Abstract
When the translation termination factor Sup35 adopts the prion state, [PSI+], the read-through of stop codons increases, uncovering hidden genetic variation and giving rise to new, often beneficial, phenotypes. Evidence suggests that prion induction involves a process of maturation, but this has never been studied in detail. To do so, we used a visually tractable prion model consisting of the Sup35 prion domain fused to GFP (PrD-GFP) and overexpressed it to achieve induction in many cells simultaneously. PrD-GFP first assembled into Rings as previously described. Rings propagated for many generations before the protein transitioned into a Dot structure. Dots transmitted the [PSI+] phenotype through mating and meiosis, but Rings did not. Surprisingly, the underlying amyloid conformation of PrD-GFP was identical in Rings and Dots. However, by electron microscopy, Rings consisted of very long uninterrupted bundles of fibers, whereas Dot fibers were highly fragmented. Both forms were deposited at the IPOD, a biologically ancient compartment for the deposition of irreversibly aggregated proteins that we propose is the site of de novo prion induction. We find that oxidatively damaged proteins are also localized there, helping to explain how proteotoxic stresses increase the rate of prion induction. Curing PrD-GFP prions, by inhibiting Hsp104’s fragmentation activity, reversed the induction process: Dot cells produced Rings before PrD-GFP reverted to the soluble state. Thus, formation of the genetically transmissible prion state is a two-step process that involves an ancient system for the asymmetric inheritance of damaged proteins and heritable changes in the extent of prion fragmentation.
Keywords: yeast prion, fiber fragmentation, IPOD (Insoluble Protein Deposit), prion inheritance, asymmetric damage distribution
Prions are self-perpetuating protein conformations that store and transmit phenotypic information independently of nucleic acids. In fungi, they act as protein-based elements of heredity, stably propagating their altered protein conformations and associated phenotypes (1, 2).
In Saccharomyces cerevisiae, seven prions are known (3) and evidence indicates that numerous other yeast proteins are capable of forming prions (4). The proteins have different molecular functions and produce different prion phenotypes. Although they share no sequence homology, their prion domains (PrDs) are enriched in asparagine and glutamine residues. These PrDs can adopt self-perpetuating prion conformations that are amyloids. They template the conversion of soluble prion proteins of the same type to the same conformation (1, 2). The AAA+ ATPase Hsp104 shears amyloid fibers to generate prion seeds, also referred to as propagons (5), facilitating inheritance of the prion state from generation to generation (6).
The protein determinant for the prion [PSI+] is the translation termination factor Sup35. Sup35’s PrD, NM, has an N-terminal amyloidogenic domain (N) and a solubilizing middle domain (M) (1, 2). In vitro, purified PrD can form amyloid fibrils on its own (7). In vivo, assembly of the prion reduces the availability of soluble, functional termination factor. This causes stop codon read-through (1, 2) and results in a large array of diverse phenotypes depending on the genetic background (8). Stressful conditions alter protein folding homeostasis and increase the rate at which cells switch to the prion state, creating a bet-hedging strategy for survival in fluctuating environments (3, 4, 9).
[PSI+] propagation is well studied. However, the spontaneous de novo formation of [PSI+] is rare and poorly understood. Overexpression of Sup35, or its PrD, dramatically increases the frequency of [PSI+] induction (10), offering an opportunity to investigate the process. During prion induction, two distinct aggregation patterns are observed: ribbon and ring structures that extend throughout the cell and round dot structures (11, 12). Dots correspond to the mature [PSI+] prion state. The ring structures appear to be a hallmark of de novo [PSI+] induction, but their nature remains a mystery. This is the subject of our investigation.
Results
Expression of PrD-GFP in a Sup35 PrD Deletion Strain Results in Rings and Dots.
To study the de novo formation and inheritance of prion amyloid in vivo on a single-cell level, we employed a fusion of the PrD (NM) with GFP, which is soluble in [prion−] cells but faithfully propagates with the wild-type Sup35 amyloid in [PRION+] cells (13). To determine whether this visually tractable protein is fully capable of forming a prion in its own right, we expressed it from the Sup35 promoter to maintain normal expression. When these cells were mated to [psi−] cells, the protein remained soluble. When mated to [PSI+] cells, it acquired the prion state (Fig. S1), was dominant in matings, and segregated 4:0 in meiosis. Thus, PrD-GFP fully recapitulates prion behavior. However, as expected, spontaneous switches to the prion state were too rare to be observed.
To provide a robust system for studying prion induction, we placed the PrD-GFP fusion under the control of the GPD promoter, which increased its expression ~15-fold, and integrated a single copy into the genome, to keep expression uniform in each cell. We also deleted the PrD from the endogenous SUP35 gene, making the translation termination domain immune from sequestration into the prion, which can be toxic when it happens excessively (10, 14).
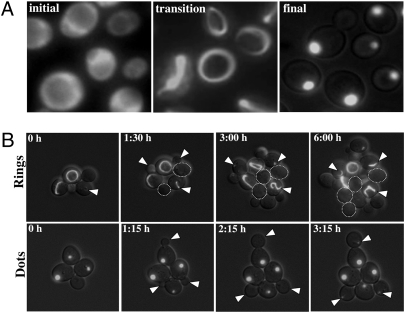
Soon after transformation, while cells were still in microcolonies, they had the diffuse GFP-fluorescence of [prion-] cells. In more mature colonies, a large fraction of cells displayed a large ring-, rod-, or ribbon-like structure of PrD-GFP fluorescence (collectively referred to as “Rings” throughout this article). This Ring pattern was inherited for many generations. After two to eight restreaks, each representing ~25 generations, the aggregation pattern changed and PrD-GFP was exclusively found in one large focus per cell, hereafter referred to as Dots (Fig. 1A). Rings and Dots were previously described during prion induction in cells transiently overexpressing PrD-GFP in the presence of wild-type Sup35 (11, 12).
Fig. 1.
Expression of PrD-GFP at high levels produces self-propagating Ring assemblies that transition to a Dot assembly only after many generations. (A) Cells constitutively expressing PrD-GFP under control of the GPD promoter are shown at different times. Initial, immediately after transformation of the expression construct; transition, transformants after 3–5 days; final, transformants after ~10 days (hundreds of generations). (B) Haploid cells propagating either PrD-GFP Rings (Upper) or Dots (Lower). Cell divisions (arrows) were followed microscopically. Images represent a merge of several focal planes of GFP fluorescence overlaid with single DIC focal plane. A dotted line encircles progeny of Ring mothers that did not form any aggregates during the course of the experiment.
Propagation of Rings and Dots Is Independent of [RNQ+].
When PrD-GFP was overexpressed in cells carrying a deletion of RNQ1, a factor required for efficient prion induction (15–17), it remained diffuse (11) (Fig. S2). When cells containing Rings or Dots were mated to cells with a RNQ deletion and sporulated, haploid progeny with the deletion maintained their Rings and Dots (Fig. S3). Thus, as for the [PSI+] prion itself (18), Dots and Rings require [RNQ+] for their induction but not their propagation.
Time-Lapse Microscopy Establishes Stable Asymmetric Inheritance of Both Aggregation States.
We followed the propagation of PrD-GFP Rings and Dots for several cell divisions using time-lapse microscopy (Fig. 1B). Both were faithfully propagated, but Rings were less stable, frequently giving rise to progeny with diffuse fluorescence (Fig. 1B Upper, cells encircled by dotted line). Surprisingly, during cell division, Rings and Dots were retained in mother cells. Visible assemblies only became detectable in daughter cells after the dividing septum had formed. Cells with Rings initially gave rise to daughters with a single small focus, much like the initiating focus in Dot cells, but these generally expanded to form a typical Ring structure by the time the cell was ready for a new division (Fig. 1B Upper).
PrD-GFP Dots Can Transmit the [PSI+] Phenotype but Rings Cannot.
Genetically, a defining feature of prions is dominance in crosses to mating partners. We first mated Ring and Dot cells to isogenic partners whose RNQ1 gene had been deleted before PrD-GFP expression. As noted above, this kept their PrD-GFP protein in the diffuse nonprion state. After mating, diffuse PrD-GFP converted to the Ring or Dot form of its mating partner. When these diploids were sporulated, all haploid progeny of Dot matings contained Dots. Rings were also inherited, albeit less faithfully (Fig. S3).
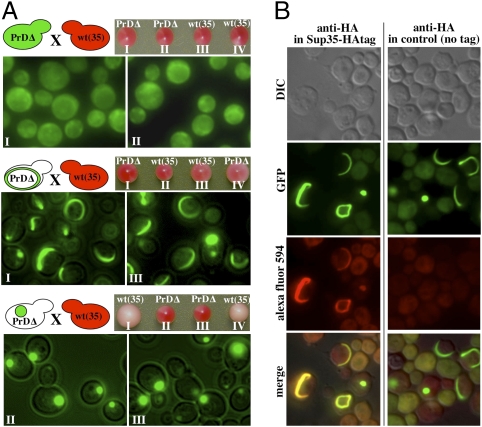
Next, we mated Ring and Dot cells to wild-type [psi-] cells to determine whether they could transmit the [PSI+] phenotype. Both Rings and Dots were maintained in the diploids. All cells carried the ade1-14 stop codon mutation (1), but because one Sup35 allele carried a PrD deletion, the [PSI+] read-through phenotype could not be detected in the diploid. However, after sporulation, haploid progeny that received the wild-type SUP35 gene and whose protein had acquired the [PSI+] state switched colony color from red to pink (Fig. 2A).
Fig. 2.
Cells with Rings do not induce the [PSI+] prion state in a mating partner. (A) Haploid strains carrying a PrD deletion of the endogenous SUP35 gene (PrDΔ) displaying diffuse PrD-GFP fluorescence, Rings or Dots were mated with a wild-type SUP35 [psi-] strain. The resulting diploids were sporulated and tetrads were dissected. Spores with the PrD-GFP fusion were analyzed by fluorescence microscopy. Colony colors of the progeny ([psi-], red; [PSI+], light pink) revealed that Dots, but not Rings, induced the prion state of wild-type SUP35. (B) Sup35-HA colocalizes with PrD-GFP Rings and Dots. A [psi-] strain carrying an inducible copy of PrD-GFP and a C-terminally HA-tagged Sup35 (SUP35-HA) was analyzed by immunofluorescence and fluorescence microscopy. Colocalization is shown in yellow in merged images.
In control matings, parents with diffuse PrD-GFP fluorescence produced only red [psi-] progeny (Fig. 2A Top). Progeny of Dot parents that received only the wild-type Sup35 gene invariably gave rise to pink [PSI+] colonies as expected of faithful transmission of the prion state from PrD-GFP to wild-type Sup35 (Fig. 2A Bottom). Also, progeny carrying both wild-type Sup35 and the PrD-GFP overexpression construct were never recovered, because of excessive, toxic sequestration of Sup35 (10, 14). Surprisingly, mating with Ring cells produced no wild-type Sup35 progeny with the [PSI+] phenotype (Fig. 2A Middle). Furthermore, cells carrying both the PrD-GFP construct and wild-type Sup 35 were readily obtained (e.g., Fig. 2A Middle, spore III). In these cells, PrD-GFP maintained its initial Ring pattern, demonstrating that Rings generally propagated faithfully during meiosis.
A simple explanation might be that the PrD-GFP in Ring cells does not interact with WT Sup35. To test this, we induced PrD-GFP Rings and Dots in cells with HA-tagged Sup35 protein as the only source of this essential protein. HA colocalized with both Rings and Dots (Fig. 2B), eliminating this possibility.
In Ring and Dot Cells, PrD-GFP Is in the Same Amyloid Conformation.
A common feature of prions is the ability to exist in several related, but distinct, amyloid conformations, known as “prion strains” (19, 20). These propagate with different polymerization and fragmentation efficiencies. Because they reach different equilibria between the soluble functional form and the amyloid, they produce distinct phenotypes (in ade1-14 cells, colonies with different shades of pink) (19–21). Thus, another explanation for our results might be that Rings represent a different prion strain than Dots, one that is too “weak” to elicit a detectable phenotype. This occurs, for example, with the [ETA+] variant of [PSI+] (22).
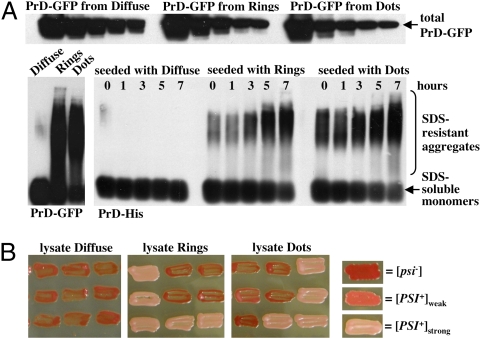
When crude cell lysates of all cell types were boiled in SDS, they all produced a PrD-GFP band of the same intensity by SDS/PAGE, confirming that the protein was expressed at the same level (Fig. 3A Upper). When analyzed without boiling on semidenaturing agarose gels, the PrD-GFP protein of both Ring and Dot cells migrated as high-molecular-weight SDS-resistant complexes typical of prion amyloids (Fig. 3A Lower Left).
Fig. 3.
Cells with Rings and Dots contain PrD-GFP in the same prion conformation. (A Upper) Crude lysates from cells displaying diffuse, Ring or Dot fluorescence were adjusted to equal protein concentrations and serial dilutions (1:2 steps) were loaded onto an SDS gel and analyzed by SDS/PAGE and Western blotting with an anti-GFP antibody to reveal the amounts of PrD-GFP in the different lysates. (A Lower Left) The three different lysates were analyzed by semidenaturing agarose gel analysis (SDD-AGE) followed by Western blotting with an anti-GFP antibody. Lysates from cells with Rings and Dots, but not diffuse PrD-GFP, contain SDS-resistant high-molecular-weight aggregates of PrD-GFP. (A Lower Right) Purified PrD-His was seeded with the three different lysates in vitro and analyzed by SDD-AGE and Western blotting with an anti-His-tag antibody. SDS-resistant high-molecular-weight aggregates were formed with comparable kinetics. (B) Protein transformations of a [psi-] tester strain (red colony color) were performed with crude lysates from cells with diffuse PrD-GFP fluorescence or Rings and Dots. The prion status and strain of the transformants was determined by colony color. Both types of aggregates induced a strong [PSI+] strain (light pink color) with similar efficiencies.
Next, we tested the ability of Dots and Rings to seed polymerization of purified soluble PrD, a defining characteristic of prions. Lysates of cells with diffuse fluorescence did not seed polymerization, but lysates of Ring and Dot cells had very similar seeding capacities (Fig. 3A Lower Right): SDS-resistant species could be detected almost immediately, and their sizes increased similarly during the course of the experiment.
Proteins from different prion strains transform [psi-] cells to [PSI+] in a phenotype-specific manner. To minimize manipulations, crude extracts were introduced directly into [psi-] cells to test for such differences (19, 20). To our surprise, lysates from cultures with Rings and Dots both gave rise to the same strong [PSI+] strain (Fig. 3B): Dot lysates induced the strong prion phenotype in 71 transformants, the weak phenotype in only 3. Ring lysates induced the strong phenotype in 101 transformants, the weak in only 6.
Although the stability of amyloid structures makes it extremely unlikely that a weak conformation might convert to a strong one during our procedure (19, 20), we considered the possibility. We eliminated the sonication step before transformation and used several different lysis procedures without changing the outcome. More definitively, we performed many similar experiments with lysates from bona fide weak prion strains but never observed conversion of weak prion strains to strong. Thus, Rings and Dots do not represent different prion conformations or strains.
Aggregate Formation Occurs at a Site Specific to the Deposition of Insoluble Aggregates.
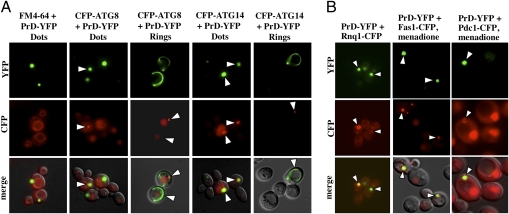
We asked whether the differences in transmissibility of the [PSI+] phenotype with Rings and Dots could be due to different cellular localizations. The vacuolar dye FM4-64 established that PrD-GFP Dots were localized adjacent to the vacuole (Fig. 4A, leftmost column). A single focus at the vacuole is characteristic of the pre-autophagosomal structure (PAS), which coordinates autophagosome formation and cytoplasm-to-vacuole vesicle trafficking (23). Colocalization of a PrD-YFP fusion and the CFP-tagged PAS markers ATG8 and ATG14 confirmed this localization (Fig. 4A). Dots were directly adjacent to the PAS. Rings intersected it (Fig. 4A).
Fig. 4.
Prion aggregates localize to the IPOD compartment as shown using markers for the pre-autophagosomal structure (PAS) and additional IPOD substrates. Cells carrying Dots were generated using a genomic copy of GPD-controlled PrD-YFP. Rings were induced using an analogous galactose-inducible construct. (A) FM4-64 visualization of the vacuolar membrane shows the perivacuolar localization of PrD-YFP Dots (leftmost column). The localization of Rings and Dots to the pre-autophagosomal structure was visualized using centromeric plasmids expressing N-terminal CFP fusions of ATG8 or ATG14. (B) Colocalization of PrD-YFP Dots with known and newly identified substrates of the IPOD compartment (indicated by arrows). Genomic copies of PDC1 or FAS1 were tagged with CFP (first and second columns), whereas the Rnq1-CFP fusion was expressed from a plasmid under control of the Cup1 promoter (third column). Aggregation of Pdc1-CFP or Fas1-CFP was induced using 0.1 or 2 mM menadione, respectively, for 4 h before microscopy. The Rnq1-CFP expression was induced using 100 μM copper sulfate. Images represent one single optical plane.
Recently, a perivacuolar PAS-associated site for the deposition of irreversibly aggregated proteins was discovered and termed the IPOD for Insoluble PrOtein Deposit (24). It also accumulates the Ure2 and Rnq1 proteins in their prion forms (24). To extend this characterization, we used mass spectrometry to identify proteins that were co-immunocaptured with PrD-GFP Dots but not with soluble PrD-GFP (Fig. S4). Rnq1 was among them (Table S1), confirming the capture of bona fide IPOD substrates. We also found proteins known to be very sensitive to oxidative damage (25).
CFP-tagged versions of Rnq1 and the two oxidation-sensitive proteins, Fas1 (fatty acid synthase) and Pdc1 (pyruvate decarboxylase 1) (25), were used to confirm cellular colocalization by microscopy. RNQ1-CFP colocalized with PrD-YFP Dots constitutively (Fig. 4B). The partial colocalization of Fas1 and Pdc1 with PrD-YFP Dots was most apparent after cells were treated with the oxidizing agent menadione (25) (Fig. 4B). Thus, the previously reported polarisome-dependent asymmetric inheritance of oxidatively damaged proteins (26, 27) occurs in part at the IPOD, which is also a site for prion accumulation.
Number of PrD-GFP Amyloid Fibers Differs Between Ring and Dot Aggregates.
Next, we investigated the difference between Rings and Dots by transmission electron microscopy (TEM). We easily identified both forms by their highly ordered fibrous appearance, which closely resembled fibrils formed by Sup35 or PrD in vitro (7, 28). We confirmed that these assemblies were indeed PrD-GFP Rings and Dots by immuno-EM using an antibody against GFP (Fig. S5). We also found the typical electron-dense foci formed by amorphous aggregates of damaged proteins at this site (Fig. 5A Lower and Fig. S6), further confirming the colocalizations described above.
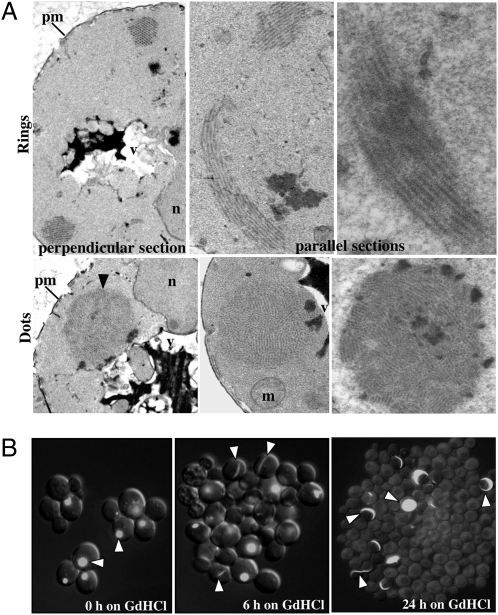
Fig. 5.
Electron microscopy of Ring and Dot cells reveals the different degrees of PrD-GFP fibril fragmentation. (A Upper) Different magnifications of ultrathin cell section with a large Dot aggregate next to the vacuole. (A Lower) Perpendicular or parallel sections of a cell displaying PrD-GFP Rings. m, mitochondrion; n, nucleus; v, vacuole; pm, plasma membrane. (B) Time-lapse microscopy of constitutively PrD-GFP expressing strain carrying a Dot aggregate. Cells were monitored for several hours in the presence of 5 mM GdHCl to inhibit Hsp104 activity. Upon inhibition of Hsp104, cells with Dot aggregates gave rise to progeny forming Ring aggregates. Progeny thereof ultimately displayed diffuse fluorescence. Images show different representative groups of cells at different time points (0, 6, 24 h) and are an overlay of a single focal plane DIC image and a merge of fluorescence z-stack images.
Rings and Dots were not separated from the surrounding cytoplasm by any compartmentalizing elements, such as membranes or cytoskeletal structures. Fibrils formed in vitro by the Sup35 PrD alone have an average diameter of ~11.5 nm, whereas fibrils of full-length Sup35 are ~25 nm (7, 28). In cross-sections, Ring and Dot PrD-GFP fibrils had identical doughnut-like morphologies with a diameter of ~25–30 nm and an inner core of 6–12 nm (Fig. S5). This is consistent with the prion domain forming the inner core of the fiber, surrounded by GFP molecules.
However, there was one profound difference between the fibrils in the Rings and Dots: their length. Rings contained bundles of very long uninterrupted fibrils (Fig. 5A Upper). Dots contained a profusion of short fibril bundles with diverse orientations in the same structure (Fig. 5A Lower). Thus, Ring and Dot aggregates displayed an extreme difference in fragmentation and, consequently, in the number of fibril ends. The cell lysis procedure used in our protein transformation or seeding assays (Fig. 3) would certainly fragment the very long fibrils of Ring cells, eliminating the sole difference between Rings and Dots.
In vivo, the only known protein with the capacity to sever PrD fibrils is the AAA+ ATPase Hsp104. Inhibition of Hsp104 increases the number of ring-bearing cells when PrD-GFP is overexpressed in the presence of wild-type Sup35 (11). However, the relationship between Hsp104 activity and Dots has never been examined. We inhibited Hsp104 activity in Dot cells by two different methods: incubation with 5 mM GdHCl, which selectively inhibits Hsp104’s ATPase cycle (29) (Fig. 5B), and expression of a dominant negative mutant of Hsp104 (Fig. S7), which is incorporated into Hsp104 hexamers and blocks their activity. In both cases, time-lapse microscopy revealed that preexisting Dots never changed, but mother cells with Dots rapidly and invariably produced daughter cells with Rings. These Rings also remained intact. However, within just a few divisions, all Ring-containing cells gave rise to progeny with diffuse PrD-GFP fluorescence. Thus, the curing of Dots by Hsp104 inhibition mirrors their formation. The distinct states of prion maturation result from differences in the number of fiber fragments generated by Hsp104 and hence the number of prion seeds.
Discussion
We find that prion induction by the Sup35 PrD involves a maturation process with a distinct, stable, and heritable intermediate. Surprisingly, this intermediate (Ring state) has all of the biochemical characteristics of the mature prion (Dot state) but distinct phenotypic properties. It does not confer a [PSI+] phenotype on meiotic progeny whose only source of Sup35 is the wild-type protein, even when PrD-GFP Rings coexist in the same cell. How might we resolve this seeming paradox and also decipher the nature of the prion induction process? Two other observations, we think, provide the key.
First, by electron microscopy, Ring fibrils are long and uninterrupted, and Dot fibrils are highly fragmented. Indeed, the degree of fiber fragmentation is the defining characteristic for the different structures, because blocking of Hsp104’s fragmentation activity reverts Dots to Rings before the assemblies are lost entirely. Second, the deposition of Rings and Dots occurs at a site specific for the compartmentalization of damaged proteins, known as the IPOD (24). In cells undergoing mitosis, Rings and Dots are retained in mother cells but reform in their daughters at the IPOD immediately after septum formation. The most parsimonious explanation is that prion seeds, too small to be detected by fluorescence (30), are liberated by the activity of Hsp104 and transmitted to daughter cells.
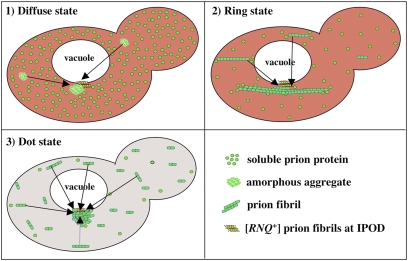
Taking these observations together, we propose a model for prion induction (Fig. 6) that also explains the distinct phenotypes and heritable nature of Rings and Dots. Upon initial expression, PrD-GFP is soluble, but the intrinsically unstructured Sup35 PrD has a high propensity to misfold. This misfolded protein is targeted to the IPOD. Because nucleated conformational conversion to the prion state requires an oligomeric intermediate (31), the high local concentration of PrD-GFP at the IPOD increases the likelihood of prion induction (12). [RNQ+] prions, which are required for [PSI+] induction (15–17), are also located at the IPOD and ready to facilitate nucleation (this study and refs. 24 and 32).
Fig. 6.
Schematic model for the induction and maturation of the PrD-GFP prion via a long-lived poorly fragmented fibril state.
Initially, only a few prion seeds form. Therefore, the rate at which soluble full-length Sup35 is sequestered from translation is limited. Rings formed in this way can coexist with the wild-type protein without causing toxicity or creating a [PSI+] phenotype (Fig. 2A Middle, spore III). The small number of seeds creates long bundles of fibers, which appear as Rings confined only by the boundary of the cell. Inheritance of a small number of seeds by daughter cells perpetuates the Ring state.
Dots consist of short highly fragmented fibers. Increased fragmentation causes a larger number of prion seeds to be inherited by daughters, explaining the more stable inheritance of Dots relative to Rings and the ability to reliably transmit the prion state to all meiotic progeny. In this case, polymerization is limited by the amount of soluble prion protein and not by the number of prion seeds. Thus, meiotic progeny inheriting PrD-GFP Dots, whose only source of the essential Sup35 is the wild-type protein, are inviable.
A remaining question is how cells switch from the transitional Ring state to the mature prion state. One possibility is that a Ring-containing mother rarely but stochastically transmits a sufficiently large number of seeds to its daughter to create a profusion of shorter fibrils. Indeed, the number of wild-type [PSI+] propagons is known to be clonally diverse (5). However, Ring and Dot cells contain the same quantity of the fibril fragmenting protein Hsp104 (Fig. S8), which plays a critical role in [PSI+] inheritance (6, 29, 33–35). With just a few initiating fibrils, one might expect them to be highly susceptible to Hsp104, but Hsp104’s remodeling functions are tightly regulated in unstressed cells (36). Thus, another explanation is a heritable switch in the activity of Hsp104. Speculatively, an inhibitor of Hsp104 might itself be a prion—the concentration of aggregated proteins at the IPOD could favor its switch to a heritable, nonfunctional state, converting Rings to highly fragmented mature prions.
However that specific detail may be resolved, many lines of evidence suggest that normal [PSI+] induction, as well as the induction of other prions, involves a transitional state like the one we describe. For example, propagons of wild-type [PSI+] are subject to biased retention in mother cells by a previously unknown mechanism (5). Both Rings and Dots are observed in cells with wild-type Sup35 during “typical” laboratory prion inductions with overexpressed PrD (11, 12), and Rings behave as an immature transitional state with unstable inheritance. Furthermore, in those cases where it has been examined, the induction of [PSI+], [URE3], and [RNQ+] by overexpression of their PrDs clearly involves a genetically unstable intermediate. Cells initially segregate both [prion-] and [PRION+] clones and only later stabilize to reliably produce [PRION+] colonies (16–18, 37, 38). Finally, in the absence of PrD overexpression, proteotoxic stresses, including oxidative stresses, increase the spontaneous induction of [PSI+] (9, 39). The more severe the stress, the more efficient the induction. Colocalization of damaged proteins at the IPOD could provide an opportunity for cross-seeding akin to that provided by [RNQ+]. Subsequently, in the absence of PrD overexpression, the fragmentation activity of Hsp104 would be sufficient to free most of the assembled protein from the IPOD, allowing direct cytoplasmic inheritance of prion propagons.
The concerted asymmetric retention of prion fibrils is tied to an ancient (but only recently discovered) system for the asymmetric inheritance of misfolded damaged proteins. Mitotic cells, ranging from bacteria to yeast to human embryonic stem cells, systematically retain and sequester damaged and misfolded proteins to one cell during division (26, 40, 41). This is thought to ensure the fitness of the next generation by allowing one of the two mitotic products to start afresh with respect to protein damage—segregating an aging (soma-like) lineage from a rejuvenated (germ-like) lineage. We propose that the IPOD is not only a depository for damaged proteins, but plays a critical positive role in prion biology as the cellular site of de novo prion induction.
Materials and Methods
Yeast Strains, Media, and Constructs.
Yeast strains BY4741 (Euroscarf) and 74D-694 (6) were grown on standard synthetic media lacking particular amino acids/bases and containing either D-glucose (SD) or D-galactose (SGal) as carbon source. Sporulation was performed on 1% potassium acetate, 0.05% dextrose, 0.1% yeast extract, and 0.01% complete amino acid mix (BIO 101). Expression of PrD-GFP (9) was controlled by either the Gal1 or GPD promoter. The expression plasmid for RNQ1-CFP is described in ref. 15. Gene deletions used primer sequences listed at the yeast deletion project web site (www-sequence.stanford.edu/group/yeast_deletion_project/Deletion_primers_PCR_sizes.txt). Genomic C-terminal Cerulean fusions were confirmed by PCR. Plasmids coding for N-terminal Cerulean ATG8 or ATG14 fusions were generated using Gateway shuttle vectors (42) and the ORFs from the Open Biosystems mORF collection.
Seeding and Transformation.
The seeding efficiency of crude lysates from cells with diffuse fluorescence, Rings, or Dots was tested with purified His-tagged protein using SDD-AGE as described in ref. 43. The same lysates were tested for transformation activity with a 74D-694-derived strain containing an ADE1 mutation suppressible by [PSI+] and a URA3 plasmid as described in ref. 20. Details are provided in SI Materials and Methods.
Microscopy.
Cells were grown in liquid culture to midlog phase or on agar plates overnight and examined with an Axioplan microscope with a ×100 objective (Zeiss) and narrow band-pass filters for colocalization studies with different fluorescent proteins. Monoclonal HA antibody (Cell Signaling) and a secondary anti-mouse IgG antibody were coupled to Alexa Fluor 594 (Invitrogen). Unless indicated, images were taken in one representative focal plane. Photoshop was used for colorization, linear adjustments of brightness and contrast, and creation of composite and merged images. Time-lapse microscopy was performed on agarose pads of ~20 × 20 × 1 mm, prepared by pouring ultrapure agarose (2% wt/vol) in SD media directly onto a microscope slide. After addition of the cells, the pad was covered with a cover slide and sealed with melted VLAP wax (1:1:1 Vaseline:lanolin:paraffin). Every 60–90 min, we collected a stack of 12–15 optical sections spaced 0.4 μm apart. Representative fluorescence images were de-blurred using a simple deconvolution algorithm and then projected to one image. For conventional and immunoelectron microscopy details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank K. Allendoerfer, K. K. Frederick, R. Krishnan, and S. Alberti for critical reading of the manuscript; E. Spooner (Whitehead Institute) for mass spectrometry; and M. J. Sa and A. W. Murray (Harvard University) for assistance with time-lapse microscopy. This work was supported by National Institutes of Health Grant GM025874 (to S.L.), a European Molecular Biology Organization Long-Term Fellowship (to J.T.), a Human Frontiers Science Program Organization Long-Term Fellowship (to J.T.), and an American Heart Association fellowship (to J.D.). S.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003895107/-/DCSupplemental.
References
- 1.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 2.Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20:125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 7.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 8.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 9.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:2605–2613. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 11.Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
- 12.Ganusova EE, et al. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 14.Vishveshwara N, Bradley ME, Liebman SW. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol. 2009;73:1101–1114. doi: 10.1111/j.1365-2958.2009.06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 16.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 17.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 18.Derkatch IL, et al. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 22.Zhou P, et al. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 26.Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 27.Liu B, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25:822–833. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawai-Noma S, et al. Dynamics of yeast prion aggregates in single living cells. Genes Cells. 2006;11:1085–1096. doi: 10.1111/j.1365-2443.2006.01004.x. [DOI] [PubMed] [Google Scholar]
- 31.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 32.Derkatch IL, et al. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borchsenius AS, Müller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet. 2006;49:21–29. doi: 10.1007/s00294-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 34.Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J. 2001;20:6683–6691. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schirmer EC, Homann OR, Kowal AS, Lindquist S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol Biol Cell. 2004;15:2061–2072. doi: 10.1091/mbc.E02-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Bellot E, Guillemet E, Cullin C. The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. EMBO J. 2000;19:3215–3222. doi: 10.1093/emboj/19.13.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- 39.Sideri TC, Stojanovski K, Tuite MF, Grant CM. Ribosome-associated peroxiredoxins suppress oxidative stress-induced de novo formation of the [PSI +] prion in yeast. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1000347107. 10.1073/pnas.1000347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rujano MA, et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:2325–2335. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.