affinity chromatography, size-exclusion column assay, and sedimentation equilibrium analysis—are provided in SI Materials and Methods. The bacterial strains and plasmids used in this study are shown in Table S1. X-ray diffraction data were collected at the synchrotron beamline BL41XU of SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (Proposals 2006B1058 and 2008A1402). The model was refined at 3.2-Å resolution to an R factor of 25.2% and a free R factor of 28.6%. The refinement statistics are summarize in Table S2.
Supplementary Material
Acknowledgments
We thank M. Kihara for a critical reading of the manuscript and helpful comments; N. Shimizu, M. Kawamoto, and K. Hasegawa at SPring-8 for their technical help with the use of beamlines; and K. Kutsukake for his kind gifts of polyclonal antibodies against Salmonella FlhC and FlhD. This work was supported in part by Grants-in-Aid for Scientific Research (18074006 to K.I. and 16087207 and 21227006 to K.N.) and the Targeted Proteins Research Program of the Ministry of Education, Science and Culture of Japan (K.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinate has been deposited in Protein Data Bank, www.pdb.org (PDB ID code 3A7M).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001866107/-/DCSupplemental.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 3.Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol Biosyst. 2008;4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- 4.Minamino T, Macnab RM. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minamino T, MacNab RM. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 6.Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 7.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 8.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 10.Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 11.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy: Not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 13.Niehus E, et al. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol. 2004;52:947–961. doi: 10.1111/j.1365-2958.2004.04006.x. [DOI] [PubMed] [Google Scholar]
- 14.Fraser GM, Bennett JCQ, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- 15.Bennett JCQ, Thomas J, Fraser GM, Hughes C. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol Microbiol. 2001;39:781–791. doi: 10.1046/j.1365-2958.2001.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerisation control by a cytosolic export chaperone. J Mol Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas J, Stafford GP, Hughes C. Docking of cytosolic chaperone–substrate complexes at the membrane ATPase during flagellar type III protein export. Proc Natl Acad Sci USA. 2004;101:3945–3950. doi: 10.1073/pnas.0307223101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans LDB, Stafford GP, Ahmed S, Fraser GM, Hughes C. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc Natl Acad Sci USA. 2006;103:17474–17479. doi: 10.1073/pnas.0605197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlinsey JE, Lonner J, Brown KL, Hughes KT. Translation/secretion coupling by type III secretion systems. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 20.Yokoseki T, Iino T, Kutsukake K. Negative regulation by fliD, fliS, and fliT of the export of the flagellum-specific anti-sigma factor, FlgM, in Salmonella typhimurium. J Bacteriol. 1996;178:899–901. doi: 10.1128/jb.178.3.899-901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto S, Kutsukake K. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol. 2006;188:6703–6708. doi: 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsot C, Hamiaux C, Page A-L. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol. 2003;6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 23.Wilharm G, Dittmann S, Schmid A, Heesemann J. On the role of specific chaperones, the specific ATPase, and the proton motive force in type III secretion. Int J Med Microbiol. 2007;297:27–36. doi: 10.1016/j.ijmm.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita M, et al. Purification, crystallization and preliminary X-ray analysis of FliT, a bacterial flagellar substrate–specific export chaperone. Acta Crystallogr F. 2009;65:825–828. doi: 10.1107/S1744309109026736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imada K, Vonderviszt F, Furukawa Y, Oosawa K, Namba K. Assembly characteristics of flagellar cap protein HAP2 of Salmonella: Decamer and pentamer in the pH-sensitive equilibrium. J Mol Biol. 1998;277:883–891. doi: 10.1006/jmbi.1998.1662. [DOI] [PubMed] [Google Scholar]
- 26.Kutsukake K, Ikebe T, Yamamoto S. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet Syst. 1999;74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- 27.Evdokimov AG, et al. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat Struct Biol. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- 28.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 29.Stebbins CE, Galán JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- 30.Luo Y, et al. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat Struct Biol. 2001;8:1031–1036. doi: 10.1038/nsb717. [DOI] [PubMed] [Google Scholar]
- 31.Birtalan S, Ghosh P. Structure of the Yersinia type III secretory system chaperone SycE. Nat Struct Biol. 2001;8:974–978. doi: 10.1038/nsb1101-974. [DOI] [PubMed] [Google Scholar]
- 32.Büttner CR, Sorg I, Cornelis GR, Heinz DW, Niemann HH. Structure of the Yersinia enterocolitica type III secretion translocator chaperone SycD. J Mol Biol. 2008;375:997–1012. doi: 10.1016/j.jmb.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Yip CK, Finlay BB, Strynadka NC. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat Struct Mol Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 34.Quinaud M, et al. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci USA. 2007;104:7803–7808. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J Mol Biol. 2008;377:819–830. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.